Abstract
Viral infection triggers innate immune sensors to produce type I interferons (IFN). However, HIV infection of T cells and macrophages does not trip these alarms. How HIV avoids activating nucleic acid sensors is unknown. The cytosolic exonuclease TREX1 suppressed IFN triggered by HIV. In Trex1−/− mouse cells and human CD4+ T cells and macrophages in which TREX1 was inhibited by RNA interference, cytosolic HIV DNA accumulated, and HIV infection induced type I IFN that inhibited HIV replication and spreading. TREX1 bound to cytosolic HIV DNA and digested excess HIV DNA that would otherwise activate IFN expression via a TBK1, STING and IRF3 dependent pathway. HIV-stimulated IFN production in cells deficient in TREX1 did not involve known nucleic acid sensors.
HIV introduces its single-stranded RNA (ssRNA) genome into a cell within the reverse transcription complex (RTC), which matures into the preintegration complex (PIC). The PIC delivers reverse-transcribed HIV DNA to the nucleus for chromosomal integration. Few copies of HIV DNA integrate, leaving behind HIV DNA in the cytosol to be cleared by host enzymes. Although nucleic acids within the RTC might be shielded from nucleic acid sensors, viral DNA within the PIC is accessible to exogenous endonucleases1 and thus potentially to cytosolic sensors of innate immunity. We previously found that the ER-associated SET complex, which contains 3 DNases (APE1, NM23-H1, TREX1) and other proteins (SET, pp32, HMGB2), binds to the HIV PIC and protects the integrase (IN)-activated DNA ends from self-attack in suicidal autointegration2. Suppressing expression of any SET complex gene increases autointegration and interferes with chromosomal integration. TREX1 is the most abundant 3′-5′ DNase in cells2. Treatment with TREX1 small interfering RNAs (siRNA) more profoundly inhibits HIV replication than siRNAs against other SET complex components, reducing viral production by a log2. TREX1 mutations are associated with inflammatory and autoimmune diseases, including Aicardi-Goutieres syndrome, chilblain lupus, and systemic lupus erythematosus (SLE), some of which have increased type I IFN3-5. TREX1 binds to transfected immunostimulatory DNA (ISD), and Trex1−/− cells accumulate cytoplasmic DNA derived from endogenous retroelements, which activates interferon regulatory factor 3 (IRF3)-dependent IFN expression6,7. Like HIV, endogenous retroelements undergo cytoplasmic reverse transcription. We therefore investigated whether HIV might use TREX1 to avoid triggering antiviral innate immunity.
RESULTS
TREX1 inhibits IFN production in response to HIV
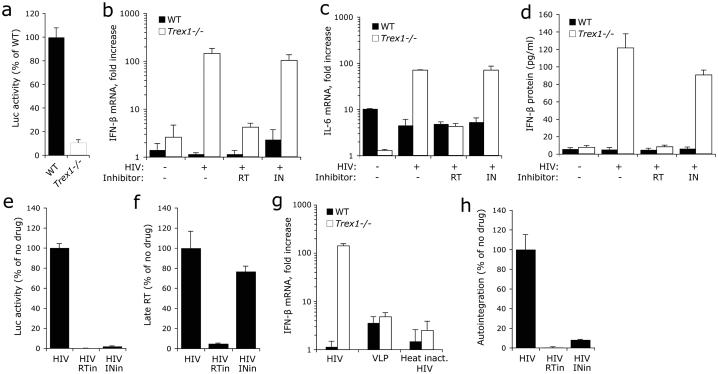
We first compared HIV replication and IFN and inflammatory cytokine expression and secretion after infection of Trex1+/+ (WT) or Trex1−/− mouse embryonic fibroblasts (MEFs) with vesicular stomatitis virus G (VSV-G)-pseudotyped single-round HIV-Luc virus that contains a near full-length HIV genome (ΔEnv, Luc replaces Nef) and can infect MEFs, integrate and execute LTR-driven luciferase (Luc) reporter expression8. As for TREX1 siRNA treatment2, Luc activity in Trex1−/− MEFs was reduced to one-tenth that of WT MEFs (Fig. 1a). Uninfected Trex1−/− MEFs constitutively expressed slightly more IFN-β mRNA than WT MEFs (Fig. 1b). In Trex1−/− cells HIV infection induced both IFN-β mRNA, which increased ~100-fold, and interleukin 6 (IL-6) mRNA, which increased ~10-fold, compared to uninfected cells (Fig. 1b,c). Neither IFN-β nor IL-6 was induced by HIV infection of WT MEFs. HIV did not induce IL-1β, IFN-α or IFN-γ in WT or Trex1−/− MEFs (data not shown). IFN-β was secreted as assessed by ELISA of cultured supernatants (Fig. 1d). Nevirapine, which inhibits HIV reverse transcription, but not the IN inhibitor raltegravir, which acts after HIV DNA synthesis, inhibited IFN-β and IL-6 expression in response to HIV in Trex1−/− cells (Fig. 1e,f), suggesting that HIV reverse-transcribed DNA, rather than genomic RNA (gRNA), was triggering the response. DNase treatment of the virus did not alter the IFN-β response to HIV-Luc in Trex1−/− cells (data not shown), eliminating concerns that plasmid DNA carryover was responsible for inducing IFN-β. Virus-like particles (VLPs) lacking gRNA and heat-inactivated HIV also did not trigger IFN-β expression (Fig. 1g), suggesting that viral nucleic acid and an infectious virus were required. Because non-productive autointegrant DNA accumulates when TREX1 is inhibited by siRNA2, autointegrant DNA might be triggering IFN-β. However, although only the RT inhibitor suppressed IFN-β production, both the RT and IN inhibitor blocked the production of autointegrants (Fig. 1h). These results suggest that HIV-stimulated IFN-β production in Trex1−/− MEFs is activated by HIV DNA other than autointegrant DNA.
Fig. 1. TREX1 deficiency inhibits HIV replication and activates IFN-β in response to HIV infection.
WT or Trex1−/− primary MEFs were infected with VSV-G-pseudotyped single round HIV virus. (a) HIV infection, measured by Luc activity 48 hpi, is reduced in Trex1−/− cells. Luc reporter expression was driven by the HIV LTR in the context of a near full length viral genome (ΔEnv, Luc replacing Nef). (b-d) Cytokine induction, measured by qRT-PCR (b, c) and ELISA (d) 22 hpi, is inhibited by RT, but not IN, inhibitors. (e,f) RT and IN inhibitors both reduce HIV-Luc replication 48 hpi, but only the RT inhibitor suppresses late reverse transcripts (late RT) measured 10 hpi. Both inhibitors were added at the same time as the virus. (g) Virus-like particles (VLP) and heat-inactivated HIV (95°C, 5 min) do not induce substantial IFN-β in either WT or Trex1−/− cells. Equivalent amounts of HIV (heat inactivated or not) or VLP were used for infection based on p24 ELISA measurements. (h) HIV autointegration is reduced by inhibiting either RT or IN. Error bars indicate S.D. of at least three independent experiments. Data from WT MEF are indicated by black bars; from Trex1−/− MEF by white bars.
