pRb is required to inhibit apoptosis in myoblasts and autophagy in myotubes but not for activation of the myogenic differentiation program.
Abstract
The retinoblastoma tumor suppressor (pRb) is thought to orchestrate terminal differentiation by inhibiting cell proliferation and apoptosis and stimulating lineage-specific transcription factors. In this study, we show that in the absence of pRb, differentiating primary myoblasts fuse to form short myotubes that never twitch and degenerate via a nonapoptotic mechanism. The shortened myotubes exhibit an impaired mitochondrial network, mitochondrial perinuclear aggregation, autophagic degradation, and reduced adenosine triphosphate production. Bcl-2 and autophagy inhibitors restore mitochondrial function and rescue muscle degeneration, leading to formation of long, twitching myotubes that express normal levels of muscle-specific proteins and stably exit the cell cycle. A hypoxia-induced glycolytic switch also rescues the myogenic defect after either chronic or acute inactivation of Rb in a hypoxia-inducible factor-1 (HIF-1)–dependent manner. These results demonstrate that pRb is required to inhibit apoptosis in myoblasts and autophagy in myotubes but not to activate the differentiation program, and they also reveal a novel link between pRb and cell metabolism.
Introduction
The retinoblastoma tumor suppressor (pRb) is lost by mutation or functional inactivation in virtually all human cancer (Burkhart and Sage, 2008; Bremner and Zacksenhaus, 2010). Its disruption leads to ectopic cell proliferation, apoptotic cell death, and incomplete differentiation and may cause cancer in many cell types (Zacksenhaus et al., 1996; Chen et al., 2004; Mantela et al., 2005; Jiang et al., 2010). Rb exerts these effects by modulating the activity of transcription factors such as activating E2Fs (Chen et al., 2009). These E2Fs activate cell cycle progression and DNA synthesis genes as well as proapoptotic, BH3-only factors that induce mitochondrial outer membrane permeabilization (MOMP) and trigger the intrinsic apoptotic machinery (Guo et al., 2001; Hershko and Ginsberg, 2004; Ho et al., 2004, 2007). Activating E2Fs also induce p53 via ARF and MDM2, various apoptogenic factors, and autophagic genes (Nahle et al., 2002; Tracy et al., 2007; Polager et al., 2008). pRb is also thought to stimulate lineage-specific transcription factors, including MyoD, myogenin, and MEF2C (Gu et al., 1993; Schneider et al., 1994; Novitch et al., 1999), and sequester inhibitors of differentiation such as Id2, HDAC1, EID-1, and RBP2 (Lasorella et al., 2000; MacLellan et al., 2000; Puri et al., 2001; Benevolenskaya et al., 2005). However, given that ectopic proliferation and apoptosis precludes differentiation, the exact role for pRb in differentiation remains ill defined. In this study, we addressed the function of Rb during myogenesis by uncoupling its effect on cell survival or proliferation from its effect on differentiation.
Results and discussion
Rb deficiency during myogenic differentiation induces autophagy
To identify cell-autonomous functions of pRb during skeletal myogenesis, we analyzed the differentiation of primary Rb−/− myoblasts isolated from limb muscles of embryonic day (E) 16.5 mini-Rb gene (mgRb):Rb−/− fetuses. These mutant embryos harbor an mgRb that directs Rb expression to the placenta and nervous system but not muscles, thereby extending the life span of Rb−/− embryos, which otherwise die at E13.5–14.5, to birth (Zacksenhaus et al., 1996; Jiang et al., 2001; unpublished data). In mgRb:Rb−/− fetuses, myotubes are initially formed at E14.5–15.5 and express early muscle–specific markers, but they continue to synthesize DNA, fail to express late markers, and degenerate (Zacksenhaus et al., 1996; Jiang et al., 2000). As expected, primary myoblasts derived from mgRb:Rb−/− muscles were devoid of pRb (Fig. 1 A).
Figure 1.
Rb−/− myoblasts transiently differentiate to form short myotubes that do not twitch, express autophagy markers, and slowly degenerate. (A) Western blot analysis for pRb in DM-2 cultures. (B) Immunostaining for MHC in DM-3 and DM-6 differentiating myotubes. Nuclei were stained with DAPI (blue). (C) Mean number of myotubes on the indicated days after differentiation. Each time point is mean ± SD of six fields from six independent experiments. (D) TUNEL staining. Arrowheads indicate TUNEL-positive nuclei (green). (E) LC3-RFP expression in control (ctrl) and Rb−/− myotubes. Insets show nuclear DAPI stain (blue). (F) Representative Western blot (n = 3) for LC3. Chloroquine was added 12 h before cell harvest. (G) Representative Western blot for LC3 in E16.5 control and mgRb:Rb−/− muscles. (H) Quantification of LC3-II/tubulin ratio in E16.5 mgRb:Rb−/− skeletal muscles relative to control as depicted in G (mean ± SD; n = 3). *, P = 0.016 by Student’s t test. (I) Representative Western blot for LC3-I and LC3-II expression in whole cell lysates from the indicated myotubes transduced with Ad.GFP or Ad.RbΔK11. (J) Quantification of LC3-II/tubulin ratio by Western blots as shown in I. Data represent mean ± SD (n = 7). *, P = 0.047; **, P = 0.005 by analysis of variance.
When confluent cultures of control primary myoblasts were incubated in differentiation medium (DM), they spontaneously fused to form long multinucleated myotubes that started to twitch by day 3–4 and persisted for weeks in culture. Primary Rb−/− myoblasts also fused to form short myotubes that contained three to six nuclei and expressed near-normal levels of myosin heavy chain (MHC; Fig. 1 B and Fig. S1 A). However, at day 3 after differentiation (DM-3), these short myotubes degenerated; by day 6, virtually all had collapsed (Fig. 1, B and C; and Fig. S1 A). Thus, Rb−/− myoblasts underwent abortive differentiation in vitro that recapitulated the muscle defects seen in mgRb:Rb−/− embryos (Zacksenhaus et al., 1996; Jiang et al., 2000).
