Low expression levels of Cdc20 result in chromatin bridging and chromosome misalignment, revealing a requirement for Cdc20 in efficient sister chromosome separation and chromosome–microtubule attachment.
Abstract
Cdc20 is an activator of the anaphase-promoting complex/cyclosome that initiates anaphase onset by ordering the destruction of cyclin B1 and securin in metaphase. To study the physiological significance of Cdc20 in higher eukaryotes, we generated hypomorphic mice that express small amounts of this essential cell cycle regulator. In this study, we show that these mice are healthy and not prone to cancer despite substantial aneuploidy. Cdc20 hypomorphism causes chromatin bridging and chromosome misalignment, revealing a requirement for Cdc20 in efficient sister chromosome separation and chromosome–microtubule attachment. We find that cyclin B1 is newly synthesized during mitosis via cytoplasmic polyadenylation element–binding protein-dependent translation, causing its rapid accumulation between prometaphase and metaphase of Cdc20 hypomorphic cells. Anaphase onset is significantly delayed in Cdc20 hypomorphic cells but not when translation is inhibited during mitosis. These data reveal that Cdc20 is particularly rate limiting for cyclin B1 destruction because of regulated de novo synthesis of this cyclin after prometaphase onset.
Introduction
Orderly progression through M phase requires appropriately timed inactivation of mitotic regulatory proteins by ubiquitin-dependent proteolysis, with the anaphase-promoting complex/cyclosome (APC/C) serving as the E3 ligase (Peters, 2006). The most critical function of the APC/C is to initiate sister chromatid separation at the metaphase to anaphase transition. It does so by ordering the destruction of two key anaphase inhibitors, securin and cyclin B1 (Sullivan and Morgan, 2007). Earlier in mitosis, immediately after initiation of nuclear envelope breakdown (NEBD), APC/C mediates the destruction of cyclin A2 and Nek2a (Geley et al., 2001; Hames et al., 2001), although it is not fully clear why these regulators need to be destroyed in prometaphase. After anaphase onset, APC/C remains active, mediating the degradation of various proteins that regulate mitosis or cytokinesis, including Plk1, aurora A, and aurora B (Pines, 2006). APC/C has two activating subunits, Cdc20 and Cdh1, which function in substrate recruitment and are thought to induce changes in APC/C conformation that increase activity (Peters, 2006). Cdc20 is APC/C’s early coactivator responsible for driving metaphase cells into anaphase (Yu, 2007). Cdh1 is kept inactive during the early stages of mitosis by Cdk1-dependent phosphorylation (Zachariae et al., 1998) but becomes active when Cdk1 activity subsides after APC/CCdc20-mediated destruction of cyclin B1 in late metaphase. Cdh1-activated APC/C targets Cdc20 for destruction in anaphase, allowing Cdh1 to become the sole APC/C activator during late mitosis and cytokinesis.
Timing of cyclin B1 and securin ubiquitination by APC/C is controlled by the mitotic checkpoint, a surveillance network that functions to promote accurate chromosome separation by delaying anaphase onset until proper spindle attachment of all mitotic chromosomes has been achieved (Musacchio and Salmon, 2007; for review see Holland and Cleveland, 2009). At mitosis onset, core mitotic checkpoint proteins, including members of the Bub and Mad protein families, start to accumulate at kinetochores of unattached chromosomes to generate a diffusible signal that acts to prevent polyubiquitination of securin and cyclin B1 by APC/C. It is assumed that the inhibitory signal consists of Mad2 that has been primed by kinetochore-associated Mad1–Mad2 to stably interact with the APC/C-activating subunit Cdc20 (Yu, 2006; Musacchio and Salmon, 2007; Kulukian et al., 2009). Mad2-bound Cdc20 is then loaded onto soluble BubR1–Bub3 complexes, which presumably present Cdc20 as a substrate to the APC/C, targeting it for degradation (Nilsson et al., 2008). Upon attachment of the last chromosome pair, the inhibitory signal is quenched and APC/C is activated by Cdc20, triggering the destruction of cyclin B1 and securin. Separase, a protease that is held in an inactive state by securin binding and cyclin B1/Cdk1–dependent phosphorylation, cleaves the Scc1 subunit of the cohesin complex that holds sister chromatids together, inducing the physical separation of sister chromatids by spindle forces (Nasmyth and Haering, 2005; Peters et al., 2008).