HIV-stimulated IFN expression is IRF3-dependent
IFN-β expression induced by transfected ISD or endogenous retroelements in Trex1−/− cells is mediated by the transcription factor IRF36,7. To investigate whether IRF3 also activates HIV-induced IFN-β expression, we compared IFN-β mRNA and HIV infectivity in WT, Trex1−/− and Trex1−/− Irf3−/− MEFs. Lack of IRF3 completely inhibited IFN-β induction (Fig. 2a). HIV-Luc activity was also partially rescued in Trex1−/− Irf3−/− cells (Fig. 2b). Therefore, IFN-β induction in response to HIV in Trex1−/− cells is mediated by IRF3. Autointegration in the absence of Trex1 was indistinguishable in Trex1−/− and Trex1−/−Irf3−/− cells (Fig. 2c), suggesting that autointegration is not altered by endogenous IFN-β production and that the two effects of TREX1 on HIV infection (blocking autointegration and inhibiting IFN-β induction) operate independently. Similarly in human 293T cells, TREX1 siRNA increased HIV-induced Luc expression from the IFN-β promoter, whereas siRNAs against some other SET complex genes had no effect on IFN-β expression (Supplementary Fig. 1). Therefore protection from IFN-β activation involves TREX1, but not the entire SET complex.
Fig. 2. HIV-stimulated IFN expression is IRF3-dependent.
Wild type (WT), Trex1−/− (KO) or Trex1−/−Irf3−/− (DKO) primary MEFs were infected as in Fig. 1. (a) HIV-stimulated IFN-β induction, assessed by qRT-PCR 22 hpi, is eliminated in the absence of IRF3. (b) Irf3-deficiency partially rescues HIV infection in Trex1-deficient cells. HIV infection was measured by Luc activity 48 hpi. (c) Increased HIV autointegration in Trex1-deficient cells is not affected by Irf3-deficiency. Error bars indicate S.D. of at least three independent experiments. (d) IRF3 translocates to the nucleus in HIV-infected Trex1−/− cells. WT and Trex1−/− MEFs that were either uninfected or infected with HIV were stained 22 hpi for IRF3 (red) and DAPI (blue). (e) Expression of GFP from an HIV reporter virus (HIV-GFP) is reduced in Trex1−/− MEF compared to WT MEF. Flow cytometry analysis of GFP expression was measured 24 hpi. A representative plot from 3 independent experiments is shown.
IRF3 activation triggers its nuclear translocation. To verify the role of IRF3 in HIV-induced IFN-β expression in Trex1−/− cells, we monitored IRF3 localization by confocal microscopy. IRF3 was cytoplasmic in uninfected WT and Trex1−/− MEFs (Fig. 2d). Following infection with VSV-G-pseudotyped virus expressing green fluorescent protein (GFP) at a multiplicity of infection (MOI) of 1, 68% of WT MEFs were infected as assessed by GFP expression. At the same MOI, viral entry was comparable in Trex1−/− cells (assessed by qRT-PCR for HIV gRNA, data not shown), but GFP expression was greatly reduced (Fig. 2e). IRF3 remained cytoplasmic in 100% of WT cells, but translocated into the nucleus in 44% of Trex1−/− cells (Fig. 2d). These data confirm that the HIV-stimulated IFN-β response is IRF3-dependent and imply the involvement of a cytosolic detection pathway. Likewise, these findings suggest that HIV DNA is not sensed by Toll-like receptor 9 (TLR9), which is largely contained within endosomes and activates type I IFN through IRF7 rather than IRF3 (refs. 9,10).
HIV reverse transcripts accumulate in Trex1−/− cells
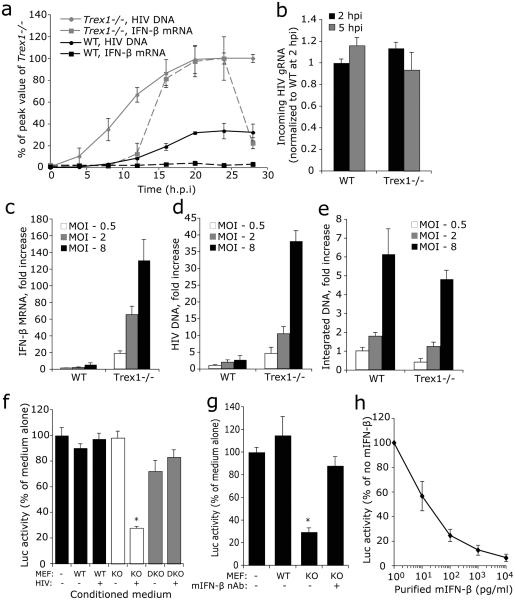
To gain insight into how TREX1 suppresses HIV-stimulated IFN-β induction, we measured cytosolic HIV DNA and IFN-β mRNA in WT and Trex1−/− cells during single-round HIV-Luc infection (MOI =1) (Fig. 3a). We also measured incoming HIV gRNA 2 and 5 h after infection to verify that WT and Trex1−/− cells were infected with the same amount of virions (Fig. 3b). Cytosolic HIV DNA steadily increased for the first 20 h post infection (hpi) and then achieved a 3-4 fold higher plateau in Trex1−/− compared to WT cells. In WT cells, IFN-β mRNA remained at baseline. In Trex1−/− cells, IFN-β mRNA was induced, but lagged behind HIV DNA accumulation, first increasing 12 hpi. IFN-β mRNA peaked 20-24 hpi and then plummeted to close to baseline, even though cytosolic HIV DNA remained elevated. IFN-β induction and accumulation of cytosolic HIV DNA increased in tandem in Trex1−/− cells as the amount of virus used for infection was increased, but even the highest viral dose (MOI=8) did not stimulate IFN-β expression in WT cells (Fig. 3c,d). Although cytosolic HIV DNA was ~10-fold more abundant in Trex1−/− than WT MEFs, HIV DNA integration was reduced in Trex1−/− vs WT cells, suggesting that most HIV DNA that accumulated in Trex1−/− cells did not contribute to productive infection (Fig. 3e).
Fig. 3. Cytosolic HIV DNA in Trex1−/− cells is the trigger for IFN expression.