Cleaved (activated) caspase-3 and cleaved poly(ADP-ribose) polymerase, markers of intrinsic apoptosis, were elevated in differentiating Rb−/− cultures relative to control (Fig. S1 B). However, TUNEL-positive nuclei were only detected in unfused myoblasts but not within myotubes at DM-2 and DM-3 (Fig. 1 D), indicating that myotube degeneration was not mediated by bona fide apoptotic cell death. Therefore, we asked whether degeneration of Rb−/− myotubes was accompanied by autophagy, which, depending on the context, can lead to cell death or survival (Morselli et al., 2009). During autophagy, bulk cytoplasm, including whole organelles, is sequestered in vesicles termed autophagosomes that fuse to lysosomes (forming autolysosomes) for degradation (Levine and Kroemer, 2008; Narendra et al., 2008; Mizushima, 2009; Tolkovsky, 2009; Mizushima et al., 2010). To detect autophagy, myoblasts were transduced with an adenoviral vector expressing microtubule-associated protein 1A/1B light chain 3 (LC3) fused to red fluorescent protein (Ad.LC3-RFP; Bampton et al., 2005). LC3 accumulates on autophagosomes as a phosphatidylethanolamine (PE)-conjugated form (LC3-II; Mizushima and Yoshimori, 2007). In control myotubes, LC3 exhibited a diffuse red fluorescent pattern (Fig. 1 E, left). In contrast, in Rb−/− myotubes, it was detected in perinuclear aggregates (Fig. 1 E, right), indicating autophagosome accumulation.
Autophagosome-bound LC3-II migrates faster than unconjugated LC3-I through denaturing protein gels (Mizushima and Yoshimori, 2007). At DM-2, LC3-II level was more than twofold higher in Rb−/− myotube cultures relative to control (n = 3; Fig. 1 F). Chloroquine, an inhibitor of autophagosome–lysosome fusion, enhanced the level of LC3-II in both Rb−/− and control cultures, demonstrating active autophagic flux. We also detected a 1.5 ± 0.12-fold (P = 0.016) increase in LC3-II expression in E16.5 mgRb:Rb−/− limb muscle (n = 3) relative to control littermates (n = 3; Fig. 1, G and H). As control, transduction of a constitutively active, phosphomutant pRb (Ad.RbΔK11; Jiang and Zacksenhaus, 2002) prevented collapse of Rb−/− myotubes (not depicted) and, concomitantly, the accumulation of LC3-II by ∼1.8-fold (n = 7; P = 0.005; Fig. 1, I and J).
Rb deficiency during myogenic differentiation induces mitochondrial loss via autophagy
To detect mitochondria, we used MitoTracker, a live cell probe that accumulates in mitochondria with intact membrane potential. In control myotubes, MitoTracker revealed a uniform, netlike distribution of mitochondria throughout the cytoplasm (Fig. 2 A, top left). In contrast, at DM-2, Rb−/− myotubes exhibited strong perinuclear MitoTracker-positive aggregates and relatively sparse cytosolic staining (Fig. 2 A, bottom left). On average, ∼70% of Rb−/− myotubes contained perinuclear aggregates compared with <10% control myotubes (P < 0.001; Fig. 2 B). Cytochrome c also exhibited differential staining: it exhibited uniform distribution throughout control myotubes but irregular punctate/perinuclear staining in Rb−/− myotubes (Fig. S1 C). To determine whether mitochondria accumulated within autophagosomes in Rb−/− myotubes, we performed double staining with MitoTracker and LC3 antibody in the presence of chloroquine. Control myotubes contained very few cytoplasmic aggregates that stained positive for both MitoTracker and LC3 (Fig. 2 C, left). In contrast, 70% of Rb−/− myotubes contained large perinuclear aggregates that stained positive for both markers (Fig. 2 C, middle). The level of overlap between LC3 and MitoTracker was 3.4-fold higher in the entire myotube population and fivefold higher in the 70% fraction of Rb−/− myotubes that exhibited perinuclear mitochondrial aggregation (Fig. 2 D). EM analysis revealed overall reduced mitochondrial content and 2.2-fold increase in perinuclear accumulation of mitochondria in the entire Rb−/− myotube population relative to control (Fig. 2 E). Some whole or partially degraded mitochondria engulfed by membrane, representing autophagosomes and autolysosomes (Mizushima et al., 2010), were observed around nuclei (Fig. 2 E, asterisk) and in cytosol (Fig. S1 D). Consistent with these results, there was a 20% reduction in mitochondrial to nuclear DNA ratio, as determined by real-time PCR (Amthor et al., 2007), in Rb−/− versus control cultures at DM-2 (P < 0.001; Fig. 2 F) and a 50% reduction in E16.5 limb muscle (P = 0.009; Fig. 2 G). Moreover, there was a 20% decrease in ATP level in Rb−/− myotube cultures (not depicted) and a 3.2-fold reduction in ATP level in E16.5 limb muscle relative to control (n = 3, P = 0.011; Fig. 2 H). The observed difference between the in vitro and in vivo results likely reflects the fact that in vitro cultures contain a smaller ratio of myotubes/unfused cells than skeletal muscle.
Figure 2.
The Rb−/− muscle defect is characterized by perinuclear mitochondrial/autophagosome aggregates, reduced mitochondrial DNA content, and reduced ATP production. (A, left) MitoTracker staining. (right) MitoTracker staining after bezafibrate treatment. (B) Quantification of myotubes with perinuclear MitoTracker-positive aggregates in untreated (vehicle) or bezafibrate-treated myotubes at DM-2. Data represent mean ± SD (n = 3). *, P < 0.001 by Student’s t test. (C) MitoTracker (red) and LC3 immunostaining (green) of myotubes at DM-2. Chloroquine and 3-MA (where indicated) were added 12 h before staining. Note the extensive overlap of MitoTracker and LC3 in Rb−/− myotubes (yellow), which is inhibited by 3-MA. (D) Mean percentage of MitoTracker (red) to LC3 (green) overlap in 25 myotubes from two independent cultures in control versus Rb−/− myotubes (total) or Rb−/− myotubes with perinuclear mitochondrial aggregation (high). *, P = 7 × 10−5 by Student’s t test. (E) Ultrastructural analysis of myotubes treated or not with 3-MA at DM-2. Arrowheads point to mitochondria. Asterisks indicate electron-dense autolysosomes. N, nucleus. (F) Ratio of mitochondrial DNA/nuclear DNA in the indicated cultures (n = 3). (G) Ratio of mitochondrial DNA/nuclear DNA in E16.5 skeletal muscle (n = 3; *, P = 0.009). (H) Quantification of ATP in E16.5 skeletal muscle (n = 3; *, P = 0.011). (F–H) Error bars represent SD. *, P < 0.001.