Although previous studies have firmly established that Cdc20 is an essential activator of sister chromatid separation at metaphase to anaphase transition in mammals (Li et al., 2007; Wolthuis et al., 2008), the full range of Cdc20 functions is unclear at present. In addition to being localized in the mitotic cytosol, Cdc20 associates with centrosomes throughout the cell cycle and accumulates at unattached kinetochores early in mitosis (Kallio et al., 2002). However, little is known about the functional significance of centrosome- and kinetochore-associated Cdc20 in mitosis. Interestingly, centrosome-associated Cdc20 has recently been implicated in dendrite morphogenesis of postmitotic neurons, presumably by triggering the polyubiquitination and subsequent degradation of Id1, a centrosome-associated protein that is known to inhibit dendrite growth (Kim et al., 2009). The subsequent discovery that neurons also require Cdc20 for the formation of presynaptic sites (Yang et al., 2009) underscores the hypothesis that Cdc20 has important functions outside of mitosis. However, whether Cdc20 depletion is associated with any neuropathology remains an open question. It is also unknown whether Cdc20 dysfunction leads to inaccurate chromosome segregation and aneuploidy, and if so, whether this would increase the risk for neoplastic transformation.
In-depth analysis of the Cdc20 functions in mitosis, normal development, and oncogenesis has been precluded by early embryonic lethality of Cdc20 knockout mice (Li et al., 2008). We bypassed this problem by generating mutant mouse strains in which the dose of Cdc20 is reduced in graded fashion. Using mice and mouse embryonic fibroblasts (MEFs) with low amounts of Cdc20, we have obtained important new insights into the in vivo functions of Cdc20 and the regulation of cyclin B1 expression in mitosis.
Results
Generation of mice with graded reduction in Cdc20
Null mice for the Cdc20 gene die as two–cell stage embryos because of failed anaphase onset (Li et al., 2007). To further explore the physiological significance of Cdc20, we generated a series of mutant mouse strains in which expression of Cdc20 is gradually reduced by using various combinations of wild-type (Cdc20+), hypomorphic (Cdc20H), and knockout (Cdc20−) alleles. The Cdc20H allele was produced by targeted insertion of a neomycin resistance cassette into the third intron of the Cdc20 gene (Fig. 1, A–C). The Cdc20− allele was obtained from gene trap mouse embryonic stem (ES) cell clone XE368 (Fig. 1, B and D). It has been shown that this gene trap allele is the equivalent of a null allele (Li et al., 2007). In contrast to Cdc20−/− mice, Cdc20+/H, Cdc20+/−, Cdc20H/H, and Cdc20−/H mice were born alive and were overtly indistinguishable from their Cdc20+/+ counterparts. Western blot analysis demonstrated that Cdc20+/H, Cdc20+/−, Cdc20H/H, and Cdc20−/H MEFs had a graded reduction of Cdc20 protein (Fig. 1 E). Similar gradations of Cdc20 reduction were observed in the testis and ovary (not depicted). Western blot analysis of extracts from thymus, liver, lung, and lung fibroblasts of Cdc20+/+ and Cdc20−/H mice further indicated that the observed Cdc20 protein reductions are generalized (Fig. 1 F).
Figure 1.
Generation of mice with graded reduction in Cdc20 dosage. (A) Cdc20 gene targeting strategy. Part of the Cdc20 locus (+), the targeting vector, the hypomorphic allele (Cdc20H), EcoR1 restriction sites, and the Southern probe are indicated. (B) Cdc20− allele from gene trap ES cell clone XE368. (C) Southern blot analysis of mice with the indicated Cdc20 genotypes. (D) PCR-based genotype analysis of Cdc20 mutant mice. Positions of PCR primers (a–e) are indicated in A and B. (E) Western blot analysis of MEF extracts for Cdc20. Cdc20 protein levels in MEFs of Cdc20−/H mice were ∼15–20% of normal. (F) Western blot analysis of extracts of the indicated tissues and cell types for Cdc20. Actin served as a loading control.
