Abstract
There is a need to develop inhibitors of mosquito-borne flaviviruses, including WNV (West Nile virus). In the present paper, we describe a novel and efficient recombinant-antibody technology that led us to the isolation of inhibitory high-affinity human antibodies to the active-site region of a viral proteinase. As a proof-of-principal, we have successfully used this technology and the synthetic naive human combinatorial antibody library HuCAL GOLD® to isolate selective and potent function-blocking active-site-targeting antibodies to the two-component WNV NS (non-structural protein) 2B–NS3 serine proteinase, the only proteinase encoded by the flaviviral genome. First, we used the wild-type enzyme in antibody screens. Next, the positive antibody clones were counter-screened using an NS2B–NS3 mutant with a single mutation of the catalytically essential active-site histidine residue. The specificity of the antibodies to the active site was confirmed by substrate-cleavage reactions and also by using proteinase mutants with additional single amino-acid substitutions in the active-site region. The selected WNV antibodies did not recognize the structurally similar viral proteinases from Dengue virus type 2 and hepatitis C virus, and human serine proteinases. Because of their high selectivity and affinity, the identified human antibodies are attractive reagents for both further mutagenesis and structure-based optimization and, in addition, for studies of NS2B–NS3 activity. Conceptually, it is likely that the generic technology reported in the present paper will be useful for the generation of active-site-specific antibody probes for multiple enzymes.
Keywords: antibody, Fab, flavivirus, IgG, serine proteinase, West Nile virus (WNV)
INTRODUCTION
There is a need to develop inhibitors of mosquito-borne and tick-borne flaviviruses, including WNV (West Nile virus). There have already been multiple cases of mosquito-borne WNV in the United States. Unfortunately, there are no effective countermeasures or vaccines as yet. WNV is an enveloped positive-stranded 11-kb RNA virus. The genomic RNA of WNV encodes a polyprotein precursor that consists of three structural proteins, C (capsid protein), prM (precursor membrane protein) and E (envelope protein), and seven NS proteins (non-structural proteins), NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5, arranged in the order C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5.
Polyprotein processing by the viral two-component NS2B–NS3pro (proteinase domain of NS3) and also by the host-cell secretase and furin is required to generate individual viral proteins [1–3]. The full-length NS3 is a multifunctional protein in which the N-terminal 184 amino-acid residues represent the NS3pro and the C-terminal sequence codes for the helicase, nucleoside triphosphatase and RNA triphosphatase activities, all of which are co-ordinately regulated through localization within membrane compartments in the infected cell. NS2B functions as an essential cofactor of NS3pro. The cofactor activity of the 48-amino-acid central portion of NS2B is roughly equivalent to that of the entire NS2B sequence [4–7]. Structural studies have determined that NS2B wraps around NS3pro, completing, in a precise and well-defined fashion, the structure of the active site [8,9]. In agreement, deletion of the NS2B sequence inactivates the functional activity of NS3pro [10,11]. These unique cofactor–protease-domain interactions are common for multiple flavivirus types [7,12–15].
NS3pro is responsible for the cleavage of the internal sites within C, NS3 and NS4A, and also at the NS2A/NS2B, NS2B/NS3, NS3/NS4A and NS4B/NS5 boundaries. Because inactivating mutations of the NS3pro cleavage sites in the polyprotein precursor abolished viral infectivity [12,14–28], it is clear that the NS3pro function is vitally important for the virus and that NS3pro antagonists have merit as viral drugs. Multiple competitive and non-competitive peptide and small-molecule inhibitory scaffolds of NS3pro have recently been identified [29–36]. There are, however, obstacles, including inefficient cell penetration and poor solubility and stability, to their development as an antiviral therapy. Additional, unconventional, approaches are required to overcome these obstacles.
Because of their unique ability to bind specifically to nearly any antigen, antibodies provide a much sought after and excellent scaffold for designing inhibitors targeted to the individual members of a family of homologous enzymes. Over the past two decades, recombinant technology has enabled the production of engineered antibody fragments such as Fab or scFv (single-chain variable fragment) antibodies [37–41]. An scFv antibody is a small engineered antibody, in which the VH (variable heavy chain) and VL (variable light chain) of the antibody molecule are connected by a short flexible polypeptide linker.
Phage-display technology has been used successfully for the isolation of specific scFvs or Fabs from human repertoire libraries [42]. For phage-display selection, the Fab format is preferred because scFv fragments usually oligomerize and, as a result, generate multimers [43,44]. Fabs usually do not oligomerize and are more stable compared with scFvs. The identified Fabs can be genetically manipulated further and linked to different tags or enzymes for detection or labelling purposes or equipped with new diversity in the CDRs (complementarity-determining regions) in order to improve affinity and specificity [45,46].
In the present paper, we report an innovative and effective antibody-selection technology. As a proof-of-principal, we used this technology and the phage Fab antibody library [47] to select the inhibitory active-site-targeting antibodies to a viral proteinase (WNV NS2B–NS3pro). This generic technology, by analogy, may be useful for the efficient generation of active-site-specific antibody probes for enzymes.
MATERIALS AND METHODS
Reagents
Reagents were purchased from Sigma–Aldrich (Milwaukee, WI, U.S.A.), unless indicated otherwise. The Pyr-RTKR–AMC (pyroglutamic acid-Arg-Thr-Lys-Arg–7-amino-4-methylcou-marin) fluorescence-quenched cleavage peptide was purchased from Bachem (King of Prussia, PA, U.S.A.). Goat anti-human F(ab′)2 fragment conjugated with HRP (horseradish peroxidase) was from AbD Serotec (Oxford, UK). Aprotinin was purchased from Serological Corporation (Norcross, GA, U.S.A.) and then additionally purified by using gel-exclusion chromatography on a Superdex-75 column (GE Healthcare, Piscataway, NJ, U.S.A.).