Inhibition of autophagy rescues Rb differentiation defect
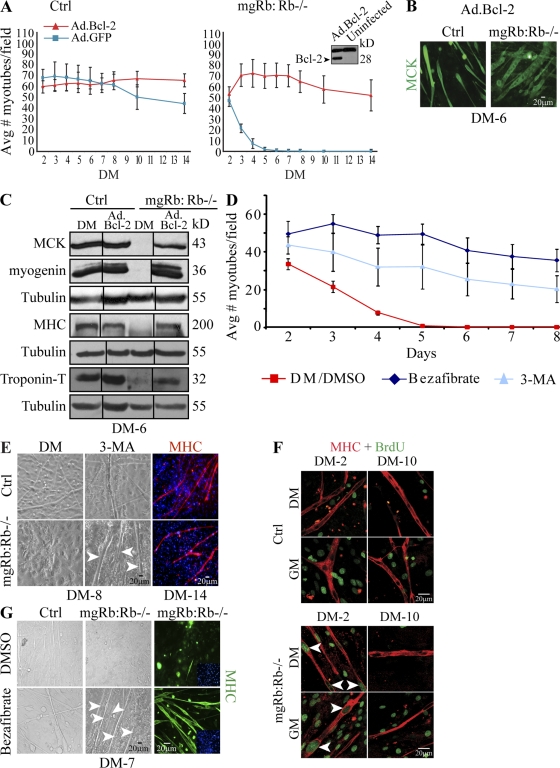
The aforementioned results prompted us to ask whether inhibition of cell death or autophagy would rescue Rb−/− myotube degeneration. Strikingly, adenovirus-mediated transduction of the survival factor Bcl-2, which inhibits both apoptosis and autophagy (Pattingre and Levine, 2006), rescued the differentiation defect, leading to long multinucleated myotubes that expressed MHC and muscle creatine kinase (MCK), and twitched like control myotubes for >3 wk in culture (Fig. 3, A and B; and Videos 1 and 2). Western blot analysis revealed that myogenin, MHC, MCK, and troponin T were undetectable in Rb−/− cultures but expressed at comparable levels in Ad.Bcl-2–transduced Rb−/− myotubes and control (Fig. 3 C).
Figure 3.
Rescue of Rb-deficient myotube degeneration by Bcl-2 and autophagy inhibitors. (A) Mean number of myotubes per field in indicated myoblast cultures induced to differentiate after transduction with Ad.GFP or Ad.Bcl-2. Each point represents the mean ± SD of six fields from six independent experiments. Bcl-2–rescued, twitching Rb−/− myotubes are shown in Video 2. (inset) Western blot for Bcl-2 in Ad.Bcl2-transduced versus uninfected primary myoblasts. (B) Immunostaining for MCK (green) in Ad.Bcl-2–transduced control (ctrl) and Rb−/− myotubes at DM-6. (C) Western blot for MCK, myogenin, MHC, and troponin T in indicated cultures at DM-6 after transduction with Ad.Bcl-2. Tubulin was used as a loading control. Black lines indicate that intervening lanes have been spliced out. (D) Mean number of Rb−/− myotubes after 3-MA treatment, bezafibrate, or DMSO/vehicle as indicated. Counts are mean ± SD of six fields (n = 4). (E, left and middle) Brightfield images of the indicated myotubes treated or not with 3-MA at DM-8. (right) Immunostaining for MHC (red) at DM-14 in the presence of 3-MA. Arrowheads label 3-MA–rescued Rb−/− myotubes at DM-8. 3-MA–rescued, twitching Rb−/− myotubes are shown in Video 4. (F) Immunostaining for BrdU (green) and MHC (red) of 3-MA–treated cultures at DM-2 or DM-10. Rb−/− myotubes incorporated BrdU at DM-2 (arrowheads) but not DM-10. (G) Brightfield images (left and middle) and MHC staining (right) of the indicated cultures treated or not with bezafibrate. Arrowheads point to bezafibrate-rescued Rb−/− myotubes.
Next, we treated cultures with 3-methyladenine (3-MA), an inhibitor of a class III phosphatidylinositol 3-kinase, Vps34, which is required for nucleation of the autophagic vesicle (Levine and Kroemer, 2008). Remarkably, a single dose of 3-MA given just before the induction of differentiation prevented formation of LC3-positive perinuclear aggregates and perinuclear mitochondrial accumulation (Fig. 2, C and D, right) and blocked the degeneration of Rb−/− myotubes, leading to myotubes that twitched for weeks in culture (Fig. 3, D and E; and Videos 3 and 4).
The number of myotubes formed was threefold lower after 3-MA treatment than after Bcl-2 transduction, but the extent of twitching was virtually the same (Videos 2 and 4). This is consistent with our observation that Bcl-2 expression increased survival of both myoblasts and myotubes, whereas 3-MA rescued myotube degeneration but increased apoptotic death of unfused myoblasts (unpublished data). Thus, different survival pathways operate in myoblasts and myotubes, and Rb−/− myotube degeneration is cell autonomous, not an indirect consequence of high level of death in unfused cells. Adenoviral-mediated transduction of a dominant-negative Vps34 allele (Ad.Vps34dn; Trincheri et al., 2008) also rescued the Rb myogenic defect, leading to twitching myotubes (Fig. S1 F). Collectively, these results indicate that degeneration of Rb−/− myotubes is associated with autophagy of impaired mitochondria and that inhibition of this process rescues the Rb myogenic defect.
Rescued Rb−/− myonuclei become stably postmitotic
In addition to increased autophagy, DM-2 Rb−/− myotubes also exhibited ectopic DNA synthesis (Fig. S1 G; Schneider et al., 1994; Chen and Wang, 2000). To determine the cell cycle status of 3-MA–rescued Rb−/− myotubes, myoblasts were induced to differentiate in the presence or absence of 3-MA. After 1 or 9 d, the cultures were either maintained in DM or shifted to GM and treated with BrdU for 16 h. At DM-2, both 3-MA–treated and untreated Rb−/− myotube nuclei but not control myotube nuclei labeled positive for BrdU (Fig. 3 F, left). Strikingly, by DM-10, 3-MA–treated Rb−/− myotubes ceased to incorporate BrdU under either DM or GM conditions (Fig. 3 F, right). Thus, rescued Rb−/− myotubes can eventually enter a stable postmitotic state.
In contrast to the effect of autophagy inhibitors, suppression of DNA synthesis with aphidicolin, or cell cycle progression with p27kip1, worsened the differentiation defect of Rb−/− myoblasts (Fig. S2). Transduction of activated mTOR (Briaud et al., 2005) or dominant-negative Foxo3a (Skurk et al., 2005), which inhibit autophagy induced by nutrient or insulin/IGF-1 deprivation (Mammucari et al., 2007; Zhao et al., 2007), also failed to prevent Rb−/− myotube degeneration, although both factors increased the number and size of control myotubes (Fig. S3, A–D). Thus, Foxo3a and mTOR affect survival-associated autophagy in response to starvation but not death-associated autophagy that accompanies Rb−/− myotube degeneration.