Graded Cdc20 reduction causes progressive aneuploidy
Previous studies have documented that complete (Li et al., 2007) or near-complete (<5% of normal; Wolthuis et al., 2008) depletion of Cdc20 precludes chromosome separation and anaphase onset. At less-profound reductions of Cdc20 expression (∼10% of normal Cdc20), cultured cells can execute anaphase and complete mitosis (Wolthuis et al., 2008). However, whether subnormal Cdc20 levels affect the accuracy of the chromosome separation process has not been investigated. To address this key question, we performed chromosome counts on metaphase spreads of Cdc20+/+, Cdc20+/−, Cdc20H/H, and Cdc20−/H MEFs. We found that 12% of Cdc20+/+ MEFs were aneuploid (Table I). In contrast, Cdc20+/−, Cdc20H/H, and Cdc20−/H MEFs had 17%, 23%, and 27% aneuploidy, respectively, uncovering an inverse correlation between the level of Cdc20 protein and the incidence of numerical chromosome abnormalities in these cells. In addition, we observed modest yet statistically significant increases in premature sister chromatid separation (PMSCS) at all levels of Cdc20 reduction, with Cdc20+/−, Cdc20H/H, and Cdc20−/H MEFs showing PMSCS in 10%, 11%, and 12% of mitotic figures, respectively, compared with 3% in Cdc20+/+ mitotic figures (Table I). Differences among Cdc20+/−, Cdc20H/H, and Cdc20−/H MEFs were marginal, suggesting that there is no clear relationship between PMSCS and the level of Cdc20 reduction in MEFs. MG132 treatment of Cdc20−/H MEFs prevented PMSCS (Table I), indicating that the PMSCS phenotype is unlikely to be caused by a defect in Sgo1-mediated protection of centromeric cohesion.
Table I.
Graded reduction of Cdc20 in MEFs induces progressive aneuploidy
| Mitotic MEF genotypea | Aneuploid figs. | Karyotypes w/ indicated chromosome No. | Mitotic figs. w/ PMSCS | Mitotic figs. w/ PMSCS in MG132 | ||||||
| 37 | 38 | 39 | 40 | 41 | 42 | 43 | ||||
| % | % | |||||||||
| +/+ | 12 (2) | 1 | 2 | 5 | 132 | 7 | 3 | NA | 3 (1) | 0 (0) |
| Cdc20+/− | 17 (1) | 1 | NA | 11 | 124 | 10 | 4 | NA | 10 (1) | NA |
| Cdc20H/H | 23 (2) | NA | 5 | 9 | 115 | 18 | 3 | NA | 11 (2) | NA |
| Cdc20−/H | 27 (1) | 3 | 4 | 9 | 110 | 14 | 7 | 3 | 12 (1) | 0 (0) |
150 mitotic figures were inspected. NA, not applicable. Numbers in parentheses indicate SD. Karyotyping was performed at passage 5. MG132 treatment was for 2 h in 10 µM of the drug.
n = 3.
Next, we performed chromosome counts on splenocytes of Cdc20+/+, Cdc20+/−, Cdc20H/H, and Cdc20−/H mice at 5 mo of age. These splenocytes were cultured only 5 h so that detected chromosome number aberrations represent in vivo–acquired aneuploidies. Although Cdc20+/+ splenocytes had 40 chromosomes, subnormal Cdc20 expression caused substantial aneuploidization, with Cdc20+/−, Cdc20H/H, and Cdc20−/H showing 15%, 17%, and 21% aneuploidy, respectively (Table II). The incidence of PMSCS in mutant splenocytes was modestly increased (Table II), but again, there was no clear correlation between PMSCS and Cdc20 dosage. Chromosome counts on cultured primary lung fibroblasts from Cdc20+/+ (8% aneuploidy) and Cdc20−/H mice (19% aneuploidy) further suggested that aneuploidy is a common consequence of Cdc20 hypomorphism, irrespective of cell type.
Table II.