(a) Cytosolic HIV DNA accumulates at higher levels early after infection in Trex1−/− (KO) than in WT primary MEFs. IFN-β expression lags behind DNA build up and only occurs in Trex1−/− cells. HIV DNA and IFN-β mRNA were measured by qPCR and qRT-PCR, respectively. (b) Trex1 deficiency does not affect viral entry. Comparable numbers of virions enter WT and Trex1−/− cells as determined by qRT-PCR of HIV genomic RNA (gRNA) assessed 2 and 5 hpi. (c-e) Increasing multiplicity of infection (MOI) leads to more cytosolic HIV DNA accumulation and IFN-β expression in Trex1−/− MEF, but not to more chromosomal integration, compared to WT MEF. WT and Trex1−/− MEFs were infected with HIV at indicated MOI. IFN-β mRNA (c) and cytosolic HIV DNA (d) were measured as in (a). Integrated DNA (e) was measured by a two-step semi-quantitive PCR assay (8, see ONLINE METHODS). (f) Conditioned medium from Trex1−/− MEF, but not from WT and Trex1−/−Irf3−/− (DKO) MEF, inhibits de novo HIV infection. Conditioned medium, obtained from cells that were either uninfected or infected with HIV-GFP, was added to WT MEFs that were then infected with HIV-Luc. Luc activity was measured 48 hpi. (g) IFN-β contributes to most of the antiviral activity secreted by Trex1−/− MEF. Conditioned medium was pre-incubated with or without mouse IFN-β neutralizing antibody before being added to WT MEFs with HIV-Luc. (h) The antiviral effect of varying concentrations of purified recombinant mIFN-β on HIV-Luc infection in WT MEF. *, P <0.01, Student's t-test. Error bars indicate S.D. of three independent experiments.
HIV-stimulated IFN-β from Trex1−/− cells inhibits HIV
Type I IFNs inhibit replication of most viruses by both cell autonomous and nonautonomous effects and block both early and late stages of the HIV life cycle11-14. To determine whether IFN-β (and potentially other cytokines) secreted during HIV infection of Trex1−/− cells suppresses de novo HIV infection, we infected WT MEFs with VSV-G-pseudotyped HIV-Luc in the presence of conditioned medium harvested from HIV-GFP- or mock-infected WT, Trex1−/− or Trex1−/−Irf3−/− MEFs. Only the conditioned medium from HIV-infected Trex1−/− MEFs inhibited luciferase activity (by a factor of 4) (Fig. 3f). Preincubating the conditioned medium from Trex1−/− cells with mIFN-β neutralizing antibody abrogated the antiviral effect (Fig. 3g). HIV-Luc infection was inhibited similarly when cells were incubated with 100 pg/ml mIFN-β or with conditioned medium from HIV-infected Trex1−/− MEFs, which contained 120 pg/ml IFN-β (Fig. 1d, 3h). These findings suggest that IFN-β is the main secreted antiviral factor. To pinpoint which stage(s) of HIV replication IFN-β blocks, we treated WT MEFs with mIFN-β and measured HIV DNA synthesis, 2-LTR circle formation, and integration. To assess LTR-mediated transcription, we incubated a well characterized human HeLa-CD4-derived cell line (TZM-bl) that harbors an integrated copy of an LTR-driven Luc reporter gene with hIFN-β. At the IFN-β concentration in conditioned medium from infected Trex1−/− cells, IFN-β strongly blocked HIV integration and LTR-mediated transcription (Supplementary Fig. 2). Higher doses of IFN-β blocked nearly all early stages of HIV replication in single-round infection. Thus, secreted IFN-β inhibits multiple steps in the early phase of HIV-1 infection.
TREX1 metabolizes HIV RT products to suppress IFN induction
To identify which HIV nucleic acids trigger IFN-β expression, we transfected WT and Trex1−/− MEFs with synthetic 100 bp gag sequence oligonucleotides that correspond to HIV nucleic acids in the cytosol during reverse transcription (ssRNA to represent gRNA, RNA:DNA hybrid, ssDNA, dsDNA) and measured IFN-β mRNA 6 h later by qRT-PCR (Fig. 4a,b). All DNA-containing oligonucleotides, but not ssRNA, induced more IFN-β in Trex1−/− compared to WT MEFs. ssDNA evoked the largest difference (~200-fold more IFN-β vs 20-40-fold for dsDNA and 2-4-fold for RNA:DNA duplexes). None of the oligonucleotides induced IFN-β in Trex1−/−Irf3−/− MEFs (data not shown), supporting IRF3's role in signaling the presence of these cytosolic nucleic acids. Although synthetic oligonucleotides may not completely mimic native HIV RT products, these data are consistent with the known enzymatic preference of TREX1 for ssDNA vs dsDNA substrates15. To determine whether transfected oligonucleotides accumulate in Trex1−/− cells as HIV DNA does during infection, we quantified cytosolic DNA after transfection of gag sequence ssDNA or dsDNA or HIV infection. Cytosolic DNA was 4-6-fold more abundant when measured 3 h post transfection or 10 hpi in Trex1−/− vs WT MEFs (Fig. 4c). We repeated this experiment using WT and TREX1 mutant (D18N) human fibroblasts (derived from a chilblain lupus patient). D18N is a mutation in a highly conserved Mg2+-binding site within the Exo1 domain of TREX1 that eliminates exonuclease activity and interferes with enzymatic activity of the WT/mutant TREX1 heterodimer3,16. Cells expressing the D18N mutant also accumulate all DNA species introduced by infection or transfection (Fig. 4c). The accumulation of HIV DNA in Trex1−/− or TREX1 mutant cells is consistent with a previous report that ssDNA derived from endogenous retroelements is increased in the cytosol of Trex1−/− cells7. These results strongly suggest that TREX1 suppresses IFN-β induction by digesting cytosolic DNA.
Fig. 4. Recognition of HIV RT products by enzymatically active TREX1 suppresses IFN induction.
(a) Schematic of synthetic nucleic acids used in (b) as they are generated during reverse transcription. (b) DNA-containing RT products trigger a stronger IFN response in Trex1−/− compared to WT cells. IFN-β expression was measured 6 h post transfection of indicated nucleic acids. (c) TREX1 metabolizes reverse-transcribed HIV DNA and transfected ssDNA and dsDNA in both mouse and human fibroblasts. WT and KO MEFs or WT and TREX1 mutant (D18N) human fibroblasts were either infected with HIV or transfected with ssDNA and dsDNA gag sequence oligonucleotides (100 bp), and cytosolic DNA was measured by qPCR using gag primers 10 hpi or 3 h post transfection. *, P <0.01, Student's t-test. Error bars indicate S.D. of three independent experiments. (d) HIV DNA, but not RNA, is pulled down with FLAG antibody from cytosolic lysates of HeLa-CD4 cells expressing FLAG-TREX1 that were infected with HIVIIIB for 10 h. Lysates were immunoprecipitated (IP) with FLAG or IgG antibodies. DNA and RNA extracted from the IP were quantified by qPCR or qRT-PCR, respectively. *, P <0.05, Student's t-test. Error bars indicate S.D. of triplicate replicates of two independent experiments. (e,f) TREX1 enzymatic activity is required to support HIV infection. (e) WT and Trex1−/− (KO) MEFs expressing GFP alone or wild type or enzymatically inactive mutant (D18N) GFP-tagged TREX1 were infected with HIV-Luc, and luciferase activity was measured 48 hpi. TREX1 expression was comparable for each construct by immunoblot. (f) GFP-TREX1, but not the D18N mutant, eliminates excess HIV DNA in Trex1−/− MEFs. *, P <0.01, Student's t-test. Error bars indicate S.D. of triplicate replicates of two independent experiments.
