Abstract
During recent years, the composition of the gut microbiota (GM) has received increasing attention as a factor in the development of experimental inflammatory disease in animal models. Because increased variation in the GM might lead to increased variation in disease parameters, determining and reducing GM variation between laboratory animals may provide more consistent models. Both genetic and environmental aspects influence the composition of the GM and may vary between laboratory animal breeding centers and within an individual breeding center. This study investigated the variation in cecal microbiota in 8-wk-old NMRI and C57BL/6 mice by using denaturing gradient gel electrophoresis to profile PCR-derived amplicons from bacterial 16S rRNA genes. Comparison of the cecal microbiotas revealed that the similarity index of the inbred C57BL/6Sca strain was 10% higher than that of the outbred Sca:NMRI stock. Comparing C57BL/6 mice from 2 vendors revealed significant differences in the microbial profile, whereas the profiles of C57BL/6Sca mice raised in separate rooms within the same breeding center were not significantly different. Furthermore, housing in individually ventilated cages did not lead to intercage variation. These results show that denaturing gradient gel electrophoresis is a simple tool that can be used to characterize the gut microbiota of mice. Including such characterizations in future quality-control programs may increase the reproducibility of mouse studies.
Abbreviations: DGGE, denaturing gradient gel electrophoresis; GM, gut microbiota; IVC, individually ventilated caging
Various rodent models are used for studying human chronic inflammatory and autoimmune diseases. A positive correlation between the composition of the gut microbiota (GM) and disease expression has been observed for several inflammatory disease models.2,11,15,18,21,32,34,36,39 Increased variation in disease expression might reflect increased variation in the GM, consequently necessitating larger group sizes. In the interest of reducing animal numbers as a principle of in vivo scientific experimentation, the extent to which the composition of the GM varies in laboratory animals—particularly rodents—should be examined. Even though inflammatory diseases, such as inflammatory bowel disease, atherosclerosis, and diabetes, are among the diseases studied most extensively in rodent models, standardization of the GM in laboratory mouse production has been given scant attention.
Microbiologic examination based on classical cultivation of cecal samples of rats has revealed a substantial interindividual variation.10 Culture-independent studies based on gas chromatography37 and terminal restriction fragment polymorphism19 show a uniform microbial profile in mice that is dependent primarily on strain and age and independent of sex. Studies based on PCR followed by denaturing gradient gel electrophoresis (DGGE) likewise report different profiles in the GM related to mouse strains,24 whereas sex does also seem to affect the composition of the fecal microbiota in outbred rats1 and inbred mice.8 Human as well as animal studies have shown that genotype has a strong influence on the gastrointestinal bacterial community, whereas environmental influence seems less important.16,17,35,37,38,42 The GM of human monozygotic twins are more similar to each other than those of dizygotic twins.40 Cross-fostering of mice revealed a strain-specific fecal profile regardless of the foster strain.16 However, microbial exposure from the environment also plays a role in the colonization of the gastrointestinal community. A recent study6 has shown that embryo transfer with genetically distinct embryos reveals similar microbial profiles between mother and offspring, regardless of the genetic background of the embryos implanted. Furthermore, relocation of C3H mice at 4 wk of age leads to changes in the fecal profiles.6
Laboratory rats and mice are available as both outbred stocks and inbred strains from various vendors. Viable counts of the total bacterial load have revealed large differences in the cecal microbiota among animals from different vendors.14 Moreover, vendors frequently raise colonies of the same strain in multiple rooms staffed by different personnel. Therefore, the same strain might be exposed to different environmental microorganisms. Furthermore, over the years, rodent housing has gradually changed from open cages, in which animals share exposure to microorganisms in the surroundings, to individually ventilated caging (IVC) with limited shared environmental exposure.
The gastrointestinal tract is a complex microbial community with high density and diversity. Most of the bacterial community in the gut is obligate and facultative anaerobic bacteria and culture dependent techniques are restricted to cover only the aerobic heterotrophic fraction of the total bacterial population capable of forming colonies on solid media.33 As such, culture independent approaches are more applicable in characterizing the GM. Denaturating gradient gel electrophoresis is a PCR based method widely applied to study complex microbial communities as in soil,28 milk products5 and the gut microbiota.9 DGGE separates DNA fragments of the same lengths on the basis of differences in base-pair sequences in a polyacrylamide gel containing a linear increasing concentration of denaturant as described by Myzer and Smalla.28
The aim of our study was to examine the extent to which the composition of the GM varies in laboratory mice in relation to differences in environment (for example, deriving from different vendors or rooms, use of IVC systems) and genetic background (for example, inbred compared with outbred mice). To this end, we used DGGE to profile PCR-derived amplicons from bacterial 16S rRNA genes to determine the variation in the composition of the GM of laboratory NMRI and C57BL/6 mice.