The WNV and DV2 (Dengue virus serotype 2) NS2B–NS3pro expression constructs
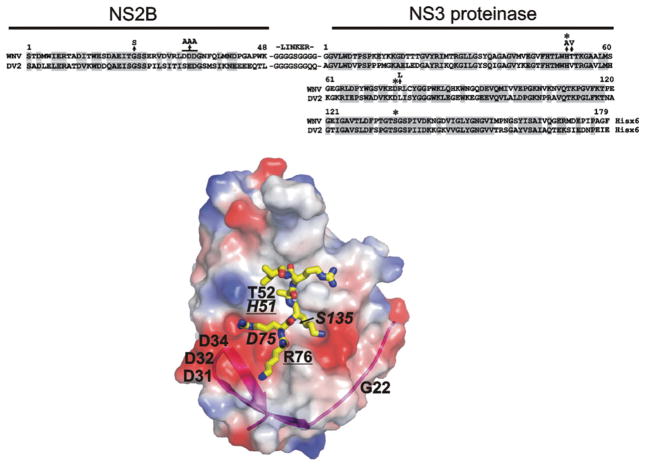
The WNV (strain NY99) and the DV2 (strain 16681) cDNAs were generously given by Dr Richard Kinney (Centers for Disease Control and Prevention, Fort Collins, CO, U.S.A.) and Dr Michael Diamond (Department of Internal Medicine, Washington University, St Louis, MO, U.S.A.) respectively. The WT (wild-type) DV2 cDNA was used in PCR reactions followed by routine gene-engineering manipulations to generate the DV2 NS2B–NS3pro constructs, which included the 48-residue NS2B cofactor (amino acids 1393–1440) linked via a nona-peptide linker GGGGSGGQQ to the NS3pro portion (amino acids 1476–1687). Similarly, the WNV cDNA was used to generate the WNV NS2B–NS3pro constructs, which included the 48-residue NS2B cofactor (amino acids 1423–1470) linked via a GGGGSGGGG linker to the NS3pro sequence (amino acids 1506–1689) [6]. The design and purification of the autolytic-site-deficient WNV NS2B–NS3pro K48A construct, the H51A construct, the G22S construct and D32A/D33A/D34A mutants (DDD/AAA) with the mutations in the NS2B cofactor sequence have been described previously [4–6,48]. The T52V and R76L mutations in the NS3pro portion were inserted into the WT WNV NS2B–NS3pro construct. As a result, the NS2B/NS3pro boundary was autocleaved in these constructs, albeit NS2B then remained non-covalently associated with the NS3pro domain [7]. The sequence of the constructs is shown in Figure 1. The constructs were re-cloned into the pET101/D-TOPO expression vector (Invitrogen, San Diego, CA, U.S.A.).
Figure 1. Structure and sequence alignment of the NS2B cofactor and the NS3 proteinase domain of WNV and DV2.
Upper panel: sequence alignment of the NS2B–NS3pro constructs; homologous amino-acid-residue positions are shaded. The asterisks above the sequences indicate His51, Asp75 and Ser135 of the catalytic triad. The arrows indicate mutations (G22S, DDD/AAA, H51A, T52V and R76L). The G22S and DDD/AAA constructs also contained the K48A mutation that inactivated the Lys48↓Gly autolytic cleavage site where glycine is the N-terminal residue of the linker. Lower panel: the structure of WNV NS2B–NS3pro (PDB accession code 2IJO) [8]. The original position of the His51, Asp75 and Ser135 of the catalytic triad (italicized) and the mutant positions are shown. The mutations that affect the binding of the proteinase with the antibodies are underlined. The surface is coloured by electrostatic potential (negative, red; positive, blue). The magenta ribbon shows NS2B contributing to the NS3pro active site. The positioning of a substrate-mimetic peptide (KRKARI) is shown in yellow. The model was presented using PyMol software (DeLano Scientific; http://www.pymol.org).
Proteinase expression and purification
Competent Escherichia coli BL21 CodonPlus® (DE3)-RIPL cells (Stratagene, San Diego, CA, U.S.A.) were transformed with the individual recombinant pET101/D-TOPO vectors. Transformed cells were grown in Luria–Bertani broth at 37 °C to reach D600 = 0.6. The protein expression was then induced at 37 °C using 1 mM isopropyl β-D-thiogalactoside for an additional 6 h. The cells were collected by centrifugation, resuspended in 20 mM Tris/HCl, pH 8.0, containing 1 M NaCl and 1 mg/ml lysozyme and disrupted by sonication (30s pulse, 30s interval, six pulses) on ice using a Misonix 3000 sonicator (Farmingdale, NY) at a power setting according to the manufacturer’s instructions for ‘liquid sample volume’. Cell debris was removed by centrifugation. The WNV and DV2 constructs were purified from the supernatant fraction using HiTrap Co2+-chelating chromatography on a 5 ml column (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s instructions. The His6-tagged NS2B–NS3pro constructs were eluted using a 0–500 mM gradient of imidazole concentrations. The fractions were analysed using SDS/PAGE followed by Coomassie staining, and also by Western blotting with a His6-tagged antibody (Clontech, Mountain View, CA, U.S.A.).
Proteinase assays with fluorogenic peptide
The assay for NS2B–NS3pro peptide-cleavage activity was performed in 0.2 ml of 20 mM Tris/HCl buffer, pH 8.0, containing 20 % (v/v) glycerol and 0.005 % Brij 35. The cleavage-peptide (Pyr-RTKR–AMC) and enzyme concentrations were 25 μM and 10 nM respectively. The reaction velocity was monitored continuously at λex = 360 nm and λem = 465 nm on a Spectramax Gemini EM fluorescence spectrophotometer (Molecular Devices, Sunnyvale, CA, U.S.A.). All assays were performed in triplicate in wells of a 96-well plate. The assays were repeated multiple times and the results were reproducible from day to day.
The concentrations of catalytically active NS2B–NS3pro in the purified samples were always quantified by active-site titration prior to kinetic studies [7]. Active-site titration of NS2B–NS3pro was performed with aprotinin (Ki = 20 nM). Briefly, 10 nM NS2B–NS3pro was incubated with increasing concentrations of aprotinin. Residual activity of NS2B–NS3pro was then measured by determining the rate of cleavage of Pyr-RTKR–AMC. The results were plotted against the amounts of aprotinin and a line was fitted through the data points. The intercept on the x-axis is equal to the concentration of active enzyme. The concentration of active NS2B–NS3pro was close to 100 % when compared with the protein concentration.