For further evidence that TREX1 is responsible for removing extraneous cytosolic HIV DNA, we assessed whether TREX1 interacts with HIV DNA during infection with wild-type HIVIIIB. Cytosolic extracts from HeLa-CD4 cells expressing FLAG-TREX1 infected with HIVIIIB for 10 h were immunoprecipitated with anti-FLAG or IgG control antibody. The enrichment for HIV gag DNA and RNA in the precipitates was assessed by qPCR or qRT-PCR, respectively. The FLAG antibody immunoprecipitated 3-fold more HIV DNA than the IgG control, but there was no enrichment for HIV RNA, confirming that HIV DNA binds to TREX1 and is its preferred target (Fig. 4d).
To test whether the enzymatic activity of TREX1 is required to enhance HIV infection, we determined whether expressing WT or D18N TREX1 in Trex1−/− cells could rescue HIV-Luc infectivity (Fig. 4e). WT TREX1 partially rescued HIV infection, but the enzymatically inactive D18N mutant had no effect, indicating that TREX1's exonuclease activity is needed for both inhibiting autointegration and blocking IFN-β induction. We also measured HIV DNA accumulation in WT and Trex1−/− cells transfected with GFP-TREX1 or GFP-TREX1(D18N). As previously shown (Fig. 3), Trex1−/− cells accumulated ~5 times more cytosolic HIV DNA than WT cells. The excess HIV DNA was completely eliminated by expressing GFP-TREX1, but not GFP-TREX1(D18N) (Fig. 4f). These results strongly suggest that TREX1 metabolizes cytosolic HIV DNA. Because over-expressing WT GFP-TREX1 did not reduce HIV DNA abundance below that seen in an infected WT cell, TREX1 may not have access to all HIV DNA products.
HIV activates IFN in TREX1-deficient human cells
Most previous experiments used mouse Trex1−/− cells to take advantage of their complete lack of TREX1 compared to the incomplete inhibition afforded by siRNAs. To investigate whether our findings are physiologically relevant during HIV infection of primary human immune cells, we used siRNAs to suppress TREX1 or TREX1 and IRF3 in monocyte-derived macrophages (MDMs) from two donors. siRNA-treated cells, which had 50-70% less TREX1 and/or IRF3 mRNA, were infected 3 d later with HIVBaL (Fig. 5a-c). After treatment with TREX1 or TREX1 plus IRF3 siRNAs, cytosolic HIV DNA increased ~4-fold 24 hpi, compared to control samples. HIV replication and spreading in the macrophage culture, assessed by measuring p24 Gag antigen in the medium, was reduced in TREX1 siRNA-treated macrophages to ¼-½ of values in control cells. When IRF3 and TREX1 expression were both suppressed, HIV replication was partially rescued, suggesting that IRF3-dependent IFN-β induction contributes to inhibiting HIV replication caused by TREX1 siRNA (Fig. 5d). Consistent with this notion, both IFN-α and IFN-β mRNA expression increased up to 10-fold in macrophages treated with TREX1 siRNA, but not with control or TREX1 and IRF3 siRNAs (Fig. 5e). Similar results were obtained following TREX1 RNAi in human peripheral blood CD4+ T cells infected with HIVIIIB (Fig. 5f-l). In TREX1 siRNA-treated T cells, HIV DNA accumulated in the cytosol and both IFN-α and IFN-β expression were induced. TREX1 siRNA similarly reduced HIV production, measured by flow cytometry analysis of intracellular p24. Both the number of p24+ cells and p24 mean fluorescence intensity were reduced. Therefore, TREX1 deficiency decreased HIV replication and spreading in the culture. The magnitude of these effects increased with the amount of transfected siRNA and the extent of TREX1 suppression. Therefore, during wild-type HIV infection of primary human cells, TREX1 suppresses HIV-induced IFN-β activation through IRF3 to promote HIV replication.
Fig. 5. TREX1 siRNA treatment induces IFN-α and IFN-β and inhibits HIV replication in primary human immune cells.
(a) Scheme of the experiment for (b-e). Human monocyte-derived macrophages (MDM) were transfected with control, TREX1 or TREX1 plus IRF3 siRNAs and infected 3 days later with HIVBaL. (b) TREX1 and IRF3 mRNAs were measured 2 days after siRNA transfection by qRT-PCR. (c) Cytosolic HIV DNA increases in MDM treated with TREX1 siRNA, irrespective of co-transfection with IRF3 siRNA. Average values, normalized to values in cells treated with control siRNA, of 4 replicates from 2 donors are shown. (d) TREX1 siRNA inhibits HIVBaL replication in MDM measured by p24 Gag Ag ELISA of culture supernatants. No measurements were made on day 1 (not determined, nd). (e) HIV infection of MDM transfected with TREX1 siRNA induces IRF3-dependent IFN gene expression. IFN-α and IFN-β mRNA levels were measured by qRT-PCR at indicated days post infection and normalized to the control values on day 1. TREX1 siRNA treatment does not induce IFN in uninfected cells. The late induction of IFN in HIVBaL-infected cells is likely due to the slow spread of infection in macrophage cultures (only 20% of cells were infected by day 7 by flow cytometry analysis of p24 immunostaining). *, P <0.01, Student's t-test. Error bars indicate S.D. of 4 replicates from 2 independent donors. (f-l) Pretreatment with TREX1 siRNAs induces IFN in HIVIIIB-infected human CD4+ T cells. CD4+ T cells, positively selected from PBMC, were transfected with control or TREX1 siRNAs and 3 days later infected with HIVIIIB. The control (CTL) siRNA was used at 600 pmole and TREX1 siRNA was used at 200, 400 and 600 pmole per 3 × 106 cells. TREX1 mRNA was measured 2 d post transfection to assess siRNA suppression (f). Cytosolic HIV DNA was measured 10 hpi (g); IFN-α (h) and IFNβ (i) mRNA were measured 22 hpi and normalized to the CTL cells. (j) Suppression of TREX1 expression inhibits HIVIIIB replication in CD4+ T cells analyzed 3 days post infection by flow cytometry analysis of p24 immunostaining. A representative analysis is shown in (e) and mean percentage of p24+ cells (f), and p24 mean fluorescence intensity (MFI) (g) from six replicates in two independent experiments are shown. *, P <0.05, Student's t-test. Error bars indicate S.D..