Materials and Methods
Mice.
All breeding methods and experimental procedures conformed to regulations provided in the Animal Welfare Act and Animal Welfare Ordinance of Sweden26 and were authorized by The National Swedish Agriculture Department and the Swedish Regional Ethics Committee.
Inbred C57BL/6Sca and outbred Sca:NMRI mice were raised and maintained in a closed system as SPF animals in accordance with the Federation of European Laboratory Animal Science Association.29 Cages and supplies were autoclaved, if possible, or passed through a chemical lock. Caretakers entering the barrier were required to shower and change to autoclaved clothes, gloves, and mouth and hair covers provided inside the barrier. C57BL/6Sca and Sca:NMRI mice were housed in IVC (Blue Line Type II, Tecniplast, Varese, Italy) containing autoclaved aspen bedding and were offered free access to UV-treated water and autoclaved diet (RM 3, SDS, Witham, UK). All animal handling was performed in a laminar air-flow cabinet (Scanbur A/S, Karlslunde, Denmark). Housing and maintenance principles were in accordance with the Council of Europe Convention ETS 123.3 The animals were bred in trios, with 1 male and 2 female mice in each IVC cage. Offspring were weaned at the age of 3 wk and separated according to sex.
From the same breeding barrier, 2 IVC cages with female C57BL/6Sca mice (age, 8 wk) and 2 IVC cages with male C57BL/6Sca mice (age, 8 wk) were sampled randomly to analyze the effects of sex and the use of IVC on the composition of the GM. To study the genetic influence on the composition of the GM, cecal samples from 32 Sca:NMRI mice (age, 8 wk) and 30 C57BL/6Sca mice (age, 8 wk) raised in the same breeding barrier were used. In addition, 15 C57BL/6Sca mice (age, 8 wk) raised in a different room were used to examine the influence of differing environmental exposure on the composition of the GM. Finally, 16 C57BL/6NCrl mice (age, 8 wk) were used to illustrate any differences in the gut community of C57BL/6 mice from different laboratory animal breeders.
Cecum sample collection.
At the age of 8 wk, mice were euthanized by cervical dislocation, and cecal samples were collected aseptically in a laminar flow hood. The samples were immediately frozen at −80 °C until use. DNA was extracted (DNA Stool Mini Kit, Qiagen, Hilden, Germany) according to the manufacturer's instructions and stored at −40°C until analysis.
PCR amplification.
The V3 region of the bacterial 16S rRNA gene was amplified by PCR by using the universal primer set PRBA338f (5′ CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG 3′; Eurofins MWG Operon, Ebersberg, Germany) and PRUN518r (5′ ATT ACC GCG GCT GCT GG 3′; Eurofins MWG Operon). All reactions were carried out in a 50-µL volume containing 1.25 U HotMaster Taq DNA Polymerase (5 Prime, Hamburg, Germany), 5 µL 10× HotMaster Taq Buffer with 2.5 mM MgCl2 (5 Prime), 100 ng DNA, 10 pmol each primer, 0.3 mM dNTP (Bioline, Luckenwalde, Germany), and 1µg bovine serum albumin (Sigma-Aldrich, Brøndby, Denmark).
The PCR reaction was performed on a Robocycler Thermoblock (Stratagene, Aarhus, Denmark). Initial denaturation was performed at 95 °C for 5 min; followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 40 s; with a final elongation step at 72 °C for 10 min. A final product length of approximately 230 bp was checked by electrophoresis on a 2% agarose gel stained with ethidium bromide (Bio-Rad, Hercules, CA).
DGGE.