Peroxisome proliferator–activated receptor (PPAR) agonist and MOMP antagonist rescue the Rb differentiation defect
If mitochondrial loss underlies myotube degeneration in the absence of pRb, we reasoned that stimulation of mitochondrial biogenesis might also rescue the defect. Mitochondrial biogenesis is regulated by the coactivator PGC-1α and PPARs, which can be activated by bezafibrate, a pan-PPAR agonist (Bastin et al., 2008; Wenz et al., 2008). As shown in Fig. 3 (D and G), bezafibrate treatment efficiently blocked degeneration of Rb−/− myotubes, leading to twitching myotubes that survived for >8 d. Longer treatment with bezafibrate became cytotoxic to both control and Rb−/− myotubes. Bezafibrate also increased overall mitochondrial content, observed by MitoTracker or cytochrome c staining (Fig. 2 A, right; and Fig. S1 H), and reduced the percentage of myotubes with perinuclear aggregates from ∼70 to 40% (P < 0.001; Fig. 2 B). This result is in accord with a recent finding that during erythropoiesis, Rb deficiency disrupts mitochondrial biogenesis (Sankaran et al., 2008). Next, we assessed the effect of blocking MOMP on Rb−/− myotube survival using minocycline, an MOMP inhibitor (Zhu et al., 2002). Minocycline treatment delayed the collapse of mutant myotubes, which survived and twitched for 8 d (Fig. S3 E). In contrast, transduction of the x-linked inhibitor of apoptosis (XIAP), which inhibits multiple caspases (Ho et al., 2007; Leonard et al., 2007), failed to block myotube degeneration (Fig. S3, F and G), indicating that collapse of Rb−/− myotubes can only be effectively inhibited before MOMP.
Hypoxia-induced glycolytic shift rescues differentiation defect after chronic or acute inactivation of Rb
The aforementioned results indicate that during differentiation, pRb loss leads to mitochondrial dysfunction and autophagy, reduced mitochondrial content and ATP levels, and ultimately myotube degeneration. An intriguing possibility is that a metabolic shift from oxidative phosphorylation (OXPHOS) to glycolysis would bypass the requirement for mitochondrial function and rescue the defect. To test this notion, we investigated the effect of hypoxia, which induces such a metabolic shift, on myogenic differentiation. A previous study showed that hypoxia inhibits the differentiation of immortalized satellite cells, C2C12 (Di Carlo et al., 2004). However, we found that differentiation of primary myoblasts was unaffected. Remarkably, Rb−/− myotubes survived under hypoxic conditions (1% O2), leading to long MHC-expressing, twitching myotubes (Fig. 4 A). Immunoblotting confirmed high expression of both early (myogenin) and late (MCK) markers in hypoxia-rescued Rb−/− myotube cultures (Fig. 4 B, left). Densitometry analysis of these factors, normalized for MHC expression (i.e., myotube number), revealed that although expression of MCK was near normal, expression of myogenin was elevated in rescued Rb−/− myotubes relative to control, although this was not statistically significant (Fig. 4 B, right). When returned to normoxia after 5 d in hypoxia, rescued Rb−/− myotubes continued to twitch, once again highlighting the transient requirement for Rb function in the first few days after differentiation.
Figure 4.
Hypoxia rescues muscle degeneration after chronic or acute inactivation of Rb by inducing glycolysis via HIF-1α. (A) Immunostaining for MHC (green) in the indicated myoblasts induced to differentiate under normoxia or hypoxia treated or not with lonidamine. Note the shortened myotubes in the presence of lonidamine. (B) Representative immunoblot (left) and mean expression of myogenin and MCK normalized for MHC expression (right; n = 3). (C) Rbf/f myoblasts transduced with Ad.GFP or Ad.Cre and immunoblotted for pRb. Tubulin was used as a loading control. (D) Rbf/f myoblasts transduced with Ad.GFP or Ad.Cre and immunostained for pRb (red). (insets) DAPI staining for nuclei (blue). (E) Immunostaining for MHC in Rbf/f myoblasts transduced with Ad.Cre and induced to differentiate in normoxia (top and middle) or hypoxia (bottom). (F, top) Brightfield images of Rbf/f myoblasts transduced with Ad.EV or Ad.Cre and induced to differentiate under hypoxia. (bottom) Larger view of the boxed field. Arrowheads point to rescued Rb−/− myotubes. This field is shown in Video 6. (G) Quantification of myotube formation in Rbf/f myoblasts transduced with Ad.EV or Ad.Cre and induced to differentiate under hypoxia. Counts are mean of six representative fields (n = 3). *, P = 0.004. (H) Quantification of myotube formation in the indicated myoblasts induced to differentiate under the indicated conditions. Myotube counts are the mean of six fields (n = 4). (I) MHC staining (red) of the indicated myoblasts transduced with Ad.NF-κB/HIF-1α and induced to differentiate in normoxia. (J) Quantification of myotube formation in the indicated myoblasts transduced with Ad.EV or Ad.NF-κB/HIF-1α and induced to differentiate in normoxia. Myotube counts are the mean of six fields (n = 3). Error bars represent SD.
To test whether hypoxia could also rescue the Rb myogenic defect after acute inactivation of this tumor suppressor, we infected embryonic myoblasts from E16.5 Rbf/f embryos in which exon 19 of Rb is flanked by loxP sites with adenovirus encoding Cre recombinase (Vooijs et al., 2002; Huh et al., 2004). We used high multiplicity of infection that deleted Rbf/f in virtually all cells (Fig. 4, C and D). Remarkably, like after chronic inactivation, hypoxia rescued the differentiation defect after acute inactivation of Rb, leading to long, twitching myotubes that continued to contract even when transferred back to normoxia (Fig. 4, E and F; and Videos 5 and 6).
Hypoxia-rescued Rb differentiation defect is glycolysis and hypoxia-inducible factor-1α (HIF-1α) dependent
Under low oxygen tension, HIF-1α induces expression of genes responsible for glucose influx and glycolysis (Maynard and Ohh, 2007). To determine whether hypoxia rescued Rb−/− myotubes by inducing a glycolytic shift, myoblasts were induced to differentiate in hypoxia in the presence of lonidamine, an antagonist of hexokinase, which catalyzes the conversion of glucose to glucose 6-phosphate (Pelicano et al., 2006). Beginning at DM-2, a relatively low concentration of 50 µM lonidamine had little obvious effect on the survival of control myotubes in hypoxia (Fig. 4, A and G). In contrast, under hypoxia, mutant myoblasts were highly sensitive to this dose of lonidamine, forming only very short myotubes, most of which ultimately degenerated (Fig. 4, A and G).