Progressive reduction of Cdc20 in splenocytes leads to increasing aneuploidy
| Mitotic MEF genotype | Age | Aneuploid figures | Karyotypes w/ indicated chromosome No. | Mitotic figures w/ PMSCS | |||||
| 37 | 38 | 39 | 40 | 41 | 42 | ||||
| % | % | ||||||||
| +/+ | 5 mo (3) | 0 (0) | NA | NA | NA | 150 | NA | NA | 0 (0) |
| Cdc20+/− | 5 mo (3) | 15 (1) | 1 | 5 | 5 | 124 | 10 | 5 | 7 (1) |
| Cdc20H/H | 5 mo (3) | 17 (1) | NA | 3 | 8 | 125 | 7 | 7 | 10 (1) |
| Cdc20−/H | 5 mo (3) | 21 (1) | NA | 4 | 9 | 119 | 10 | 8 | 11 (1) |
150 mitotic figures were inspected. NA, not applicable. Numbers in parentheses indicate SD.
Cdc20 loss leads to chromosome misalignment and chromatin bridging
To explore how Cdc20 deficiency creates aneuploidy, we monitored the chromosome movements of Cdc20+/+ and Cdc20−/H MEFs undergoing mitosis using time-lapse live cell microscopy. Chromosomes were visualized by transduction of cells with a lentivirus encoding YFP-H2B. Our analysis revealed three types of errors: chromosome misalignment, chromosome lagging, and chromatin bridge formation (Fig. 2, A–C). Of these, chromosome lagging occurred at similar rates in Cdc20−/H and Cdc20+/+ MEFs, indicating that this defect is unlikely to be caused by Cdc20 depletion. The other two defects, however, were substantially increased at low Cdc20 levels, with 9% of Cdc20−/H MEFs showing misalignment (vs. 1% of Cdc20+/+ MEFs) and 10% having bridges (vs. 0% of Cdc20+/+ MEFs). Most cells had either a single misaligned chromosome (Fig. 2 B) or a single chromatin bridge (Fig. 2 C).
Figure 2.
Cdc20 is required for proper chromosome alignment and segregation. (A) Analysis of chromosome segregation errors in Cdc20+/+ and Cdc20−/H MEFs by live cell imaging. (B and C) Live cell images of H2B-YFP–positive Cdc20−/H MEFs showing chromosome misalignment (B) and bridging (C) during unchallenged mitosis. (D and E) Live cell images of 10 µM MG132-treated H2B-YFP–positive Cdc20−/H MEFs showing minor (D) and major chromosome misalignments. (B–E) Misaligned chromosomes are marked by arrows, and chromatin bridges are marked by arrowheads. (F) Quantification of MEFs with misaligned chromosomes in MG132. Shaded and nonshaded areas denote cells with minor (≤2) and major (≥3) chromosomal misalignments, respectively. Error bars represent SD. (G) Images of MG132-treated Cdc20+/+ and Cdc20−/H MEFs immunostained for tubulin (green), kinetochores (red), and DNA (blue). Insets highlight MT–kinetochore interactions. (H) Analysis of MT–kinetochore attachments in the cold (green, MTs; red, kinetochores). Insets show details of cold stable or cold unstable MT–kinetochore interactions (the inset of the Cdc20−/H cells includes Hoechst staining to visualize sister chromatids). (I) Analysis of the distance between sister kinetochores in Cdc20+/+ and Cdc20−/H MEFs. Kinetochores were visualized by ACA immunostaining. Cells were costained for MTs (α-tubulin) and DNA (Hoechst) to identify bioriented and misaligned chromosomes. Bars, 10 µm.
Resolution of chromatin bridges might cause structural chromosome damage such as chromosome gaps and breaks (Sotillo et al., 2007), but screening of Giemsa-stained metaphase spreads of Cdc20−/H MEFs and splenocytes revealed no evidence for such damage (unpublished data). Formation of chromatin bridges has been associated with aberrant inner centromere targeting of Topo IIα during the early stages of mitosis (Dawlaty et al., 2008), but no such defect was observed in Cdc20−/H MEFs (unpublished data).