PCR amplicons were analyzed by DGGE (phorU-2 System, Ingeny, Goes, Netherlands) according to the manufacturer's instructions. The acrylamide concentration in the gel was 9%, and the linear denaturation gradient was 30% to 65% (100% denaturant corresponds to 7 M urea and 40% deionized formamide). Before loading, 35 µL PCR product was mixed with 6 µL 6× loading dye (Fermentas GmbH, St Leon-Rot, Germany). In addition to the samples, an inhouse standard PCR product was run on the DGGE gels allowing accurate alignment of lanes and bands within and between gels. Electrophoresis was performed in 0.5× TAE (1× TAE corresponds to 40 mM Tris acetate, 1 mM EDTA, pH 8.0), at 60 °C for 16 h at 120 V. Gels were stained by using SYBR Gold (dilution factor, 1:10000; Invitrogen, Eugene, OR) in 1× TAE for 1 h and photographed under UV transillumination (302 nm; (EDAS 290, Eastman Kodak, Rochester, NY).
Data analysis.
The 16S rRNA gene amplicon-based profiles obtained by DGGE were analyzed by using the Bionumerics program (version 4.5, Applied Maths, Sint-Martens-Latem, Belgium). To compare Dice similarity coefficients with a position tolerance setting of 1% optimization and 1% position tolerance for band composition, the unweighted pair-group method with arithmetic averages clustering algorithm (UPGMA) was used, and dendrogram patterns derived from the similarity of the DGGE profiles were constructed (Applied Maths). Group separation statistics based on the Jacknife method (Applied Maths) was performed to evaluate the stability of the given clustering, in which every sample within a group was compared with the samples of the opposite group, thereby identifying the integrity of the defined group. Furthermore, 3D principal component analysis based on the DGGE data was carried out (Applied Maths). The Fisher exact test was performed for every band class in the DGGE profiles, and P values for all band classes were calculated. Range-weighted richness (Rr),25 a unitless mathematical expression for the number of bands found in a DGGE profile independent of the denaturing gradient used, was calculated. A P value of 0.05 was used to define statistical significance.
Cluster analysis based on DGGE profiles indicated the similarity between individual profiles, where 100% indicated complete similarity and 0% complete dissimilarity. Interpretation of cluster analysis served to define a level at which similarities beyond this level are incidental. The level of reproducibility in our analysis was 92% (data not shown), therefore similarities beyond 92% were defined as being incidental.
Results
Similarity indices based on DGGE profiles from inbred C57BL/6Sca and outbred Sca:NMRI mice.
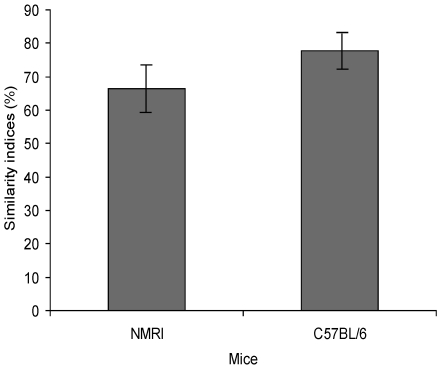
The composition of the GM of 32 Sca:NMRI and 30 C57BL/6Sca mice was analyzed. The overall similarity index (mean ± 1 SD) for Sca:NMRI mice was 66.5% ± 6.8%, whereas the overall similarity index for C57BL/6Sca mice was 77.8% ± 4.4% (Figure 1). Microbial diversity, based on Rr calculation, was high for both groups: 229 for C57BL/6Sca and 135 for Sca:NMRI mice.
Figure 1.
Similarity indices from cluster analysis based on DGGE profiles representing the similarity in the composition of the cecum microbiota from Sca:NMRI and C57BL/6Sca mice. The similarity index for Sca:NMRI mice is 66.5% ± 6.8%; that for C57BL/6Sca mice is 77.8% ± 4.4%.
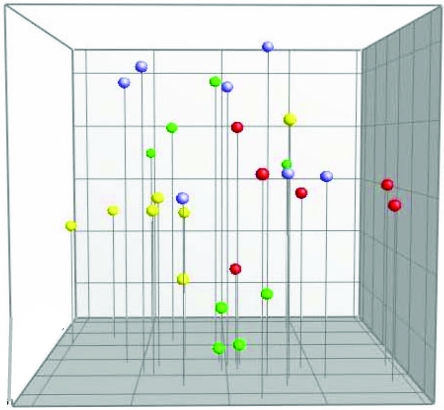
Cluster analysis and principal component analysis based on DGGE data from C57BL/6Sca mice housed in IVC.