Under normoxia, HIF-1α undergoes ubiquitination via the von Hippel–Lindau tumor suppressor protein (VHL), leading to its proteosomal degradation (Maynard and Ohh, 2007). To ask whether HIF-1α was responsible for preventing the degeneration of Rb−/− myotubes in hypoxia, we first used a VHL allele that is constitutively active both in normoxia and hypoxia (Ad.VHL-T7-HPACGV; Sufan et al., 2009). Expression of this allele dramatically reduced survival of Rb−/− myotubes under hypoxic conditions (Fig. 4 G, bottom right), implicating HIF-1α in mediating rescue of Rb−/− myotubes under low oxygen tension. Next, we tested whether HIF-1α was sufficient to prevent Rb−/− myotube degeneration using a constitutively active HIF-1α allele, HIF-1α/NF-κB, where the N terminus of HIF-1α is fused to the NF-κB transactivation domain (Vincent et al., 2000). Transduction of Ad.HIF-1α/NF-κB rescued degeneration of Rb−/− myotubes in normoxia, leading to stable myotubes for >10 d (Fig. 4, H and I). We conclude that Rb−/− myotubes survive in hypoxia because of an HIF-1α–induced glycolytic shift.
Our results demonstrate that pRb is required for survival during terminal myogenesis but not to actively stimulate the myogenic differentiation program. Although we do not rule out the possibility that interaction of pRb with MyoD may be involved in cell cycle exit or survival, the presence of pRb is clearly not required for myoblast differentiation in vitro. Our results are consistent with observations that Rb−/− cells can efficiently contribute to most adult tissues in Rb−/−:Rb+/+ chimeric mice (Maandag et al., 1994; Williams et al., 1994; Tracy et al., 2007). In muscle, Rb+/+ cells can complement the Rb−/− defect after cell fusion. However, in other tissues, it appears that most Rb−/− cells are inherently capable of functional differentiation. This has direct implications for differentiation-induced therapy for Rb mutant cancer cells.
We show that pRb is required during myogenesis to suppress mitochondrial destruction. In most Rb−/− cell types, including unfused myoblasts, induction of differentiation at low mitogens leads to MOMP and apoptotic cell death. In Rb−/− myotubes, we suggest that the intrinsic apoptotic machinery is inactive and that mitochondrial dysfunction leads to disruption of the mitochondrial network, perinuclear aggregation, and autophagic degradation. A similar interplay between inhibition of caspase-dependent apoptosis and autophagy has been documented in other contexts (Xue et al., 2001; Shimizu et al., 2004). We propose that abortive cell differentiation in Rb−/− cells is caused by mitochondrial dysfunction and survival factors that can restore mitochondrial function or redirect metabolism toward glycolysis can maintain Rb−/− cell survival and differentiation.
A switch from OXPHOS to glycolysis (Dang et al., 2009; Vander Heiden et al., 2009) and Rb inactivation (Burkhart and Sage, 2008; Bremner and Zacksenhaus, 2010) represents two major hallmarks of cancer. Our results indicate that Rb loss leads to inappropriate cell division, which is oncogenic, and also to impaired mitochondrial function, which promotes cell death. Therefore, in the context of neoplastic transformation, Rb loss must cooperate with mutations that effectively suppress mitochondrial damage or autophagy or shift ATP production from OXPHOS to glycolysis. Some of the latter mutations may directly induce glycolysis, whereas others may cooperate with deregulated E2F to transcriptionally activate glycolytic genes (Darville et al., 1995). Thus, we have identified a novel role for pRb in maintaining mitochondrial integrity that is important for normal differentiation and has implications for cancer metabolism.
Materials and methods
Mouse analysis
Experiments were performed in accordance with guidelines of the Canadian Council on Animal Care and Animal Ethics Committee (University Health Network, Toronto, Ontario, Canada). Mice were genotyped using DNA extracted from tail biopsies and the following primers: mgRb/Rblox (forward), 5′-ATTTCAGAAGGTCTGCCAAC-3′ and (reverse) 5′-AGAGCAGGCCAAAAGCCAGGA-3′; Rb mutant (forward), 5′-AATTGCGGCCGCATCTGCATCTTTATCGC-3′ and (reverse) 5′-GAAGAACGAGATCAGCAG-3′; Rb wild type (forward), 5′-AATTGCGGCCGCATCTGCATCTTTATCGC-3′ and (reverse) 5′-CCCATGTTCGGTCCCTAG-3′; Rbfloxed (Rb18 + Rb19E) (forward), 5′-GGCGTGTGCCATCAATG-3′ and (reverse) 5′-CTCAAGAGCTCAGACTCATGG-3′. For timed pregnancies, mice were mated overnight, and the day of vaginal plug observation was considered E0.5.
Primary myoblast cell cultures
Over 1,000 embryos from >200 timed pregnancies were used in this study to generate primary mouse embryonic myoblast cultures. To maintain consistency between experiments, primary myoblasts were induced to differentiate at passage 2. Myoblasts were isolated from skeletal muscle of limbs of E.16.5–17.5 embryos. Tissues were digested for 20 min at 37°C in 80 µl solution containing 1.5 U/ml collagenase IV (Sigma-Aldrich), 2.4 U/ml dispase (Roche), and 5 mM CaCl2 gently triturated and plated onto a 60-mm collagen I–coated culture dish. Myoblasts were maintained in growth medium (GM), HAM’s-F10 (Lonza) supplemented with 20% FBS (PAA), and 2.5 ng/ml basic FGF (Sigma-Aldrich), in a humidified incubator at 5% CO2 and 37°C. To induce differentiation, myoblasts were rinsed once in 1× PBS and shifted to DM, DME, high glucose, and sodium pyruvate (Sigma-Aldrich), supplemented with 3% horse serum (PAA; Ho et al., 2004). Adenoviruses were amplified in 293T cells maintained in DME plus 10% FBS and penicillin/streptomycin (Sigma-Aldrich). For drug treatment, a single dose of 5 mM 3-MA was added upon differentiation. 500 µM bezafibrate and 200 µM minocycline were refreshed every other day. 40 µM chloroquine was added for 12 h before harvesting the cells. Aphidicolin was used at 2.5 µg/ml. For hypoxia experiments, confluent myoblast cultures were fed DM and transferred hypoxic conditions (1% O2). 50 µM lonidamine (refreshed every other day; Sigma-Aldrich) and Ad.VHL-T7-HPACGV were added at DM-1.