Determination of the Ki values of the inhibitory antibodies
The NS2B–NS3pro constructs (50 nM) were pre-incubated with increasing concentrations of the antibodies for 30 min at room temperature (22 °C) in 0.1 ml of 20 mM Tris/HCl buffer, pH 8.0, containing 20 % glycerol and 0.005 % Brij 35. The Pyr-RTKR–AMC substrate (25 μM) was then added in 0.1 ml of the same buffer. All assays were performed in triplicate in wells of a 96-well plate. IC50 values were calculated for each antibody by determining the antibody concentration needed to inhibit half of the maximum cleavage activity of NS2B–NS3pro against Pyr-RTKR–AMC. The measured IC50 values were used to calculate the Ki values. Ki values of the competitive antibody inhibitors were derived using the Cheng–Prusoff equation, Ki = IC50/(1 + [S]/Km), where Ki is the binding affinity of the inhibitor, IC50 is the functional strength of the inhibitor, [S] is the substrate concentration and Km is the affinity of the substrate for the enzyme. GraphPad Prism was used as a fitting software. Because the [S] and Km values of the Pyr-RTKR–AMC substrate that we used were 25 μM and 43–71 μM respectively [5,7], the actual difference between the IC50 and Ki values of the inhibitory antibodies was less than 25 %.
ELISA
Wells of a Nunc-Immuno MaxiSorp 96-well flat-bottomed plate (Thermo Fisher Scientific, Rochester, NY, U.S.A.) were coated in triplicate for 18 h at 4 °C using the WNV or DV construct (0.1 ml/well; 1 μg/ml). Control wells were coated with 1 μg/ml BSA. After washing with PBS, non-specific binding sites were saturated by incubation with 0.2 ml of 1 % BSA for 2 h at room temperature. The wells were then washed using PBS/0.05 % Tween 20. The 2 μg/ml antibody solution (0.1 ml) in PBS/1 %BSA/0.05 %Tween 20 was added to the wells. After a 2 h incubation at room temperature, the wells were washed using PBS/0.05 % Tween 20. Goat anti-human F(ab′)2 fragment (1:5000 dilution in 1 % BSA/0.05 % Tween 20 in PBS; 0.1 ml) was then added to the wells and the incubation was continued for 2 h at room temperature. After washing with PBS/0.05 % Tween 20 and incubation with 0.1 ml of TMB/M (TMB Super Sensitive One-Component HRP Microwell Substrate solution; BioFX, Owings Mills, MD, U.S.A.), the reaction was stopped by the addition of 0.05 ml of 1 M HCl, and the intensity of the colour reaction was measured at 450 nm using a Spectra Fluor Plus fluorescence plate reader (Tecan, Mannedorf, Switzerland).
Generation of human NS2B–NS3-specific antibodies from the HuCAL (human combinatorial antibody library) GOLD® library
The synthetic phage-display HuCAL GOLD® library was designed, developed and tested by MorphoSys (Martinsried/Planegg, Germany). Recombinant antibodies were isolated from a one-billion-gene sub-library of the HuCAL GOLD® collection of 15 billion antibody genes [47] by three rounds of selection (panning) using the immobilized recombinant WT NS2B–NS3pro protein as a bait [46,47,49–52]. Prior to selection, the phage library was blocked with the NS2B–NS3pro H51A construct in order to deplete the antibodies to the common epitopes present in both the immobilized NS2B–NS3pro and the H51A mutant. BSA and the ubiquitin–His6 and CD33–His6 constructs were used as controls to eliminate false-positive antibodies and antibodies to the His6 tag. After three rounds of selection, the enriched Fab gene pool was sub-cloned into an expression vector that leads to the expression of bivalent Fab mini-antibodies [53]. Single E. coli colonies were screened through ELISAs using both the NS2B–NS3pro K48A and the H51A mutant, and the antibodies specific for the WT protein were expressed in E. coli and then purified using metal-chelating chromatography.
Western blotting
Following the transfer to the Immobilon P membrane (Millipore, Bedford, MA, U.S.A.), the membrane was incubated for 16 h at 4 °C with the primary antibodies AbD05320, AbD05321, AbD05322, AbD05444, AbD05445 and AbD05446 (0.25 μg/ml) or the AbD05323 antibody (0.5 μg/ml). The secondary antibody (1:1000 dilution) was goat anti-human IgG F(ab′)2 fragment conjugated with HRP. TMB/M was used as a substrate.
RESULTS
The structure of the two-component flaviviral proteinase
Multiple previous studies have showed that the 48-residue central NS2B domain linked to the N-terminus of NS3 via an artificial flexible linker (GGGGSGGGG) is active in the in vitro protease assays [54,54a]. However, the NS3pro activity usually cleaves the initial K48G↓GGGSGGGG linker sequence, leading to the presence of the non-covalently associated NS2B cofactor and the NS3pro domain in the samples. The K48A mutation of the C-terminal amino-acid residue of the NS2B sequence inactivated the autolytic cleavage site. As a result, the NS2B–NS3pro K48A mutant is resistant to autoproteolysis and is represented by the intact single-chain NS2B–NS3pro construct in the samples. In the additional WNV mutant, called H51A, an alanine residue was substituted for the catalytically essential His51 of the NS3pro active site. As a result of this mutation, the H51A construct became catalytically inert and was not autocleaved.
NS3pro from DV and WNV share 50 % sequence identity. Despite the limited number of amino-acid substitutions proximal to the catalytic triad, the two proteinases display significant differences in their substrate-cleavage preferences and, accordingly, in the structure of the active-site region. Active-site differences between WNV and DV exist at Thr52 (Val52 in DV) and Arg76 (Leu76 in DV). To explore the potential role of the Thr52 and Arg76 residues, we constructed chimaeric proteins with replacements of DV residues into the WNV protein, leading to the construction of the T52V and R76L mutants.