There was no clear separation between the different IVC units housing female and male C57BL/6Sca mice (Figure 2). Neither IVC cage nor sex was a statistically significant factor for the composition of the cecal microbiota.
Figure 2.
Principal component analysis based on DGGE profiles of 16S rRNA gene-derived amplicons of cecal samples collected from C57BL/6Sca mice housed in IVC. Each cage is marked with a different color; mice from the same cage have the same colored dots. Yellow and red dots represent female mice, and green and blue dots represent male mice.
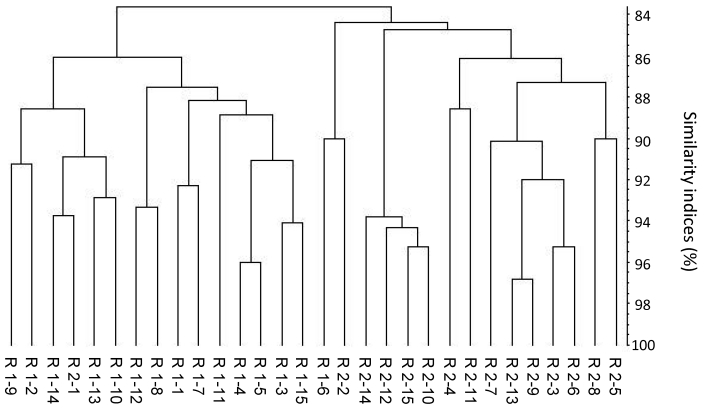
Comparison of DGGE profiles from C57BL/6Sca mice raised in different rooms at the same breeding center.
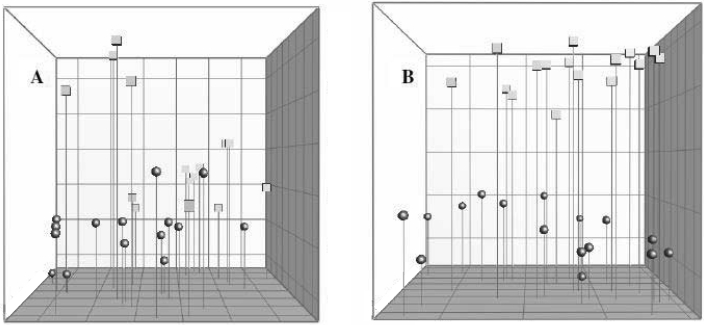
This comparison yielded 2 main clusters among the C57BL/6Sca mice that were raised in 2 different rooms at the same breeding center (Figure 3). The overall similarity index (regardless of room) was 83.7% ± 3.8%, that for mice in room 1 was 86.0% ± 3.2%, and that for mice from room 2 was 84.4% ± 3.4%. Group violation (Table 1) indicates that in 31.4% and 22.9% of the cases, the given DGGE profiles could have been assigned to the opposite group. The 3D principal component analysis plot (Figure 4 A) shows no clear separation between the samples from the 2 rooms, and no significant difference in the composition of the cecal microbiota between C57BL/6Sca mice raised in separate rooms could be identified based on PCR/DGGE.
Figure 3.
Dendrogram based on DGGE profiles representing 16S rRNA gene-derived amplicons of cecal samples collected from C57BL/6Sca mice originating from 2 breeding rooms (R1 and R2) at the same laboratory animal vendor. Each figure within the groups indicates a sample from a single mouse (numbered from 1 to 15). The scale bar represents the similarity index as a percentage (100 indicates complete similarity and 0 complete dissimilarity). The overall similarity index is 83.7% ± 3.8%. The similarity index for room 1 is 86.0% ± 3.2%; that for room 2 is 84.4% ± 3.4%.
Table 1.
Group separation statistics based on cluster analysis obtained from DGGE profiles comparing the cecal microbiota in C57BL/6 mice from different rooms within the same laboratory animal vendor or from different laboratory animal vendors
| Room 1 | Room 2 | Breeding center 1 | Breeding center 2 | |
| Room 1 | 77.1% | 31.4% | ||
| Room 2 | 22.9% | 68.6% | ||
| Breeding center 1 | 94.1% | 6.0% | ||
| Breeding center 2 | 5.9% | 94.0% |
Figure 4.