HIV DNA signals via STING, TBK1 and IRF3
To investigate the pathway triggered by HIV DNA, we examined the effect on HIV-stimulated IFN-β expression of treatment of Trex1−/− MEFs with siRNAs targeting selected genes implicated in DNA-stimulated IFN induction (Fig. 6a,b). Inhibiting a gene required for HIV-stimulated IFN signaling in Trex1−/− cells should reduce IFN-β expression. As expected, Irf3 siRNA reduced HIV-stimulated IFN-β mRNA. Similarly, siRNA targeting Tbk1, which encodes an IRF3 kinase, also strongly inhibited HIV-stimulated IFN-β expression (Fig. 6b). siRNAs against DNA sensor genes (Tlr917, Aim218, Irrfip119, Hmgb220), RIG-I (Ddx58), which recognizes RNA transcribed from cytosolic DNA21,22, or its adaptor Mavs (also known as IPS-1, VISA, CARDIF)23-26, did not suppress HIV-stimulated IFN-β expression, suggesting that an unknown sensor detects HIV DNA, or that multiple known DNA sensors might function redundantly. Additional experiments confirmed that the POL-III–RIG-I–MAVS DNA detection pathway21,22 was not involved in HIV-stimulated IFN induction (Supplementary Fig. 3). However, siRNA against another membrane-associated adaptor Sting (also known as MITA, ERIS27-29), recently identified to mediate innate immune responses to cytosolic DNA27,28, strongly inhibited HIV-stimulated IFN-β expression (Fig. 6b). STING is phosphorylated by TBK128,30. siRNAs against Sting, Irf3 or Tbk1 did not affect cytosolic HIV DNA accumulation in Trex1−/− MEFs, suggesting that they act downstream of DNA sensing (Fig. 6c). Thus, HIV DNA is detected by a pathway that signals though STING, TBK1 and IRF3 and does not involve any known DNA sensor (Supplementary Fig. 4).
Fig. 6. HIV-stimulated IFN induction requires IRF3, TBK1 and STING.
(a) Inhibition of selected innate immune genes by siRNA transfection of Trex1−/− MEFs assessed by qRT-PCR 48 h post transfection. *, P <0.01. Student's t-test. Error bars indicate S.D. of 3 independent experiments. (b) IRF3, TBK1 and STING are required for HIV-stimulated IFN-β induction in Trex1−/− cells. WT and Trex1−/− MEFs transfected with indicated siRNAs were infected with VSV-G-pseudotyped HIV and IFN-β mRNA was measured 24 hpi by qRT-PCR. The fold increase compared to si-CTL is displayed. *, P <0.01. Student's t-test. Error bars indicate S.D. of 3 independent experiments. (c) Innate immune factors involved in HIV-stimulated IFN response do not affect HIV DNA accumulation. Cytosolic HIV DNA in Trex1−/− MEFs transfected with the indicated siRNA was measured 10 hpi by qPCR. See Supplementary Fig. 4 for a model of the innate immune pathway stimulated by HIV DNA.
HMGB2, but not its homolog HMGB1, associates with TREX1 in the cytosolic SET complex31. Hmgb2 siRNA treatment of both HIV-infected WT and Trex1−/− MEFs enhanced IFN-β expression (Fig. 6b), suggesting that HMGB2 inhibits the response to cytosolic HIV DNA. However, Hmgb2 siRNAs did not further increase cytosolic HIV DNA in Trex1−/− MEFs more than control siRNA (Fig. 6c), suggesting that HMGB2 might act downstream of HIV DNA recognition. HMGB proteins repress transcription of many genes, including proinflammatory genes, such as TNF32-34. To test whether HMGB2 might regulate IFN-β transcription, we transfected HMGB2 siRNAs into 293T cells expressing a Luc reporter plasmid driven by the IFN-β promoter. IFN-β-Luc expression increased ~2-fold in cells treated with HMGB2 siRNA compared to control siRNA in response to poly(dA:dT) or MAVS over-expression, suggesting that HMGB2 inhibits IFN-β expression, either directly or indirectly, though its promoter, but acting downstream of DNA sensing (Supplementary Fig. 5). To test whether HMGB2 plays a role in cytosolic HIV DNA removal or HIV-stimulated IFN expression in primary human MDM, we suppressed HMGB2 alone or with TREX1. HMGB2 siRNA on its own did not significantly increase cytosolic HIV DNA. However, unlike in Trex1−/− MEFs, HMGB2 and TREX1 dual siRNA treatment in human macrophages significantly enhanced cytosolic HIV DNA accumulation compared to TREX1 siRNA alone. HMGB2 siRNA treatment led to HIV-stimulated IFN-α and IFN-β expression (but did not induce IFN expression in uninfected cells (data not shown)), which was significantly greater than with TREX1 siRNA alone and was not enhanced by both siRNAs. These results suggest that HMGB2 may cooperate with TREX1 to remove HIV DNA from the cytoplasm (Supplementary Fig. 4). Further work is needed to elucidate HMGB2's role in suppressing HIV DNA-stimulated IFN induction, but these results suggest that HMGB2 might act at multiple points (i.e., by recognizing and/or helping eliminate cytosolic DNA and suppressing the IFN-β promoter).
DISCUSSION
HIV infection of its main target cells, macrophages and CD4+ T cells, does not induce cell-autonomous IFNs35. We showed here that the host cytoplasmic exonuclease TREX1 helps HIV evade innate immunity by digesting reverse transcripts that are not imported into the nucleus and would otherwise induce IFNs. When TREX1 is inhibited by RNAi, HIV infection of primary cells triggers Type I IFN expression and secretion. The HIV-stimulated IFN response in cells deficient in TREX1, like the response to endogenous retroelement DNA and transfected DNA7, is IRF3-dependent. IFN induction can be blocked by expressing enzymatically active TREX1 or by simultaneously suppressing IRF3 expression to interrupt IFN signaling. HIV-stimulated innate immune signaling also requires the adaptor protein STING and the protein kinase TBK1. Based on siRNA experiments, none of the known DNA sensors is involved. Therefore our working model of the innate immune pathway activated by cytosolic HIV DNA starts with an unknown sensor (that may preferentially recognize ssDNA) that signals through STING, TBK1 and IRF3 to activate IFN expression.