Additional mutants used, G22S and DDD/AAA, involved the modifications of the NS2B–NS3pro K48A sequence that might affect either the folding or the interactions of NS2B with NS3pro in the proximity of the active-site region or both parameters (Figure 1).
WT DV and WNV NS2B–NS3pro, together with the WNV/DV chimaeras, were expressed in E. coli with C-terminal His6 tags and isolated from the soluble fraction by metal-chelating chromatography. The cleavage kinetics of the Pyr-RTKR–AMC fluorescent peptide substrate was measured to confirm the catalytic potency of the constructs (Table 1). The purified constructs were used as baits in the antibody selection and characterization procedures.
Table 1. Kinetic parameters of the Pyr-RTKR–AMC cleavage by the DV2 and WNV constructs.
The G22S, DDD/AAA and H51A WNV mutants have no activity because the G22S and DDD/AAA mutations affect the interactions of the NS2B cofactor with the NS3pro domain [48] and because the H51A mutation inactivates the proteinase. Results are means ± S.D.
| NS2B–NS3pro | Km (μM) | kcat (s−1) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| WNV WT | 71 ± 15 | 6.3 ± 0.35 | 88 ± 12 |
| WNV K48A | 58.8 ± 10 | 5.25 ± 0.25 | 89.3 ± 11 |
| WNV T52V | 0.13 ± 0.006 | 0.013 ± 0.0006 | 100 ± 10 |
| WNV R76L | 89 ± 16 | 7.3 ± 0.4 | 82 ± 11 |
| DV2 WT | 3.6 ± 0.2 | 0.02 ± 0.001 | 5.5 ± 0.5 |
Identification of the active-site-targeting antibodies
To identify the Fabs capable of binding to the active-site region of WNV NS2B-NS3pro, we used a bio-panning procedure that involved both the catalytically active NS2B–NS3pro K48A construct with the intact active-site sequence and the inert NS2B–NS3pro H51A mutant with the inactivated active site (Figure 2).
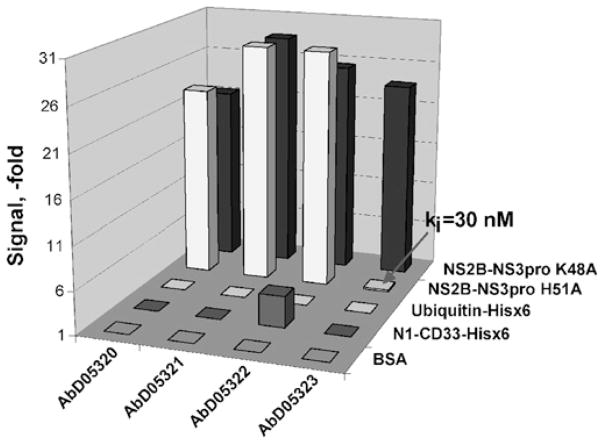
Figure 2. Selection process of the antibodies directed to the region proximal to the essential His51 of the catalytic triad.
The purified NS2B–NS3pro K48A construct was used for selection of the phage antibody library, whereas the inert H51A mutant with the mutation of the active-site His51 was used for library blocking and counter-screening. The constructs were C-terminally tagged with a His6 tag. The antibodies, which recognize the K48A enzyme and which do not recognize the mutant, were selected from the antibody library. BSA, ubiquitin–His6 and CD33–His6 were used as controls to eliminate false-positive antibodies. The AbD05323 antibody (indicated by an arrow) was selected in this particular experiment. Hisx6, His6; ki, Ki.
We tested several different panning methodologies, each of which included three consecutive rounds of phage selection. In these procedures, the NS2B–NS3pro K48A construct was immobilized on microtitre plates as bait. In the first approach, we blocked the phage library using the H51A mutant construct (20 μg/ml). The obtained antibodies (AbD05320 and AbD05321) bound both the K48A protein and the mutant, indicating that the blocking was not stringent enough. In the course of performing the second methodology, we reduced the amount of the immobilized NS2B–NS3pro K48A construct from 50 μg/ml in the first round to 5 μg/ml in the second round and, finally, to 1 μg/ml in the third round. Again, blocking of the library was performed with the H51A mutant (20 μg/ml). Using this strategy, we isolated two additional antibodies (AbD05322 and AbD05323); the latter was specific for the K48A construct. In the third approach, we blocked the library using a high concentration (100 μg/ml) of the H51A mutant. As a result, we selected three additional antibodies (AbD05444, AbD05445 and AbD05446). Overall, as a result of the bio-panning of the HuCAL GOLD® library, seven individual antibodies were identified (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/427/bj4270369add.htm). These seven antibodies were isolated from the respective recombinant E. coli cells and the purified antibody samples were characterized further.
Characterization of the antibodies
To determine whether the antibodies were resistant to NS2B–NS3pro proteolysis, the purified antibody samples (4 μg; 8 μM each) were co-incubated for 2 h at 37 °C with the purified NS2B–NS3pro K48A construct (1 μg; 1.4 μM; the enzyme/substrate molar ratio was 1:6) in 10 mM Tris/HCl buffer, pH 8.0, containing 20 %(v/v) glycerol and 0.005 %Brij 35. The digested samples were separated by gradient SDS/4–20 % PAGE. All of the antibodies were completely resistant to NS2B–NS3pro proteolysis (results not shown).
To determine the inhibitory potency of the antibodies, increasing concentrations of the purified AbD05320, AbD05321, AbD05322, AbD05323, AbD05444, AbD05445 and AbD05446 samples were each co-incubated for 60 min at room temperature with NS2B–NS3-K48A (10 nM). The residual proteolytic activity of the protease was then measured using the Pyr-RTKR–AMC fluorescent peptide substrate. A high-level inhibition of the proteolytic activity of NS2B–NS3-K48A was observed at a 300 nM concentration of AbD05320, AbD05322, AbD05323, AbD05444 and AbD05445, whereas AbD05321 and AbD05446 were less inhibitory (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/427/bj4270369add.htm).