Principal component analysis based on DGGE profiles of 16S rRNA gene-derived amplicons of cecal samples collected from C57BL/6 mice. (A) C57BL/6Sca mice originating from 2 breeding rooms within the same laboratory animal vendor. The 2 rooms are marked with circles and squares. (B) C57BL/6 mice originating from 2 different laboratory animal vendors. The 2 breeders are marked with circles and squares.
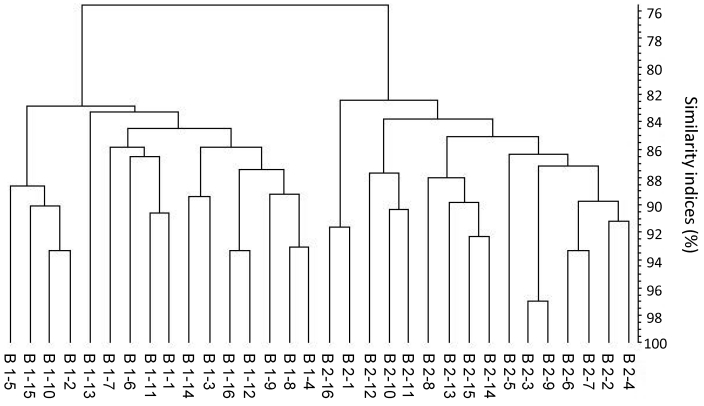
Comparison of DGGE profiles from C57BL/6 mice originating from 2 different laboratory animal vendors.
The dendrogram in Figure 5 clearly reveals 2 main clusters, each reflecting the C57BL/6 mice raised at each of the 2 laboratory animal vendors. The overall similarity index (regardless of vendor) was 75.6% ± 4.6%, that for vendor 1 was 82.8% ± 3.1%, and that for vendor 2 was 82.5% ± 4.2%. Group violation (Table 1) shows that in only 6% and 5.9% of the cases, the given DGGE profiles could have been assigned to the opposite group. The 3D principal component analysis plot (Figure 4 B) shows a clear separation between the samples from the 2 vendors, and the P value (P < 0.05) demonstrates a statistically significant difference in the composition of the cecal microbiota of C57BL/6 mice purchased from 2 laboratory animal vendors.
Figure 5.
Dendrogram based on DGGE profiles representing 16S rRNA gene-derived amplicons of cecal samples collected from C57BL6 mice originating from 2 breeding centers (B1 and B2). Each figure within the groups indicates a sample from a single mouse (numbered from 1 to 16). The scale bar represents the similarity index as a percentage (an index of 100% indicates complete similarity). The overall similarity is 75.6% ± 4.6%; that for animals from B1 is 82.8% ± 3.1%, that for B2 is 82.5.0% ± 4.2%.
Discussion
Our results show that the GM of inbred mice are more similar between animals than are those of outbred mice, according to DGGE profiles of cecal samples, and that the similarity index of C57BL/6Sca mice was 10% higher than that of Sca:NMRI mice. This observation supports the general understanding that variation is lower in inbred compared with outbred animals. Furthermore, we have shown that the GM of mice originating from 2 commercial vendors is significantly different. If the observed variation in GM leads to variation in the parameter response in murine inflammatory models, this variation will need to be considered when planning experiments and developing future standards of quality control of laboratory animals might be applicable. This consideration is necessary both to improve the reproducibility and predictability of animal experimentation and to strive for reduction and refinement in laboratory animal science. Depending on the correlation between the diversity of the GM and the expression of disease in mouse models, decreased variation between the GM of the animals may lead to smaller group sizes or higher power in studies influenced by the GM. In the present study, we have shown that DGGE is a simple tool that may enable breeders to document the similarity of the GM of their mice in the same way as they currently document the pathogen or genetic status of their animals.