Type I IFNs inhibit HIV replication at multiple steps in the early phase of its life cycle and thereby suppress viral spreading. Failure to induce antiviral IFNs in infected T cells and macrophages may promote transmission by allowing the virus to spread from the initial nidus of infection to neighboring cells within genital tissue. However, testing TREX1's importance in transmission will require efficient methods to inhibit TREX1 expression in vivo in the immune cells that HIV infects. Such methods are not available, but are being developed. We used an MOI of 1 to achieve a reasonable frequency of infected cells. Since HIV DNA cytoplasmic accumulation and IFN triggering depend upon the MOI, the physiological relevance of our results to HIV transmission hinge upon what viral concentrations are achieved in vivo, which is unknown. Viral concentrations may reach high MOIs locally during the replicative burst that occurs within the genital tract during transmission, when a strong intrinsic antiviral immune response might prevent dissemination36. Other settings of high viral concentration might be activated lymph nodes or gut-associated lymphoid tissues.
We did not examine whether TREX1 affects IFN production by pDCs, the major source of Type I IFNs during HIV infection. HIV replication is inefficient in DCs. IFN stimulation in pDCs appears to be triggered mostly by endocytosed virus, whose gRNA is recognized by TLR7 in endosomes37. Productive HIV infection of macrophages and T cells, however, involves viral membrane fusion with the cell membrane and direct uncoating of the viral capsid into the cytosol, bypassing the endosomal compartment and TLR signaling. Nonetheless, it will be important to determine whether TREX1 modulates IFN signaling in pDC.
HIV-stimulated IFN activation in Trex1−/− cells is eliminated by treating infected cells with an RT inhibitor, but not an IN inhibitor, suggesting that HIV DNA, but not gRNA, is the nucleic acid that triggers innate immunity. ssDNA is the nucleic acid most sensitive to TREX1 activity and is therefore likely its main substrate. IFN-β mRNA is normally detected 6-8 h after transfecting ISD or infection with a DNA virus6,21,38. After HIV infection, IFN mRNA is not detected until 12 hpi; the lag in IFN-β expression is likely due to time needed to complete reverse transcription. The rapid decline in IFN-β mRNA expression after reaching its peak value suggests that a cell autonomous secondary mechanism tempers the innate immune response that, if unchecked, could be harmful to the host. Some HIV DNA accumulated in the cytoplasm of HIV-infected cells, even when TREX1 is normally expressed, but did not activate IFN expression. A cytoplasmic DNA threshold, which might vary in different cell types, may need to be exceeded to trigger innate immunity.
HMGB proteins have been proposed as innate immune sentinel proteins that facilitate nucleic acid recognition by cytosolic RNA and DNA sensors20. Here HMGB2 siRNA experiments showed an opposite effect; HMGB2 helped suppress IFN induction by HIV in human cells. Although Hmgb2 RNAi in Trex1−/− MEFs did not increase cytosolic HIV DNA, HMGB2 RNAi in human 293T cells, also treated with TREX1 siRNA, enhanced cytosolic HIV DNA and IFN-β and IFN-β induction. Therefore HMGB proteins may play a more complex role in innate immunity than originally suggested. In their role as foreign nucleic acids sentinels, they may facilitate the recognition of nucleic acids both by sensors that trigger innate immune responses as well as by proteins, such as TREX1, that inhibit IFN induction. Therefore the net effect of reduced HMGB proteins could be either to inhibit IFN induction (as in20) or to enhance it, as shown here. We also found that HMGB2 can also act downstream of nucleic acid sensing at the IFN-β promoter to suppress IFN-β transcription, adding another layer of complexity. This transcriptional effect extends to non-HIV innate immune stimuli (poly(dA:dT) and MAVS over-expression). In the published study20, poly(dA:dT)-stimulated IFN-β expression in Hmgb2−/− MEFs was reduced compared to WT MEFs, whereas we found the opposite effect with Hmgb2 RNAi. The apparent discrepancy between the previous results and ours could be due to a difference in the consequences of complete or partial Hmgb2 elimination, especially if HMGB2 operates at multiple steps in IFN induction.
Many of our experiments used genetically deficient mouse cells to demonstrate that Trex1 helps HIV evade innate immune detection and define the HIV DNA-stimulated IFN signaling pathway. Knockout mouse cells are powerful tools for HIV research8,39,40. Once the block in entry in mouse cells is overcome by VSV-G-pseudotyping, most early steps of HIV replication, including reverse transcription, integration and LTR-driven transcription, are similar in human and mouse cells. Furthermore, human TREX1 is 73.3% identical to its mouse homolog in amino acid sequence, and is 71.4%/100%/86.7% identical in its three exonuclease motifs15. Human and mouse TREX1 have the same enzymatic activity and can substitute for each other15. Therefore mouse cells are well suited for studying TREX1 function in HIV replication. Nonetheless, human immune cells susceptible to HIV can differ from MEF in their ability to activate innate immune pathways. For example, IFN-β was induced by HIV in human immune cells, but not MEFs. The differential role of HMGB2 in HIV DNA accumulation in human macrophages and MEFs may be another case in point. We validated all key findings in primary human cells, including HIV DNA accumulation, IFN induction and inhibition of HIV replication when TREX1 was inhibited by RNAi, and efficient rescue by co-suppressing IRF3 expression.
Our data shed light on the fate of non-productive or non-integrated HIV DNA in the cell. At an MOI of 1, HIV infection produces many reverse transcripts (although only one per incoming gRNA), but very few copies manage to integrate into the host chromosome. The remaining HIV DNA is cleared by TREX1, since cytosolic HIV DNA builds up when TREX1 function is deficient or inhibited and can be removed by expression of enzymatically active TREX1. As a consequence, WT TREX1 fully rescues the HIV infection block in Trex1−/− MEFs and the D18N mutant fails to rescue. Other host nucleases might also help digest cytosolic HIV DNA. It is unclear why the excess HIV DNA that accumulates in Trex1−/− cells does not lead to more chromosomal integration. Sequencing these excess HIV DNAs may reveal whether they are capable of integration and what prevents them from integrating. These excess HIV DNAs may be mostly non-productive RT products.
TREX1 promotes HIV replication in two ways – it inhibits autointegration and suppresses the IFN response. Several models might explain the dual function of TREX1 on HIV DNA. One possibility is that TREX1 might sort productive vs non-productive HIV RT products. HIV RT is error-prone and often produces incomplete products. TREX1 recognizes ssDNA or dsDNA with single strand overhangs – the kind of DNA in failed RT products. TREX1 might bind to HIV DNA nonspecifically in the cytosol, but as an exonuclease can only efficiently digest HIV DNA that contains broken ends or single strand overhangs. HIV integrase binds to the ends of reverse transcripts that are capable of chromosomal integration and might protect them from TREX1 digestion. Incomplete RT products, however, would not bind integrase and therefore would be susceptible to TREX1 degradation. Autointegration requires the full-length RT product and active DNA ends bound by integrase, which catalyzes autointegration. Although TREX1 likely binds to full-length integration-competent products, as well as to transcripts that are not competent for integration, its exonuclease activity might be inhibited in the full-length transcript by lack of some DNA feature that facilitates digestion (such as shielding by integrase). Another possibility is that TREX1 is inhibited by components of the SET complex that also bind to the HIV PIC2. Another DNase in the SET complex, NM23-H1, is inhibited by SET protein and is only activated when Granzyme A cleaves SET41. TREX1 is an abundant protein that is not exclusively in the SET complex. One could also imagine that two subpopulations of TREX1 are involved in different actions: the SET complex-associated TREX1 inhibits autointegration; while TREX1 outside the SET complex is enzymatically active and removes excess HIV DNA. This model would also explain why siRNAs against most other SET complex genes do not induce IFN, but do protect against autointegration. Further studies are needed to test these ideas.