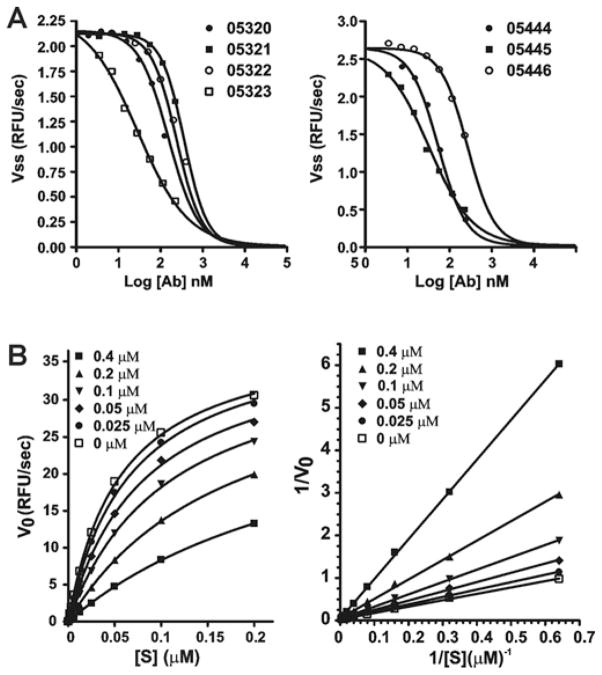
To analyse the inhibitory characteristics of the antibodies in more detail, we determined the Ki values of the antibodies (Figure 3). For these purposes, NS2B–NS3-K48A (10 nM) was pre-incubated with the increasing concentrations of the antibodies for 60 min at room temperature in 10 mM Tris/HCl buffer, pH 8.0, containing 20 % (v/v) glycerol and 0.005 % Brij 35. The Pyr-RTKR–AMC substrate (25 μM) was then added to the 0.1 ml reactions. Then, the Ki values of AbD05320, AbD05321, AbD05322, AbD05323, AbD05444, AbD05445 and AbD05446 were determined to be 264, 400, 170, 31, 58, 35 and 288 nM respectively. Because the antibodies bind to the active-site region of NS2B–NS3pro, Lineweaver–Burk plots, as a result, demonstrated strictly competition-type inhibition by the antibody inhibitors. The representative results are shown in Figure 3. Because excessively high concentrations of the antibodies were required for the V0 values to reach clear plateaus, the incomplete plots approaching the plateaus were used.
Figure 3. Catalytic parameters of the inhibitory antibodies.
(A) Before the addition of the Pyr-RTKR–AMC substrate (25 μM), the purified WNV proteinase (10 nM) was co-incubated for 30 min with increasing concentrations of the antibodies. The cleavage of the Pyr-RTKR–AMC peptide by the proteinase was monitored to determine the Ki values. (B) Left-hand panel: competitive inhibition of NS2B–NS3pro proteolysis of Pyr-RTKR–AMC NS2B–NS3pro by the AbD05444 antibody (0–4 μM). Right-hand panel: Lineweaver–Burk plot of the results. The inhibitory antibody was added to the reactions. The initial velocity of the Pyr-RTKR–AMC cleavage was then measured in triplicate. Note that the Km values, but not the Vmax values, are affected by the inhibitor. RFU, relative fluorescence unit; V0 and Vss, steady-state rate.
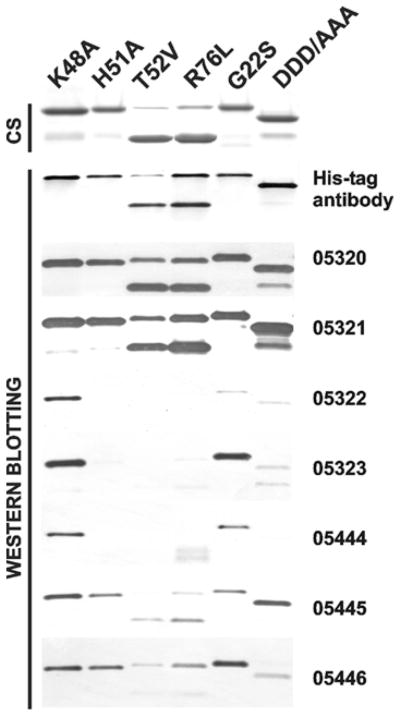
To determine whether the antibodies selectively bind to the WNV and DV constructs, we used a Western blotting procedure (Figure 4). The purified WNV K48A, H51A, T52V, R76L, G22S and DDD/AAA constructs (100 ng of each) were added to the E. coli soluble protein fraction (20 μg of total protein) and the samples were analysed by Western blotting with the AbD05320, AbD05321, AbD05322, AbD05323, AbD05444, AbD05445 and AbD05446 antibodies followed by a goat anti-human IgG F(ab′)2 fragment conjugated to HRP and a TMB/M substrate. Because the NS2B/NS3pro boundary is autocleaved in the T52V and R76L mutants, the resulting NS3pro domain has a low molecular mass compared with other mutants that were constructed using the autolytic-site-deficient K48A background.
Figure 4. The H51A, T52V and R76L WNV NS2B–NS3 mutants do not interact with the AbD05323 and AbD05444 antibodies.
The purified mutant constructs were analysed by SDS/PAGE (4 μg/lane) followed by Coomassie staining (CS) and Western blotting (100 ng/lane) with a His6-tagged antibody and the selected AbD05320, AbD05321, AbD05322, AbD05323, AbD05444, AbD05445 and AbD05446 antibodies. Note that AbD05323 and AbD05444 did not bind to the H51A, T52V and R76L constructs, which exhibit mutations in the proximity of the active site.