As documentation would make similarity a quality parameter, a further perspective would be that the vendors would actually work to increase the similarity of their mice. To do so, it seems essential to discuss the background to these dissimilarities between the GM of individual laboratory mice. Studies based on different culture-independent techniques have shown that the host's genotype affects the bacterial community of the GM.37,38,42 After transplantation of human20 or zebrafish GM31 to germ-free mice, the GM subsequently changes into a more murine microbial profile, thereby supporting genetics as a key factor for determination of the GM. In the current study, the higher similarity index of the inbred strain compared with outbred stock was not due to lower bacterial diversity, because the diversity of the cecal microbiota was high for both colonies. Instead, the higher similarity of the GM of the inbred strain likely is related to a higher genetic similarity, because the GM differs markedly between congenic mouse strains that differ only at the major histocompatibility complex.35 In general, different colonies of inbred mice (for instance, from different vendors) have the same major histocompatibility complex and match each other in other parts of the genetic profile. Mathematically, an inbred strain has a genetic similarity coefficient of as high as 99% which, if genetics has the predominant effect on GM, should lead to high similarity between the GM of these host populations. According to cellular fatty acid analysis, inbred mice within the same strain show rather uniform GM, with similarity indices of greater than 90%.17 Although we could not confirm a similarity index of 90% in our group of 30 inbred mice, we achieved levels as high as 90% when looking at subclusters representing fewer animals.17 Although the genetic background seems to have an important effect on the colonization of the GM, the genetic influence cannot be absolute, because only microorganisms present in the environment can colonize. However, literature on the actual inbreeding coefficient monitored as the percentage of loci expressing homozygosity is rather sparse and dates from before the availability of modern methods such as SNP chips.13,30 In addition the separation of rodent strains into discrete colonies is known to give rise to genetic variation after a limited number of generations.30 Therefore, it cannot be excluded that a higher inbreeding coefficient could still be achieved in future laboratory mice, and that doing so would decrease interindividual variation between the GM of mice of the same strain.
Mice examined in previous studies reporting that the genetic background plays an important role in determining the composition of the GM35,37,38, 42 were housed in open cages, most likely all in the same room, and to a large extent were exposed to the same microorganisms from the surroundings. Today, laboratory mice more typically are housed in IVC, which limit exposure to microorganisms from the surroundings and cross-contamination between cages. In our study, C57BL/6Sca and Sca:NMRI mice were housed in IVC. Nonetheless, we did not see clustering related to individual cages of the mice and therefore saw no indication that IVC as a housing system results in intercage variation with respect to the composition of the GM. In contrast, the GM of mice raised in the same room clustered. In another study, mice purchased at the age of 6 wk and maintained in different rooms likewise showed a clustering to the respective rooms.9 Furthermore, relocation of C3H mice at 4 wk of age led to markedly different fecal profiles.6 These data indicate that exposure to different environmental factors, such as different caretakers, and deviations in treatments of food, water, and bedding, may influence the composition of the GM. However, in the current study, the difference between breeding rooms identified by PCR/DGGE was not statistically significant. We investigated C57BL/6Sca and Sca:NMRI mice at a breeding center where conditions of maintenance are standardized, and inconsistency in the environmental source of microbes may therefore have been less of a factor than might otherwise be expected. The only difference in the conditions of maintenance between the 2 rooms was the caretaking staff, which nevertheless is a relevant environmental factor, in that some early studies indicate a certain sharing of bacteria between caretakers and animals.41 Purchasing BALB/c mice from different laboratory animal vendors also seems to lead to increased variation in the GM, as monitored by culture-dependent techniques.14 Our analysis of C57BL/6 mice originating from different vendors revealed 2 main clusters which were significantly different. Different environmental exposure, especially different diets,4,7,12,23,27 influences the GM and gut immunology.
Our experiments do not clarify the extent to which the genetic background or environment caused the identified variations between mice originating from different vendors. In that the similarity index for mice from vendor 1 was 82.8% and for mice from vendor 2 was 82.5%, C57BL/6 mice seem to have a fixed level of similarity. However, in view of the differences in the GM between mice from different commercial vendors, the origin of animals is an important aspect to consider with respect to the reproducibility in research using animal models. Even though the correlation between the development of chronic inflammatory diseases and the GM seems increasingly well-documented, and given that dramatic changes in the microbial status of rodent models, such as germ-free rearing or probiotic feeding, lead to demonstrable changes in disease expression,11,21 the effect of these apparently small dissimilarities on the reproducibility in research using animal models is unknown. Here we have shown that interindividual variation can be documented and that small environmental differences, such as different rooms or IVC housing, at the same vendor have less of an effect on the GM of mice, whereas obtaining mice from different vendors may lead to larger deviations in the GM.