BrdU DNA synthesis assay
DM-1 myotube cultures were restimulated in GM or maintained in DM supplemented with 20 µM BrdU for 16 h before fixation with 3.7% formaldehyde (10 min). Aphidicolin was added 5 h before addition of BrdU for 16 h. Cultures were permeabilized using 0.3% Triton X-100 for 10 min, treated with 2 N HCl for 25 min, and neutralized with two washes of 0.5 M sodium borate, pH 8.5, for 5 min. After blocking in 1.0% BSA for 20 min and anti-MHC antibody (1:50; Sigma-Aldrich) for 1 h, cells were washed three times for 3 min each with PBS. Secondary antibody fluorescein-conjugated Alexa Fluor 563 (red; Invitrogen) was used. BrdU was detected using anti-BrdU antibody conjugated to FITC (Alexa Fluor 488) and diluted as per manufacturer’s protocol (BD). Confocal images of 0.5-µm sections were captured at room temperature using a 40× or 63× 1.2 NA C Apochromat water objective lens using a confocal microscope (LSM510 META; Carl Zeiss, Inc.) and acquisition software (AIM 3.2; Carl Zeiss, Inc.). Photoshop (CS2; Adobe) was used to overlay images.
Immunofluorescence and EM
500,000 myoblasts were seeded on 22-mm round collagen I–coated coverslips (BD) and induced to differentiate. Cells were fixed in 3.7% formaldehyde, permeabilized in 0.3% Triton X-100, and blocked for 20 min in 1% BSA/PBS at room temperature. The following antibodies and dilutions were used: myogenin, 1:10 (F5D; Santa Cruz Biotechnology, Inc.); MHC, 1:50 (Sigma-Aldrich); MCK, 1:50 (Santa Cruz Biotechnology, Inc.), cytochrome c, 1:50 (clone A-8; Santa Cruz Biotechnology, Inc.). Secondary antibodies fluorescein-conjugated Alexa Fluor 563 and 488 (Invitrogen) were added for 45 min. Nuclei were counterstained with DAPI (Invitrogen) for 10 min and mounted in fluorescent mounting media (Dako). Mitochondrial LC3 (red/green) colocalization was conducted with the Colocalization plug-in for ImageJ (National Institutes of Health). Electron micrographs were captured at 80 kV using a transmission electron microscope (H-7000; Hitachi) equipped with an AMT digital camera (Hitachi) at the University of Toronto. Image contrast was enhanced using Photoshop.
Western blot analysis
Cells were lysed on ice in K4IP buffer (50 mM Hepes, pH 7.5, 0.1% Tween-20, 1 mM EDTA, 2.5 mM EGTA, 150 mM NaCl, 1.0 mM DTT, and 10% glycerol) containing protease inhibitors (Sigma-Aldrich). The following antibodies and dilutions were used for 3 h at room temperature or overnight at 4°C: α/β-tubulin, 1:4,000 (Cell Signaling Technology); MHC, 1:200 (Sigma-Aldrich); LC3b, 1:500 (Cell Signaling Technology); MCK, 1:200 (Santa Cruz Biotechnology, Inc.); pRb, 1:1,000 (BD); troponin T-FS, 1:200 (clone c-18; Santa Cruz Biotechnology, Inc.); poly(ADP-ribose) polymerase, 1:500 (Cell Signaling Technology); and caspase-3, 1:500 (Cell Signaling Technology). Secondary antibodies used were HRP-linked anti-IgG at 1:2,000 (Cell Signaling Technology) for 1.5 h in blocking buffer, and HRP activity was detected using SuperSignal West Dura chemiluminescent substrate (Thermo Fisher Scientific) and captured by x-ray film. Films were digitized using a scanner (Canon). Quantification of Western blots was conducted using Photoshop.
Mitochondrial and nuclear DNA quantitative PCR
Mitochondrial DNA copy number was quantified for an 80-bp fragment of mouse mitochondrial DNA (primers: forward, 5′-GCAAATCCATATTCATCCTTCTCAAC-3′; reverse, 5′-GAGAGATTTTATGGGTGTAATGCGGTG-3′) relative to the NADH dehydrogenase (ubiquinone) flavoprotein 1 single-copy nuclear gene (GenBank/EMBL/DDBJ accession no. NM_133666) (primers: forward, 5′-CTTCCCCACTGGCCTCAAG-3′; reverse, 5′-CCAAAACCCAGTGATCCAGC-3′) using quantitative real-time PCR (7900T lightcycler; Applied Biosystems).
ATP, MitoTracker red CMXRos, and TUNEL assays
ATP quantification was performed using a luminescence detection system according to the manufacturer’s protocol (ATPlite; PerkinElmer). Mitochondria membrane potential was detected using MitoTracker red CMXRos according to the manufacturer’s protocol (Molecular Probes). For TUNEL, differentiating myoblasts on collagen I–coated coverslips were fixed in 3.7% formaldehyde for 10 min, washed three times in PBS, permeabilized with 0.3% Triton X-100 solution, and washed three times in PBS. Subsequently, 30 U TdT (Fermentas) was added to 50 µl TUNEL label solution (Roche).
Adenovirus Cre infections
For adenovirus Cre (Ad.cre; Vector Biolabs) infection of Rbf/f myoblasts, cells were transduced at a multiplicity of infection of 1,300. At a lower multiplicity of infection (<1,000), some pRb-positive nuclei within myotubes were still detected. 48 h after transduction, myoblasts were induced to differentiate. For all other recombinant adenoviruses, transduction was performed when cultures were induced to differentiate. The following adenovirus vectors were provided by the indicated sources: Ad.Bcl-2 (M. Crescenzi, Higher Institute of Health, Rome, Italy), Ad.GFP-LC3 and Ad.RFP-LC3 (A.M. Tolkovsky, University of Cambridge, Cambridge, England, UK), Ad.HIF-1α/NF-κB and Ad.EV (K.A. Vincent, Genzyme Corporation, Framingham, MA), Ad.VHL-T7-HPACGV (M. Ohh, University of Toronto, Toronto, Ontario, Canada), Ad.Vps34DN (D. Murphy, University of Bristol, Bristol, England, UK), Ad.mTOR, Ad.KD.mTOR, and Ad.ca.mTOR (C.J. Rhodes, The University of Chicago, Chicago, IL), Ad.FOXO3a and Ad.DN.FOXO3a (K. Walsh, Boston University School of Medicine, Boston, MA), Ad.RbΔK11 and Ad.p27 (D.S. Park, University of Ottawa, Ottawa, Ontario, Canada), and Ad.XIAP and anti-XIAP antibody (R.G. Korneluk, University of Ottawa, Ottawa, Ontario, Canada).
Brightfield images and videos
Brightfield images and videos were captured at room temperature using 20× or 40× air objective lenses on a microscope (TE200; Nikon) fitted with a charge-coupled device digital camera (Hamamatsu Photonics). Images were acquired using imaging software (SimplePCI; Hamamatsu Photonics). Photoshop was used to enhance clarity and contrast using same parameters for control and experimental samples.