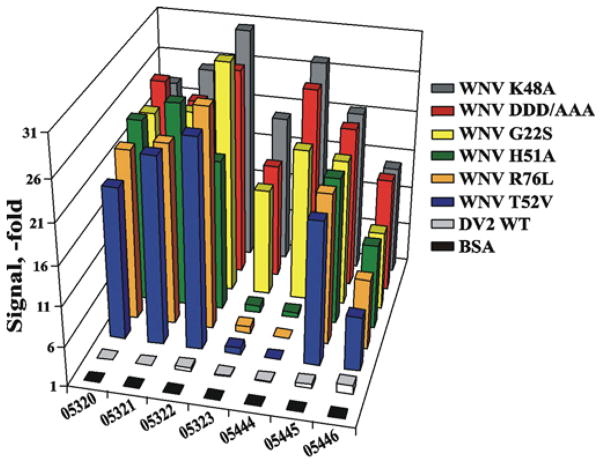
Although the G22S and DDD/AAA mutations in the NS2B cofactor did not notably affect the binding efficiency of all of the antibodies, the presence of H51A, T52V and R76L mutations in the NS3pro active-site region significantly decreased the binding efficiency of AbD05322 (Ki = 170 nM), AbD05323 (Ki = 31 nM) and AbD05444 (Ki = 58 nM). In turn, AbD05320, AbD05321, AbD05445 and AbD05446 partially cross-reacted with the H51A and additional tested mutants (Table 2).
Table 2.
The Ki values (nM) of the antibodies against the original WNV NS2B–NS3pro K48A construct and the WNV NS2B–NS3pro mutants
To corroborate these results using the native proteinase constructs (rather than partially denatured ones as in the Western-blotting experiments), we performed an assessment of the antibody binding with the WNV and DV constructs using ELISA. After coating wells of a 96-well plate with the constructs, the antibodies were allowed to bind to the immobilized material. The level of the resulting immune complexes was determined using the goat anti-human Fab fragment conjugated with HRP (Figure 5). None of the selected anti-WNV antibodies recognized the closely related DV and HCV proteinases, as well as human serine proteinases, including furin (results not shown). Because AbD05323 and AbD05444 did not interact with the H51A, R76L and T52V WNV constructs, these two antibodies were clearly different when compared with the other anti-WNV antibodies we analysed. Overall, we concluded that the AbD05323 and AbD05444 antibodies target the active-site region of the two-component WNV NS2B–NS3pro in a selective and focused manner.
Figure 5. ELISA of the NS2B–NS3pro constructs.
The antibodies were allowed to interact with the native K48A and mutant WNV co nstructs and with the WT DV2 constructs. The binding efficiency is expressed as a fold increase relative to the control (BSA).
DISCUSSION
Because of their unique ability to bind specifically to nearly any antigen, antibodies provide an excellent scaffold for designing inhibitors targeted to the individual members of a family of homologous enzymes. Over the past decade, recombinant technology has enabled the production of engineered antibody fragments such as Fabs or single-chain scFvs. Phage-display technology allows the presentation of the scFv or Fab on the phage surface and this technology has been used successfully for the isolation of specific antibodies from human repertoire (immune, non-immune or synthetic) libraries via several enrichment cycles. The identified scFvs/Fabs can be genetically manipulated further for improved specificity and affinity. The scFv–His6 antibody format can be readily modified and transformed into a two-chain Fab format tagged with FLAG or streptavidin tags or vice versa.
There is a need to develop inhibitors of flaviviruses, including Dengue haemorrhagic fever, yellow fever, Japanese encephalitis viruses and WNV. Today, there are millions of cases of Flaviviridae infections worldwide and multiple cases of WNV in the United States. The two-component NS2B–NS3pro is the only proteinase encoded by the flaviviral genome. This proteinase activity is essential for the proteolytic processing of the viral polyprotein precursor and virus infectivity. NS2B–NS3pro consists of two components of which the NS3pro portion represents the proteinase domain and the NS2B part is a cofactor. NS3pro alone is inactive. Because of its irreplaceable role in the virus life cycle, NS2B–NS3pro is a highly promising target for drug design.
We successfully used the phage Fab human synthetic antibody HuCAL library to isolate selective and potent function-blocking, active-site-targeting antibodies to WNV NS2B–NS3pro. For this purpose, we developed a novel technology, which, by analogy, allows the rapid and efficient identification of active-site-targeting antibodies to any enzyme. In our methodology, we tried to identify only those antibodies that were able to bind to the WT proteinase, but which were then incapable of binding with the proteinase mutant bearing a point mutation of the catalytically essential active-site residue (in the case of NS2B–NS3pro, the histidine residue of the catalytic triad). As a result, the active-site-targeting antibodies were readily selected from the antibody library. Conceptually, the use of the focused panning technology we described in our report may facilitate the identification of the active-site-targeting recombinant antibodies to any enzyme.
From the seven initially isolated Fabs, AbD05323 (Ki = 31 nM) and AbD05444 (Ki = 58 nM) appeared to be the most promising. These two Fabs were able to bind to the WT WNV proteinase, but not the to proteinase with the H51A, T52V and R76L mutations in the active-site region. The Fabs AbD05323 and AbD05444 were then transformed into an scFv format. These low-nanomolar-range inhibitory human antibodies are both novel molecular tools for the determination of the NS2B–NS3pro active-site parameters and a valuable guiding scaffold for the design of the anti-flaviviral inhibitors. Because the NS3pro viral enzyme activity is in close association with the endoplasmic-reticulum membrane inside the cell, a limited level of cell penetration, unfortunately, remains a major obstacle in using the anti-NS3pro antibodies for clinical purposes. However, the selected Fab antibodies may be used in vitro for the determination, in better detail, of the NS2B–NS3pro active-site parameters. In addition, the structural parameters of the antibody–protease active-site interface may guide a scaffold for the design of the selective, small-molecule, anti-flaviviral inhibitors. On the basis of the available structure of the inhibitory complex of the trypsin-like matriptase serine proteinase with the E2 Fab antibody, which is a 15 pM inhibitor of matriptase [55,56], we believe that further refinement of the 30–50-nM-range inhibitory NS2B–NS3pro antibodies is possible and that this refinement can be accomplished by using either mutagenesis of CDR-H (heavy-chain variable regions) or DNA shuffling or both.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant numbers AI061139, AI055789 (to A.S.)].