TREX1 mutations that interfere with TREX1 enzymatic function or localization are associated with SLE and other autoimmune/inflammatory diseases3-5. Patients with SLE are underrepresented in HIV-infected populations42. It would be worth evaluating whether TREX1 polymorphisms or autoimmune disease are associated with reduced HIV transmission or a more benign disease course. The innate immune pathway uncovered in this study will improve understanding of how HIV intersects with innate immunity and may also shed light on autoimmune and inflammatory syndromes linked to TREX1 mutation.
ONLINE METHODS
Cells
WT, Trex1−/− and Trex1−/−Irf3−/− MEFs7,43 were provided by D. Stetson (Univ. Washington) under agreement with D. Barnes and T. Lindahl (Cancer Research UK). RIG-I-KO (also known as Ddx58−/−) MEFs were provided by J. Jung (Univ. of Southern Cal.). The human fibroblast line carrying the D18N heterozygous mutation in TREX1 was derived from a chilblain lupus patient. Primary human macrophages and CD4+ T cells were isolated from PBMCs and maintained using standard protocols. HeLa-CD4 and 293T cells were described previously2. HeLa-CD4, TZM-bl, 293T, and human fibroblasts were grown in DMEM (Invitrogen) and MEFs were grown in DMEM/F12 (Invitrogen), both supplemented with 10% heat-inactivated fetal bovine serum (FBS). Primary CD4+ T cells were grown in RPMI (Invitrogen) supplemented with 10% heat-inactivated FBS, activated in 2 μg/ml PHA and maintained in 30 U/ml rhuIL-2. Monocyte-derived macrophages (MDM) were grown in RPMI supplemented with 10% heat-inactivated human serum. Experiments involving human and mouse materials were approved by the institutional review boards of the Immune Disease Institute, Harvard Medical School and the Children's Hospital of Technical University Dresden.
Viruses, infection and antiviral compounds
HIVIIIB and HIVBaL were propagated as described44. HIV-GFP plasmid (pNL4-3/Env−)45 was provided by D. Gabuzda (Dana-Farber Cancer Inst.). HIV-Luc plasmid (pNL4-3/Env−)46 was provided by A. Engelman (Dana-Farber Cancer Inst.). Viral supernatants were produced from transfected 293T cells as described2. VLPs were produced by transfecting 293T cells with VSV-G plasmid and a plasmid that contains only HIV gag-pol. Virus was harvested in 3 batches every 12 h from 48-72 h post transfection, and concentrated approximately 10-fold using Centricon filters (Millipore, cat#UFC910024) according to the manufacturer's instructions.
All HIV viruses were titered on WT MEFs (for mouse cell experiments) and on 293T cells (for human cell experiments) by flow cytometry analysis of p24 Ag 24 hpi. Cells were stained with p24-FITC (Beckman Coulter, cat#KC57-FITC) or not stained for HIV-GFP. MOI, calculated using the formula, % infected = 1-e−moi (e.g. 63.2% positive gives MOI=1), was determined separately for mouse and human cells using pseudotyped viruses. Infections were performed using an MOI of 1 unless otherwise indicated for 6-8 h before replacing viral supernatants with fresh medium. Primary CD4+ T cells were infected in 24 well plates (3×105 cells/well with HIVIIIB (400 ng/ml p24)) and MDM were similarly infected with HIVBaL (200 ng/ml p24). HIV-GFP and HIV-Luc infectivity were measured as described2,45. For experiments that measure stage-specific HIV-1 DNAs, viral supernatants were pretreated with 40 U/ml Turbo DNase (Ambion) at 37 °C for 30 min before infection. Cytosolic HIV DNA was isolated at indicated times post infection by first separating nuclear and cytoplasmic fractions by lysis for 10 min on ice in lysis buffer (20 mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, complete mini-protease inhibitor cocktail (EDTA-free, Roche)) followed by centrifugation at 10,000 g for 10 min. Subgenomic DNA in the supernatant (cytoplasmic fraction) was then isolated using the Hirt method47. Infection of CD4+ T cells was by spinoculation at 1500 g for 2 h.
To obtain conditioned medium, WT, Trex1−/− and Trex1−/−Irf3−/− cells were infected with VSV-G-pseudotyped HIV-GFP for 6-8 h. Viruses were then removed and replaced with fresh medium and ‘conditioned’ overnight. The ‘conditioned medium’ was collected the next morning, filtered (0.45 μm), and added to WT cells with HIV-Luc viruses. Luc activity was then measured 48 hpi. A different reporter virus was used at the second step to eliminate any concerns of virus carryover. Neutralizing antibody experiments were done by pre-incubating conditioned medium with 5 μg/ml anti-mIFN-β (Ambion) at room temperature for 1 h before adding to WT cells with HIV-Luc viruses.
RT and IN inhibitor drugs (nevirapine and raltegravir), obtained form the NIH AIDS Reagent and Reference Program, were used at 5 μM. The POL-III inhibitor ML-60218 was purchased from Calbiochem and used at indicated concentrations.
Plasmids, siRNAs, DNA/RNA oligonucleotides and transfection
The GFP-TREX1 plasmid was previously described4. GFP-TREX1(D18N) was generated by site-directed mutagenesis. Rescue experiments were performed by transfecting MEFs using the Amaxa nucleofector kit (cat# VPD-1004) and sorting for GFP+ cells 24 h post transfection. Cells expressing GFP or GFP fusion proteins were then infected with VSV-G-pseudotyped HIV. IFNB-Luc and CMV-Renilla-Luc plasmids were provided by J. Hiscott (McGill University, Canada) and L. Gehrke (Harvard Medical School)48,49. Dual Luciferase assay was performed according to standard protocols (Invitrogen). Primary MDM were transfected with control (CTL) siRNA (600 pmole per 3 × 106 cells) or si-TREX1 (600 pmole per 3 × 106 cells) using the Amaxa nuclefector kit (cat# VPA-1008). CD4+ T cells were transfected with similar siRNA concentrations (200-600 pmole per 3 × 106 cells for si-TREX1) using Amaxa nucleofector kit (cat# VPA-1002). HA-MAVS plasmid was provided by S. Nagata (Kyoto University, Japan).