Online supplemental material
Fig. S1 shows characterization of myogenic defects during abortive differentiation of Rb−/− myoblasts. Fig. S2 shows that cell cycle inhibitors aphidicolin and p27Kip1 accelerate the degeneration of Rb−/− myotubes. Fig. S3 shows differential effects of mTOR, Foxo3, minocycline, and XIAP on differentiation of Rb−/− myoblasts. Video 1 shows twitching control myotubes (Ad.Bcl-2 transduced), Video 2 shows twitching Rb−/− myotubes (Ad.Bcl-2 transduced), Video 3 shows twitching control myotubes (3-MA treated), Video 4 shows twitching Rb−/− myotubes (3-MA treated), Video 5 shows twitching control Ad.EV-transduced Rbf/f myotubes (hypoxia), and Video 6 shows twitching RbΔf myotubes (hypoxia). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005067/DC1.
Acknowledgments
We thank Andrew T. Ho and Zhe Jiang for advice, Rod Bremner and Sean Egan for comments on the manuscript, Michael Ohh for hypoxia chamber access, and the following scientists for kindly providing adenovirus vectors: Marco Crescenzi, Robert G. Korneluk, David Murphy, Michael Ohh, David S. Park, Christopher J. Rhodes, Aviva M. Tolkovsky, Karen A. Vincent, and Kenneth Walsh.
This work was supported by a grant from the Canadian Institute for Health Research (to E. Zacksenhaus).
Footnotes
Abbreviations used in this paper:
- 3-MA
- 3-methyladenine
- DM
- differentiation medium
- HIF-1α
- hypoxia-inducible factor-1α
- MCK
- muscle creatine kinase
- mgRb
- mini-Rb gene
- MHC
- myosin heavy chain
- MOMP
- mitochondrial outer membrane permeabilization
- OXPHOS
- oxidative phosphorylation
- VHL
- von Hippel–Lindau tumor suppressor protein
- XIAP
- x-linked inhibitor of apoptosis
References
- Amthor H., Macharia R., Navarrete R., Schuelke M., Brown S.C., Otto A., Voit T., Muntoni F., Vrbóva G., Partridge T., et al. 2007. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc. Natl. Acad. Sci. USA. 104:1835–1840 10.1073/pnas.0604893104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampton E.T., Goemans C.G., Niranjan D., Mizushima N., Tolkovsky A.M. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 1:23–36 10.4161/auto.1.1.1495 [DOI] [PubMed] [Google Scholar]
- Bastin J., Aubey F., Rötig A., Munnich A., Djouadi F. 2008. Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J. Clin. Endocrinol. Metab. 93:1433–1441 10.1210/jc.2007-1701 [DOI] [PubMed] [Google Scholar]
- Benevolenskaya E.V., Murray H.L., Branton P., Young R.A., Kaelin W.G., Jr 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell. 18:623–635 10.1016/j.molcel.2005.05.012 [DOI] [PubMed] [Google Scholar]
- Bremner R., Zacksenhaus E. 2010. Cyclins, Cdks, E2f, Skp2, and more at the first international RB Tumor Suppressor Meeting. Cancer Res. 70:6114–6118 10.1158/0008-5472.CAN-10-0358 [DOI] [PubMed] [Google Scholar]
- Briaud I., Dickson L.M., Lingohr M.K., McCuaig J.F., Lawrence J.C., Rhodes C.J. 2005. Insulin receptor substrate-2 proteasomal degradation mediated by a mammalian target of rapamycin (mTOR)-induced negative feedback down-regulates protein kinase B-mediated signaling pathway in beta-cells. J. Biol. Chem. 280:2282–2293 10.1074/jbc.M412179200 [DOI] [PubMed] [Google Scholar]
- Burkhart D.L., Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 8:671–682 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.T., Wang J.Y. 2000. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol. Cell. Biol. 20:5571–5580 10.1128/MCB.20.15.5571-5580.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Livne-bar I., Vanderluit J.L., Slack R.S., Agochiya M., Bremner R. 2004. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 5:539–551 10.1016/j.ccr.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Chen H.Z., Tsai S.Y., Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 9:785–797 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V., Le A., Gao P. 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15:6479–6483 10.1158/1078-0432.CCR-09-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville M.I., Antoine I.V., Mertens-Strijthagen J.R., Dupriez V.J., Rousseau G.G. 1995. An E2F-dependent late-serum-response promoter in a gene that controls glycolysis. Oncogene. 11:1509–1517 [PubMed] [Google Scholar]
- Di Carlo A., De Mori R., Martelli F., Pompilio G., Capogrossi M.C., Germani A. 2004. Hypoxia inhibits myogenic differentiation through accelerated MyoD degradation. J. Biol. Chem. 279:16332–16338 10.1074/jbc.M313931200 [DOI] [PubMed] [Google Scholar]
- Gu W., Schneider J.W., Condorelli G., Kaushal S., Mahdavi V., Nadal-Ginard B. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 72:309–324 10.1016/0092-8674(93)90110-C [DOI] [PubMed] [Google Scholar]
- Guo Z., Yikang S., Yoshida H., Mak T.W., Zacksenhaus E. 2001. Inactivation of the retinoblastoma tumor suppressor induces apoptosis protease-activating factor-1 dependent and independent apoptotic pathways during embryogenesis. Cancer Res. 61:8395–8400 [PubMed] [Google Scholar]
- Hershko T., Ginsberg D. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279:8627–8634 10.1074/jbc.M312866200 [DOI] [PubMed] [Google Scholar]
- Ho A.T., Li Q.H., Hakem R., Mak T.W., Zacksenhaus E. 2004. Coupling of caspase-9 to Apaf1 in response to loss of pRb or cytotoxic drugs is cell-type-specific. EMBO J. 23:460–472 10.1038/sj.emboj.7600039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.T., Li Q.H., Okada H., Mak T.W., Zacksenhaus E. 2007. XIAP activity dictates Apaf-1 dependency for caspase 9 activation. Mol. Cell. Biol. 27:5673–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh M.S., Parker M.H., Scimè A., Parks R., Rudnicki M.A. 2004. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166:865–876 10.1083/jcb.200403004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zacksenhaus E. 2002. Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J. Cell Biol. 156:185–198 10.1083/jcb.200106084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Liang P., Leng R., Guo Z., Liu Y., Liu X., Bubnic S., Keating A., Murray D., Goss P.E., Zacksenhaus E. 2000. E2F1 and p53 are dispensable, whereas p21(Waf1/Cip1) cooperates with Rb to restrict endoreduplication and apoptosis during skeletal myogenesis. Dev. Biol. 227:8–41 10.1006/dbio.2000.9892 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Guo Z., Saad F.A., Ellis J., Zacksenhaus E. 2001. Retinoblastoma gene promoter directs transgene expression exclusively to the nervous system. J. Biol. Chem. 276:593–600 10.1074/jbc.M005474200 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Deng T., Jones R., Li H., Herschkowitz J.I., Liu J.C., Weigman V.J., Tsao M.S., Lane T.F., Perou C.M., Zacksenhaus E. 2010. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J. Clin. Invest. 120:3296–3309 10.1172/JCI41490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A., Noseda M., Beyna M., Yokota Y., Iavarone A. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 407:592–598 10.1038/35036504 [DOI] [PubMed] [Google Scholar]