Abbreviations used
- C
capsid protein
- CDR
complementarity-determining region
- DV2
Dengue virus serotype 2
- E
envelope protein
- HRP
horseradish peroxidase
- HuCAL
human combinatorial antibody library
- NS
non-structural protein
- NS3pro
proteinase domain of NS3
- prM
precursor membrane protein
- Pyr-RTKR–AMC
pyroglutamic acid-Arg-Thr-Lys-Arg–7-amino-4-methylcoumarin
- scFv
single-chain variable fragment
- TMB/M
TMB Super Sensitive One-Component HRP Microwell Substrate Solution
- WNV
West Nile virus
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Sergey Shiryaev characterized the purified scFv and Fab antibodies. Ilian Radichev performed mutagenesis and isolated the mutant NS2B–NS3 proteinase constructs. Boris Ratnikov performed the experiments that were designed to determine the inhibitory constants of the antibodies. Alexander Aleshin and Boguslaw Stec contributed to the modelling of the NS2B–NS3 proteinase structure. Katarzyna Gawlik contributed to the initial stages of the project. Christian Frisch and Achim Knappik performed the bio-panning procedures and isolated the scFv and Fab antibodies from the HuCAL GOLD® library. Alex Strongin developed and co-ordinated the project and wrote the paper. All authors discussed the results and their implications, and commented on the manuscript at all stages.
References
- 1.Beasley DW. Recent advances in the molecular biology of West Nile virus. Curr Mol Med. 2005;5:835–850. doi: 10.2174/156652405774962272. [DOI] [PubMed] [Google Scholar]
- 2.Cahour A, Falgout B, Lai CJ. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 4.Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. Expression and purification of a two-component flaviviral proteinase resistant to autocleavage at the NS2B–NS3 junction region. Protein Expr Purif. 2007;52:334–339. doi: 10.1016/j.pep.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiryaev SA, Kozlov IA, Ratnikov BI, Smith JW, Lebl M, Strongin AY. Cleavage preference distinguishes the two-component NS2B–NS3 serine proteinases of Dengue and West Nile viruses. Biochem J. 2007;401:743–752. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiryaev SA, Ratnikov BI, Aleshin AE, Kozlov IA, Nelson NA, Lebl M, Smith JW, Liddington RC, Strongin AY. Switching the substrate specificity of the two-component NS2B–NS3 flavivirus proteinase by structure-based mutagenesis. J Virol. 2007;81:4501–4509. doi: 10.1128/JVI.02719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiryaev SA, Ratnikov BI, Chekanov AV, Sikora S, Rozanov DV, Godzik A, Wang J, Smith JW, Huang Z, Lindberg I, et al. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393:503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 10.Falgout B, Miller RH, Lai CJ. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B–NS3 protease activity. J Virol. 1993;67:2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessaud M, Grard G, Peyrefitte CN, Pastorino B, Rolland D, Charrel RN, de Lamballerie X, Tolou HJ. Identification and enzymatic characterization of NS2B-NS3 protease of Alkhurma virus, a class-4 flavivirus. Virus Res. 2005;107:57–62. doi: 10.1016/j.virusres.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Droll DA, Krishna Murthy HM, Chambers TJ. Yellow fever virus NS2B–NS3 protease: charged-to-alanine mutagenesis and deletion analysis define regions important for protease complex formation and function. Virology. 2000;275:335–347. doi: 10.1006/viro.2000.0488. [DOI] [PubMed] [Google Scholar]
- 14.Wu CF, Wang SH, Sun CM, Hu ST, Syu WJ. Activation of dengue protease autocleavage at the NS2B–NS3 junction by recombinant NS3 and GST–NS2B fusion proteins. J Virol Methods. 2003;114:45–54. doi: 10.1016/j.jviromet.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 16.Bera AK, Kuhn RJ, Smith JL. Functional characterization of cis and trans activity of the Flavivirus NS2B–NS3 protease. J Biol Chem. 2007;282:12883–12892. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- 17.Chambers TJ, Diamond MS. Pathogenesis of flavivirus encephalitis. Adv Virus Res. 2003;60:273–342. doi: 10.1016/S0065-3527(03)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers TJ, Droll DA, Tang Y, Liang Y, Ganesh VK, Murthy KH, Nickells M. Yellow fever virus NS2B–NS3 protease: characterization of charged-to-alanine mutant and revertant viruses and analysis of polyprotein-cleavage activities. J Gen Virol. 2005;86:1403–1413. doi: 10.1099/vir.0.80427-0. [DOI] [PubMed] [Google Scholar]
- 19.Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B–NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 21.Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chappell KJ, Nall TA, Stoermer MJ, Fang NX, Tyndall JD, Fairlie DP, Young PR. Site-directed mutagenesis and kinetic studies of the West Nile virus NS3 protease identify key enzyme–substrate interactions. J Biol Chem. 2005;280:2896–2903. doi: 10.1074/jbc.M409931200. [DOI] [PubMed] [Google Scholar]
- 23.Niyomrattanakit P, Winoyanuwattikun P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J Virol. 2004;78:13708–13716. doi: 10.1128/JVI.78.24.13708-13716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niyomrattanakit P, Yahorava S, Mutule I, Mutulis F, Petrovska R, Prusis P, Katzenmeier G, Wikberg JE. Probing the substrate specificity of the dengue virus type 2 NS3 serine protease by using internally quenched fluorescent peptides. Biochem J. 2006;397:203–211. doi: 10.1042/BJ20051767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Insights to substrate binding and processing by West Nile virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J Biol Chem. 2006;281:38448–38458. doi: 10.1074/jbc.M607641200. [DOI] [PubMed] [Google Scholar]
- 26.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Protein Expr Purif. 2007;53:87–96. doi: 10.1016/j.pep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J Gen Virol. 2008;89:1010–1014. doi: 10.1099/vir.0.83447-0. [DOI] [PubMed] [Google Scholar]