To further characterize the chromosome misalignment phenotype, we performed live cell imaging on Cdc20−/H MEFs in the presence of MG132, a proteasome inhibitor that extends metaphase by preventing cyclin B1 and securin degradation (Lee and Goldberg, 1998). Instead of improving the misalignment phenotype by allowing more time for establishment of kinetochore–microtubule (MT) attachment, MG132 exacerbated it as the incidence of congression failure, and the number of misaligned chromosomes were substantially higher in treated than nontreated Cdc20−/H MEFs (Fig. 2, D–F). Immunostaining of Cdc20−/H MEFs for tubulin and centromeric proteins revealed that misaligned chromosomes consist of duplicated sister chromatids captured by MTs emanating from a single spindle pole (Fig. 2 G). At a low temperature, these kinetochore MT fibers (K-fibers) depolymerized, indicating that MT–kinetochore attachments at misaligned chromosomes were unstable (Fig. 2 H). In contrast, aligned chromosomes of Cdc20−/H MEFs had K-fiber attachments that were stable in the cold. Intersister kinetochore distances of misaligned chromosomes of Cdc20−/H MEFs were greatly reduced compared with those of bioriented chromosome pairs of Cdc20+/+ MEFs, a finding that is consistent with monotelic or syntelic attachment (Fig. 2 I). In contrast, aligned chromosomes of Cdc20−/H MEFs had normal inter sister kinetochore distances, indicating that they were under proper tension. Collectively, the presence of misaligned chromosomes with monopolar attachments even after prolonged metaphase arrest suggests a requirement for Cdc20 in efficient kinetochore–MT attachment.
Accumulation of Cdc20 at unattached kinetochores is reduced in Cdc20−/H MEFs
Cdc20 is present in the mitotic cytosol, where it serves as an activator of APC/C, but it also localizes at kinetochores where its function is unknown (Kallio et al., 2002). Consistent with earlier findings in human cell lines (Kallio et al., 2002), high levels of Cdc20 accumulated at kinetochores of Cdc20+/+ MEFs during the early stages of mitosis (Fig. 3 A). In contrast, most kinetochores of Cdc20−/H MEFs showed greatly reduced Cdc20 immunostaining at corresponding stages of mitosis. This reduction does not seem to be part of a more generalized kinetochore assembly problem because Cdc20−/H MEFs accumulated normal amounts of Bub1, BubR1, Mad1, Mad2, and centromere protein E (CENP-E) at kinetochores (Fig. S1; and not depicted). Furthermore, aurora B and MCAK, two key components of a mechanism that resolves aberrant chromosome attachments, properly localized to the inner centromeric regions of Cdc20−/H metaphases (unpublished data). We noticed that kinetochores of chromosomes that were not properly aligned in the spindle equator of Cdc20+/+ metaphases accumulated higher levels of Cdc20 than kinetochores of properly bioriented chromosomes (Fig. 3 B). Kinetochores of unaligned chromosomes of Cdc20−/H MEFs also stained more intensely than kinetochores of aligned chromosomes but were less bright than their counterparts in Cdc20+/+ MEFs. These data raise the intriguing possibility that Cdc20 exerts its influence on efficient MT–kinetochore capture through its kinetochore-associated fraction.
Figure 3.
Unattached kinetochores of hypomorphic MEFs have low Cdc20 levels. (A) Kinetochore localization of Cdc20 during early mitotic stages of Cdc20+/+ and Cdc20−/H MEFs. P, prophase; PM, prometaphase; M, metaphase. (B) Images of Cdc20+/+ and Cdc20−/H metaphases with misaligned chromosomes (arrows) stained for Cdc20 and centromeres. Arrowheads mark kinetochores with high Cdc20 levels that are out of focus. Bars, 10 µm.