- Leonard K.C., Petrin D., Coupland S.G., Baker A.N., Leonard B.C., LaCasse E.C., Hauswirth W.W., Korneluk R.G., Tsilfidis C. 2007. XIAP protection of photoreceptors in animal models of retinitis pigmentosa. PLoS One. 2:e314 10.1371/journal.pone.0000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maandag E.C., van der Valk M., Vlaar M., Feltkamp C., O’Brien J., van Roon M., van der Lugt N., Berns A., te Riele H. 1994. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 13:4260–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan W.R., Xiao G., Abdellatif M., Schneider M.D. 2000. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 20:8903–8915 10.1128/MCB.20.23.8903-8915.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. 2007. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6:458–471 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Mantela J., Jiang Z., Ylikoski J., Fritzsch B., Zacksenhaus E., Pirvola U. 2005. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development. 132:2377–2388 10.1242/dev.01834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard M.A., Ohh M. 2007. The role of hypoxia-inducible factors in cancer. Cell. Mol. Life Sci. 64:2170–2180 10.1007/s00018-007-7082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2009. Physiological functions of autophagy. Curr. Top. Microbiol. Immunol. 335:71–84 10.1007/978-3-642-00302-8_3 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. 2007. How to interpret LC3 immunoblotting. Autophagy. 3:542–545 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. 2010. Methods in mammalian autophagy research. Cell. 140:313–326 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E., Galluzzi L., Kepp O., Vicencio J.M., Criollo A., Maiuri M.C., Kroemer G. 2009. Anti- and pro-tumor functions of autophagy. Biochim. Biophys. Acta. 1793:1524–1532 10.1016/j.bbamcr.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Nahle Z., Polakoff J., Davuluri R.V., McCurrach M.E., Jacobson M.D., Narita M., Zhang M.Q., Lazebnik Y., Bar-Sagi D., Lowe S.W. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859–864 10.1038/ncb868 [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch B.G., Spicer D.B., Kim P.S., Cheung W.L., Lassar A.B. 1999. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9:449–459 10.1016/S0960-9822(99)80210-3 [DOI] [PubMed] [Google Scholar]
- Pattingre S., Levine B. 2006. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 66:2885–2888 10.1158/0008-5472.CAN-05-4412 [DOI] [PubMed] [Google Scholar]
- Pelicano H., Martin D.S., Xu R.H., Huang P. 2006. Glycolysis inhibition for anticancer treatment. Oncogene. 25:4633–4646 10.1038/sj.onc.1209597 [DOI] [PubMed] [Google Scholar]
- Polager S., Ofir M., Ginsberg D. 2008. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 27:4860–4864 10.1038/onc.2008.117 [DOI] [PubMed] [Google Scholar]
- Puri P.L., Iezzi S., Stiegler P., Chen T.T., Schiltz R.L., Muscat G.E., Giordano A., Kedes L., Wang J.Y., Sartorelli V. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell. 8:885–897 10.1016/S1097-2765(01)00373-2 [DOI] [PubMed] [Google Scholar]
- Sankaran V.G., Orkin S.H., Walkley C.R. 2008. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 22:463–475 10.1101/gad.1627208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J.W., Gu W., Zhu L., Mahdavi V., Nadal-Ginard B. 1994. Reversal of terminal differentiation mediated by p107 in Rb-/- muscle cells. Science. 264:1467–1471 10.1126/science.8197461 [DOI] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6:1221–1228 10.1038/ncb1192 [DOI] [PubMed] [Google Scholar]
- Skurk C., Izumiya Y., Maatz H., Razeghi P., Shiojima I., Sandri M., Sato K., Zeng L., Schiekofer S., Pimentel D., et al. 2005. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J. Biol. Chem. 280:20814–20823 10.1074/jbc.M500528200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sufan R.I., Moriyama E.H., Mariampillai A., Roche O., Evans A.J., Alajez N.M., Vitkin I.A., Yang V.X., Liu F.F., Wilson B.C., Ohh M. 2009. Oxygen-independent degradation of HIF-alpha via bioengineered VHL tumour suppressor complex. EMBO Mol Med. 1:66–78 10.1002/emmm.200900004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky A.M. 2009. Mitophagy. Biochim. Biophys. Acta. 1793:1508–1515 10.1016/j.bbamcr.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Tracy K., Dibling B.C., Spike B.T., Knabb J.R., Schumacker P., Macleod K.F. 2007. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 27:6229–6242 10.1128/MCB.02246-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincheri N.F., Follo C., Nicotra G., Peracchio C., Castino R., Isidoro C. 2008. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis. 29:381–389 10.1093/carcin/bgm271 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324:1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent K.A., Shyu K.G., Luo Y., Magner M., Tio R.A., Jiang C., Goldberg M.A., Akita G.Y., Gregory R.J., Isner J.M. 2000. Angiogenesis is induced in a rabbit model of hindlimb ischemia by naked DNA encoding an HIF-1alpha/VP16 hybrid transcription factor. Circulation. 102:2255–2261 [DOI] [PubMed] [Google Scholar]
- Vooijs M., te Riele H., van der Valk M., Berns A. 2002. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in interphotoreceptor retinol binding protein-expressing cells. Oncogene. 21:4635–4645 10.1038/sj.onc.1205575 [DOI] [PubMed] [Google Scholar]
- Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. 2008. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 8:249–256 10.1016/j.cmet.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Williams B.O., Schmitt E.M., Remington L., Bronson R.T., Albert D.M., Weinberg R.A., Jacks T. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Fletcher G.C., Tolkovsky A.M. 2001. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 11:361–365 10.1016/S0960-9822(01)00100-2 [DOI] [PubMed] [Google Scholar]
- Zacksenhaus E., Jiang Z., Chung D., Marth J.D., Phillips R.A., Gallie B.L. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051–3064 10.1101/gad.10.23.3051 [DOI] [PubMed] [Google Scholar]
- Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6:472–483 10.1016/j.cmet.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Zhu S., Stavrovskaya I.G., Drozda M., Kim B.Y., Ona V., Li M., Sarang S., Liu A.S., Hartley D.M., Wu D.C., et al. 2002. Minocycline inhibits cytochrome c release and delays progression of amyotrophic lateral sclerosis in mice. Nature. 417:74–78 10.1038/417074a [DOI] [PubMed] [Google Scholar]