- 28.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. West Nile virus NS2B/NS3 protease as an antiviral target. Curr Med Chem. 2008;15:2771–2784. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- 29.Ekonomiuk D, Su XC, Ozawa K, Bodenreider C, Lim SP, Yin Z, Keller TH, Beer D, Patel V, Otting G, et al. Discovery of a non-peptidic inhibitor of West Nile virus NS3 protease by high-throughput docking. PLoS Negl Trop Dis. 2009;3:e356. doi: 10.1371/journal.pntd.0000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, et al. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B–NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 31.Knox JE, Ma NL, Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, et al. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J Med Chem. 2006;49:6585–6590. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- 32.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Lohr K, Knox JE, Phong WY, Ma NL, Yin Z, Sampath A, Patel SJ, Wang WL, Chan WL, Rao KR, et al. Yellow fever virus NS3 protease: peptide-inhibition studies. J Gen Virol. 2007;88:2223–2227. doi: 10.1099/vir.0.82735-0. [DOI] [PubMed] [Google Scholar]
- 34.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of West Nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoermer MJ, Chappell KJ, Liebscher S, Jensen CM, Gan CH, Gupta PK, Xu WJ, Young PR, Fairlie DP. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J Med Chem. 2008;51:5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- 36.Tomlinson SM, Malmstrom RD, Russo A, Mueller N, Pang YP, Watowich SJ. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 2009;82:110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donzeau M, Knappik A. Recombinant monoclonal antibodies. Methods Mol Biol. 2007;378:14–31. doi: 10.1007/978-1-59745-323-3_2. [DOI] [PubMed] [Google Scholar]
- 38.Filpula D. Antibody engineering and modification technologies. Biomol Eng. 2007;24:201–215. doi: 10.1016/j.bioeng.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Hust M, Dubel S. Phage display vectors for the in vitro generation of human antibody fragments. Methods Mol Biol. 2005;295:71–96. doi: 10.1385/1-59259-873-0:071. [DOI] [PubMed] [Google Scholar]
- 40.Pini A, Bracci L. Phage display of antibody fragments. Curr Protein Pept Sci. 2000;1:155–169. doi: 10.2174/1389203003381397. [DOI] [PubMed] [Google Scholar]
- 41.Wang XX, Shusta EV. The use of scFv-displaying yeast in mammalian cell surface selections. J Immunol Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Kretzschmar T, von Ruden T. Antibody discovery: phage display. Curr Opin Biotechnol. 2002;13:598–602. doi: 10.1016/s0958-1669(02)00380-4. [DOI] [PubMed] [Google Scholar]
- 43.Holliger P, Prospero T, Winter G. ‘Diabodies’: small bivalent and bispecific antibody fragments. Proc Natl Acad Sci USA. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidner KM, Denzin LK, Voss EW., Jr Molecular stabilization effects of interactions between anti-metatype antibodies and liganded antibody. J Biol Chem. 1992;267:10281–10288. [PubMed] [Google Scholar]
- 45.Marasco WA, Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat Biotechnol. 2007;25:1421–1434. doi: 10.1038/nbt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Pluckthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 47.Rothe C, Urlinger S, Lohning C, Prassler J, Stark Y, Jager U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Radichev I, Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B–NS3 proteinase. J Gen Virol. 2008;89:636–641. doi: 10.1099/vir.0.83359-0. [DOI] [PubMed] [Google Scholar]
- 49.Frisch C, Brocks B, Ostendorp R, Hoess A, von Ruden T, Kretzschmar T. From EST to IHC: human antibody pipeline for target research. J Immunol Methods. 2003;275:203–212. doi: 10.1016/s0022-1759(03)00011-5. [DOI] [PubMed] [Google Scholar]
- 50.Jarutat T, Frisch C, Nickels C, Merz H, Knappik A. Isolation and comparative characterization of Ki-67 equivalent antibodies from the HuCAL phage display library. Biol Chem. 2006;387:995–1003. doi: 10.1515/BC.2006.123. [DOI] [PubMed] [Google Scholar]
- 51.Krebs B, Rauchenberger R, Reiffert S, Rothe C, Tesar M, Thomassen E, Cao M, Dreier T, Fischer D, Hoss A, et al. High-throughput generation and engineering of recombinant human antibodies. J Immunol Methods. 2001;254:67–84. doi: 10.1016/s0022-1759(01)00398-2. [DOI] [PubMed] [Google Scholar]
- 52.Rauchenberger R, Borges E, Thomassen-Wolf E, Rom E, Adar R, Yaniv Y, Malka M, Chumakov I, Kotzer S, Resnitzky D, et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem. 2003;278:38194–38205. doi: 10.1074/jbc.M303164200. [DOI] [PubMed] [Google Scholar]
- 53.Pack P, Pluckthun A. Miniantibodies: use of amphipathic helices to produce functional, flexibly linked dimeric FV fragments with high avidity in Escherichia coli. Biochemistry. 1992;31:1579–1584. doi: 10.1021/bi00121a001. [DOI] [PubMed] [Google Scholar]
- 54.Nall TA, Chappell KJ, Stoermer MJ, Fang NX, Tyndall JD, Young PR, Fairlie DP. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem. 2004;279:48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- 54.Padmanabhan R, Strongin AY. Translation and processing of the Dengue virus polyprotein. In: Hanley K, Weaver SC, editors. Frontiers in Dengue Virus Research. Caister Academic Press; Trowbridge: 2010. pp. 13–33. [Google Scholar]
- 55.Farady CJ, Egea PF, Schneider EL, Darragh MR, Craik CS. Structure of an Fab–protease complex reveals a highly specific non-canonical mechanism of inhibition. J Mol Biol. 2008;380:351–360. doi: 10.1016/j.jmb.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farady CJ, Sun J, Darragh MR, Miller SM, Craik CS. The mechanism of inhibition of antibody-based inhibitors of membrane-type serine protease 1 (MT-SP1) J Mol Biol. 2007;369:1041–1051. doi: 10.1016/j.jmb.2007.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.