The ssDNA oligonucleotide is ‘Gag-100 forward’ (see Supplementary Table 1 for sequence). dsDNA was generated by hybridizing ‘Gag-100 forward’ and ‘Gag-100 reverse’ at equimolar concentrations (by heating at 100 °C for 3 min followed by slowly cooling to room temperature). ssRNA was generated by in vitro transcription using the T7 transcription kit (Ambion, cat#1354) and ‘Gag-100-T7’ as a template. The RNA:DNA hybrid was generated by hybridizing ssDNA and ssRNA at equimolar concentrations. Poly(dA:dT) was purchased from Sigma. Nucleic acids were transfected using Lipofectamine 2000 (Invitrogen). Cytosolic ssDNA and dsDNA were quantified by qPCR using gag primers.
siRNA sequences are provided in Supplementary Table 2. siRNAs were purchased from Dharmacon. Transfections were performed using 2 μl Lipofectamine 2000 (Invitrogen) and 100 nM siRNA in 24-well plates, or using Amaxa nucleofection for primary cells.
HIV DNA, cytokine mRNA and protein analysis
DNA primer sequences are provided in Supplementary Table 1. Stage specific HIV DNA in mouse cells was measured as described previously2,8 using mouse mitochondrial DNA for normalization; integrated DNA in mouse cells was measured using a similar nested PCR design with two mouse LINE primers8 instead of one Alu primer and normalized to mouse Gapdh DNA. Cytokine mRNA was extracted using Trizol reagent (Invitrogen), and measured by qRT-PCR using specific gene primers (Supplementary Table 1). Mouse IFN-β protein in culture supernatants was measured by ELISA (PBL, cat#42400-1).
Immunoblotting and immunostaining
Antibodies for mTREX1 (1:1000, mouse, BD Biosciences, clone 29), SET (1:1000, rabbit)2, APE1 (1:1000, rabbit)2, NM23-H1 (1:1000, rabbit, Santa Cruz), HMGB1 (1:2000, rabbit, Abcam), HMGB2 (1:2000, rabbit, Abcam), RIG-I (1:1000, rabbit, Abcam), GFP (1:500, rabbit, Abcam) and Tubulin (1:1000, mouse, Sigma, clone B-5-1-2) were used for immunoblotting using standard protocols. Anti-FLAG (mouse, Sigma, clone M2) and mouse IgG (Jackson Lab) were used for IP as previously described2. mIRF3 antiserum (1:100, rabbit) was provided by T. Fujita (Kyoto Univ., Japan) and used for staining endogenous mIRF3 in MEFs. MEFs grown on coverslips were fixed in 4% PFA and permeabilized and stained using standard protocols. Samples mounted in Vectashield mounting medium containing DAPI (Vector Laboratories) were imaged by using a Zeiss 200M inverted epifluorescence microscope (Carl Zeiss MicroImaging, Inc.) equipped with SlideBook software (Intelligent Imaging Innovations, Inc.).
Statistical methods
Statistical significance was determined by Student's t-test. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH AI45587 (JL), the Harvard CFAR and NIH T32 HL066987-10 (NY), Harvard SHURP program (ADR) and DFG grant (Le 1074/3-1; MLK). We thank D. Stetson, S. Harvey, D. Barnes, T. Lindahl, A. Engelman, D. Gabuzda, T. Fujita, S. Nagata, K. Fitzgerald, J. Hiscott, L. Gehrke for reagents, and members of the Lieberman lab for helpful discussions.
Footnotes
Author contributions statement NY conceived the study, designed and performed most experiments and helped write the paper. ADR-M and BS helped perform the experiments. MLK provided human cell lines and scientific advice. JL conceived and supervised the study and helped write the paper. Users may view, print, copy, download and text and data- mine the content in such documents, for the purposes of academic research, subject always to the full Conditions of use: http://www.nature.com/authors/editorial_policies/license.html#terms
References
- 1.Bowerman B, Brown N, Bishop K, Varmus H. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 2.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. The SET Complex Acts as a Barrier to Autointegration of HIV-1. PLoS Pathog. 2009;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 4.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 5.Crow Y, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 6.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shun M, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 11.Agy MB, Acker RL, Sherbert CH, Katze MG. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+ T-cell lines by distinct mechanisms: evidence for decreased stability and aberrant processing of HIV-1 proteins. Virology. 1995;214:379–386. doi: 10.1006/viro.1995.0047. [DOI] [PubMed] [Google Scholar]
- 12.Coccia EM, Krust B, Hovanessian AG. Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J. Biol. Chem. 1994;269:23087–23094. [PubMed] [Google Scholar]
- 13.Shirazi Y, Pitha PM. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 1992;66:1321–1328. doi: 10.1128/jvi.66.3.1321-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baca-Regen L, Heinzinger N, Stevenson M, Gendelman HE. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J. Virol. 1994;68:7559–7565. doi: 10.1128/jvi.68.11.7559-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazur D, Perrino F. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′-->5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 16.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J. Biol. Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung V, Latz E. Intracellular DNA recognition. Nat. Rev. Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 18.Schroder K, Muruve DA, Tschopp J. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr. Biol. 2009;19:R262–265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Yang P, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 20.Yanai H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 21.Chiu Y-H, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 25.Xu L-G, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber G. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:647–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong B, et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Z, Beresford P, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell Biol. 2002;22:2810–2820. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehming N, Le Saux A, Schüller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 34.El Gazzar M, et al. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol. Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldfeld AE, Birch-Limberger K, Schooley RT, Walker BD. HIV-1 infection does not induce tumor necrosis factor-alpha or interferon-beta gene transcription. J. Acquir. Immune. Defic. Syndr. 1991;4:41–47. [PubMed] [Google Scholar]
- 36.Haase AT. Targeting early infection to prevent HIV-1 mucosal transmission. Nature. 2010;464:217–223. doi: 10.1038/nature08757. [DOI] [PubMed] [Google Scholar]
- 37.Beignon A-S, et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J. Clin. Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 39.Siva A, Bushman F. Poly(ADP-ribose) polymerase 1 is not strictly required for infection of murine cells by retroviruses. J. Virol. 2002;76:11904–11910. doi: 10.1128/JVI.76.23.11904-11910.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagans S, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Z, Beresford P, Oh D, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 42.Zandman-Goddard G, Shoenfeld Y. HIV and autoimmunity. Autoimmun. Rev. 2002;1:329–337. doi: 10.1016/s1568-9972(02)00086-1. [DOI] [PubMed] [Google Scholar]
- 43.Morita M, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 45.He J, et al. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 46.Shun MC, Daigle JE, Vandegraaff N, Engelman A. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 2007;81:166–172. doi: 10.1128/JVI.01953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 48.Paz S, et al. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol. Cell Biol. 2009;29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.