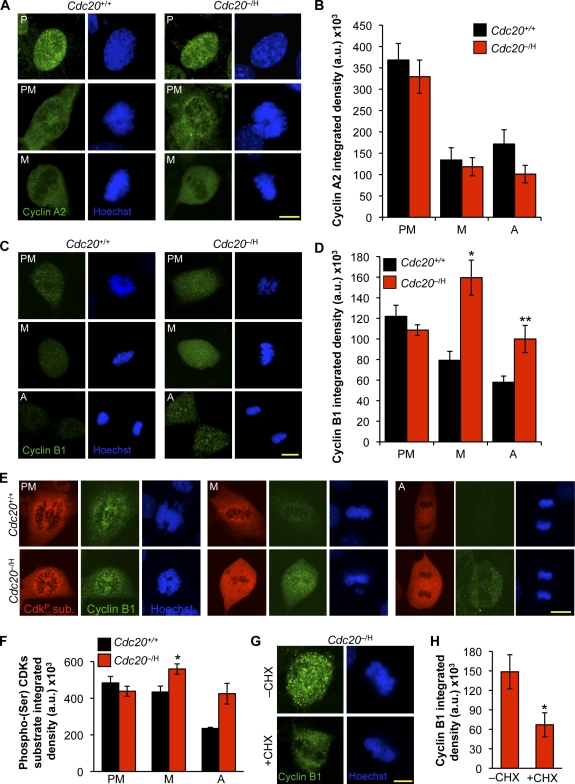
Cyclin B1 levels of Cdc20 hypomorphic cells rise in metaphase
Another possibility is that reduced levels of Cdc20 in the mitotic cytosol result in low APC/C activity, thereby perhaps stabilizing key mitotic regulators whose timely destruction might be required for accurate chromosome separation. To explore this scenario, we measured endogenous cyclin A2 and B1 levels of Cdc20+/+ and Cdc20−/H MEFs at various stages of mitosis by indirect immunofluorescence. Consistent with earlier work (van Ree et al., 2010), cyclin A2 was present at high levels in prophase of Cdc20+/+ MEFs and promptly degraded as cells progressed to prometaphase (Fig. 4, A and B). The exact same degradation pattern was observed in Cdc20−/H MEFs, indicating that Cdc20 levels of these cells meet the threshold for efficient destruction of cyclin A2.
Figure 4.
Cyclin B1 levels spike during metaphase of Cdc20 hypomorphic cells. (A) Immunostaining of Cdc20+/+ and Cdc20−/H prophases (P), prometaphases (PM), and metaphases (M) for cyclin A2. (B) Quantification of cyclin A2 signals in A (n = 26 Cdc20+/+ prophases, 10 Cdc20+/+ prometaphases, and 5 Cdc20+/+ metaphases; n = 23 Cdc20−/H prophases, 8 Cdc20−/H prometaphases, and 5 Cdc20−/H metaphases). Error bars represent SD. (C) MEFs at the indicated mitotic stages (A, anaphase) stained for cyclin B1. (D) Quantification of cyclin B1 signals (n = 11–15 cells/stage). Error bars represent SEM. *, P < 0.003 versus wild-type (unpaired t test); **, P < 0.01 versus wild-type (unpaired t test). (E) MEFs at the indicated mitotic stages stained for cyclin B1 and phosphoserine Cdk substrates. (F) Quantification of phosphoserine Cdk substrate signals. *, P < 0.05 versus Cdc20−/H prometaphase (unpaired t test). Error bars represent SEM (n = 8 cells for each group). (G) Cdc20−/H metaphases stained for cyclin B1 after treatment with 1 µg/ml CHX for 10 min. (H) Quantification of cyclin B1 signals (n = 10 cells). *, P < 0.0001 versus nontreated (unpaired t test). Error bars represent SEM. Bars, 10 µm.
As expected, cyclin B1 levels gradually declined from prometaphase to anaphase in Cdc20+/+ MEFs (Fig. 4 B). In contrast, instead of decreasing from prometaphase to metaphase, cyclin B1 levels markedly increased in Cdc20−/H MEFs (Fig. 4, C and D). Furthermore, when Cdc20−/H MEFs entered anaphase, they did so with elevated levels of cyclin B1 compared with Cdc20+/+ MEFs. These data suggest that Cdc20 seems to be rate limiting for timely destruction of cyclin B1. By immunofluorescence, Cdc20−/H metaphases and anaphases showed elevated levels of phosphorylated Cdk substrates (Fig. 4, E and F), suggesting that the rise in cyclin B1 expression resulted in increased cyclin B1–Cdk1 activity. Furthermore, these results imply that residual Cdk1 activity is not a barrier for anaphase entry.
Next, we examined whether securin destruction was also delayed in Cdc20−/H MEFs. Because endogenous securin is not detectable in Cdc20+/+ and Cdc20−/H MEFs by immunostaining (unpublished data), we decided instead to monitor the destruction of ectopically expressed securin-EYFP fusion protein by live cell microscopy. Unlike cyclin B1, securin-EYFP levels did not rise in mitotic Cdc20−/H MEFs (Fig. S2). However, although Cdc20−/H MEFs had quite normal rates of securin-EYFP destruction, the onset of degradation was substantially delayed compared with Cdc20+/+ MEFs, indicating that there is a critical Cdc20 threshold for proper timing of securin degradation.