Acknowledgments
Majbritt Ravn Hufeldt and parts of the work performed were funded by Scanbur A/S, Denmark, and a grant from The Ministry of Science, Technology and Innovation, Denmark.
References
- 1.Bernbom N, Nørrung B, Saadbye P, Mølbak L, Vogensen FK, Licht TR. 2006. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host, and gender. J Microbiol Methods 66:87–95 [DOI] [PubMed] [Google Scholar]
- 2.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, Muccioli GG, Delzenne NM. 2009. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP2-driven improvement of gut permeability. Gut 58:1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Council of the European Communities Council Directive 86/609/EEC of 24 November 1986 on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes. Off J Eur Communities L358:1–28 [Google Scholar]
- 4.Ejsing-Duun M, Josephsen J, Aasted B, Buschard K, Hansen AK. 2008. Dietary gluten reduces the number of intestinal regulatory T cells in mice. Scand J Immunol 67:553–559 [DOI] [PubMed] [Google Scholar]
- 5.Fasoli S, Marzotto M, Rizzotti L, Rossi F, Dellaglio F, Torriani S. 2003. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int J Food Microbiol 82:59–70 [DOI] [PubMed] [Google Scholar]
- 6.Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, McBain AJ. 2010. Site- and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5:e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara R, Watanabe J, Sonoyama K. 2008. Assessing changes in composition of intestinal microbiota in neonatal BALB/c mice through cluster analysis of molecular markers. Br J Nutr 99:1174–1177 [DOI] [PubMed] [Google Scholar]
- 8.Fushuku S, Fukuda K. 2008. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim 57:489–493 [DOI] [PubMed] [Google Scholar]
- 9.Fushuku S, Fukuda K. 2008. Inhomogeneity of fecal flora in separately reared laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim 57:95–99 [DOI] [PubMed] [Google Scholar]
- 10.Hansen AK. 1992. The aerobic bacterial flora of laboratory rats from a Danish breeding centre. Scand J Lab Anim Sci 19:59–68 [Google Scholar]
- 11.Hansen AK, Ejsing-Duun M, Aasted B, Josephsen J, Christensen BG, Vogensen FK, Hufeldt M, Buschard K. 2009. The impact of the post natal gut microbiota on animal models, p 95–99 Proceedings of the Tenth FELASA Symposium and the XIV ICLAS General Assembly and Conference, 11–14 Jun 2007, Cernobbio, Italy [Google Scholar]
- 12.Hansen AK, Ling F, Kaas A, Funda DP, Farlov H, Buschard K. 2006. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in nonobese diabetic mice. Diabetes Metab Res Rev 22:220–225 [DOI] [PubMed] [Google Scholar]
- 13.Hins J, Gruber FP. 1991. Genetic monitoring of inbred and outbred strains, transgenic individuals, and 3T3-cells of Mus musculus with the Probe Best Mz1.3. Zentralbl Veterinarmed A 38:61–72 [PubMed] [Google Scholar]
- 14.Hirayama K, Endo K, Kawamura S, Mitsuoka T. 1990. Comparison of the intestinal bacteria in specific-pathogen-free mice from different breeders. Jikken Dobutsu 39:263–267 [DOI] [PubMed] [Google Scholar]
- 15.Hooper LV, Gordon JI. 2001. Commensal host–bacterial relationships in the gut. Science 292:1115–1118 [DOI] [PubMed] [Google Scholar]
- 16.Itoh K, Oowada T, Mitsuoka T. 1985. Characteristic faecal flora of NC mice. Lab Anim 19:7–15 [DOI] [PubMed] [Google Scholar]
- 17.Jussi V, Erkki E, Paavo T. 2005. Comparison of cellular fatty acid profiles of the microbiota in different gut regions of BALB/c and C57BL/6J mice. Antonie Van Leeuwenhoek 88:67–74 [DOI] [PubMed] [Google Scholar]
- 18.Kelly D, King T, Aminov R. 2007. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res 622:58–69 [DOI] [PubMed] [Google Scholar]
- 19.Kibe R, Sakamoto M, Hayashi H, Yokota H, Benno Y. 2004. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbiol Lett 235:139–146 [DOI] [PubMed] [Google Scholar]
- 20.Kibe R, Sakamoto M, Yokota H, Ishikawa H, Aiba Y, Koga Y, Benno Y. 2005. Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl Environ Microbiol 71:3171–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauritzen L, Hufeldt MR, Aasted B, Hansen CHF, Midtvedt T, Buschard K, Hansen AK. 2010. The impact of a germ-free perinatal period on the variation in animal models of human inflammatory diseases. Scand J Lab Anim Sci 37:43–54 [Google Scholar]
- 22.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licht TR, Hansen M, Poulsen M, Dragsted LO. 2006. Dietary carbohydrate source influences molecular fingerprints of the rat faecal microbiota. BMC Microbiol 6:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh G, Brodziak F, Blaut M. 2008. The Toll-like receptors TLR2 and TLR4 do not affect the intestinal microbiota composition in mice. Environ Microbiol 10:709–715 [DOI] [PubMed] [Google Scholar]
- 25.Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10:1571–1581 [DOI] [PubMed] [Google Scholar]
- 26.Ministry of Agriculture. [Internet] 2009. Welfare Act and Animal Welfare Ordinance of Sweden. [Cited Dec 2009]. Available at:http://www.sweden.gov.se/content/1/c6/09/03/10/f07ee736.pdf
- 27.Montesi A, Garcia-Albiach R, Pozuelo MJ, Pintado C, Goni I, Rotger R. 2005. Molecular and microbiological analysis of caecal microbiota in rats fed with diets supplemented either with prebiotics or probiotics. Int J Food Microbiol 98:281–289 [DOI] [PubMed] [Google Scholar]
- 28.Muyzer G, Smalla K. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek 73:127–141 [DOI] [PubMed] [Google Scholar]
- 29.Nicklas W, Baneux P, Boot R, Decelle T, Deeny A, Fumanelli M, Illgen-Wilcke B. 2002. Recommendations for the health monitoring of rodent and rabbit colonies in breeding and experimental units. Lab Anim 36:20–42 [DOI] [PubMed] [Google Scholar]
- 30.Prins JB, Herberg L, Den BM, van Zutphen LF. 1991. Genetic variation within and between lines of diabetes-prone and nondiabetes-prone BB rats; allele distribution of 8 protein markers. Lab Anim 25:207–211 [DOI] [PubMed] [Google Scholar]
- 31.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartor RB. 2005. Role of commensal enteric bacteria in the pathogenesis of immune-mediated intestinal inflammation: lessons from animal models and implications for translational research. J Pediatr Gastroenterol Nutr 40 Suppl 1:S30–S31 [DOI] [PubMed] [Google Scholar]
- 33.Suau A, Bonnet R, Sutren M, Gordon JJ, Gibson GR, Collins MD, Doré J. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65:4799–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tlaskalová-Hogenová H, Stepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Zadniková R, Kozáková H, Rossmann P, Bártová J, Sokol D, Funda DP, Borovská D, Reháková Z, Sinkora J, Hofman J, Drastich P, Kokesová A. 2004. Commensal bacteria (normal microflora), mucosal immunity, and chronic inflammatory and autoimmune diseases. Immunol Lett 93:97–108 [DOI] [PubMed] [Google Scholar]
- 35.Toivanen P, Vaahtovuo J, Eerola E. 2001. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun 69:2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 37.Vaahtovuo J, Toivanen P, Eerola E. 2001. Study of murine faecal microflora by cellular fatty acid analysis; effect of age and mouse strain. Antonie Van Leeuwenhoek 80:35–42 [DOI] [PubMed] [Google Scholar]
- 38.Vaahtovuo J, Toivanen P, Eerola E. 2003. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS Microbiol Ecol 44:131–136 [DOI] [PubMed] [Google Scholar]
- 39.Vaarala O, Atkinson MA, Neu J. 2008. The ‘perfect storm’ for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57:2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Merwe JP, Stegeman JH, Hazenberg MP. 1983. The resident faecal flora is determined by genetic characteristics of the host. Implications for Crohn's disease? Antonie Van Leeuwenhoek 49:119–124 [DOI] [PubMed] [Google Scholar]
- 41.Wullenweber M, Lenz W, Werhan K. 1990. Staphylococcus aureus phage types in barrier-maintained colonies of SPF mice and rats. Z Versuchstierkd 33:57–61 [PubMed] [Google Scholar]
- 42.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. 2001. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis 13:129–134 [Google Scholar]