Cyclin B1 is de novo synthesized in mitosis
The rise in cyclin B1 levels from prometaphase to metaphase in Cdc20−/H MEFs was intriguing, which prompted us to further explore the underlying mechanism. To test whether the increase was dependent on de novo protein synthesis, Cdc20+/+ and Cdc20−/H MEFs were treated with cycloheximide (CHX) for 10 min and immediately fixed and stained for cyclin B1. Although inhibition of protein biosynthesis had limited impact on cyclin B1 protein levels in Cdc20+/+ metaphases, it prevented the increase in cyclin B1 that is characteristic for Cdc20−/H metaphases (Fig. 4, G and H). Earlier work in HeLa cells suggests that the cyclin B1 gene promoter is transcriptionally active in mitosis (Sciortino et al., 2001), but actinomycin D treatment failed to have an impact on de novo synthesis of cyclin B1 in Cdc20−/H metaphases (Fig. S3, A and B). Together, these results suggested that Cdc20−/H MEFs produce cyclin B1 in mitosis via a mechanism that involves translation of preexisting cyclin B1 transcripts.
To evaluate whether cyclin B1 synthesis in mitosis is also a feature of normal cells, we treated wild-type MEFs with nocodazole for 15 min before fixation for cyclin B1 staining. As shown in Fig. 5 (A and B), nocodazole-treated cells showed significantly higher cyclin B1 immunofluorescence than untreated prophases, prometaphases, and metaphases, implying that MT depolymerization leads to increased cyclin B1 expression. This increase was significantly blunted when cells were treated simultaneously with nocodazole and CHX, indicating that it involved de novo cyclin B1 synthesis. To complement these staining experiments with biochemical data, we collected HeLa cells by mitotic shake off (without nocodazole) and cultured the harvested mitotic cells for 15 min in medium containing either nocodazole alone or both nocodazole and CHX, after which we prepared cell extracts and performed Western blot analyses. As shown in Fig. 5 C, CHX-treated cells indeed had lower cyclin B1 levels than their nontreated counterparts. Cdc20, another protein that is de novo synthesized during mitosis (Nilsson et al., 2008), was also lower, whereas Mad2, cyclin A2, and securin were not. Consistent with cyclin B1 protein synthesis during normal mitosis, Cdc20+/+ metaphases that had been exposed to CHX for 15 min exhibited significantly lower cyclin B1 immunofluorescence than untreated counterparts (Fig. 5, D and E). Very similar results were obtained for HeLa cells (Fig. 5, F and G).
Figure 5.
Cyclin B1 is de novo synthesized during normal mitosis. (A) Cdc20+/+ mitotic MEFs increase cyclin B1 levels in response to MT depolymerization but not when protein synthesis is blocked with CHX. Cells were either untreated or treated with 100 ng/ml nocodazole (Noc) or 100 ng/ml nocodazole + 1 µg/ml CHX for 15 min. (B) Quantification of cyclin B1 signals (n > 10 cells per group). *, P < 0.02 versus prophase (P), prometaphase (PM), and metaphase (M); **, P < 0.001 versus nocodazole + CHX-treated cells (by unpaired t test). (C) Extracts of mitotic HeLa cells treated for 15 min with 100 ng/ml nocodazole or 100 ng/ml nocodazole and 1 µg/ml CHX analyzed for the indicated proteins by Western blotting. (D) Cdc20+/+ metaphases in which de novo protein synthesis is blocked by CHX show decreased cyclin B1 immunofluorescence. Cells were treated with 1 µg/ml CHX 15 min before fixation. (E) Quantification of endogenous cyclin B1 signals (n = 10 Cdc20+/+ MEFs/group). *, P = 0.0366 versus untreated cells (by unpaired t test). (F) As in D but shown for HeLa cells. (G) Quantification of endogenous cyclin B1 signals (n = 10 HeLa cells/group). *, P = 0.0005 versus untreated cells (by unpaired t test). Error bars represent SD. Bars, 10 µm.
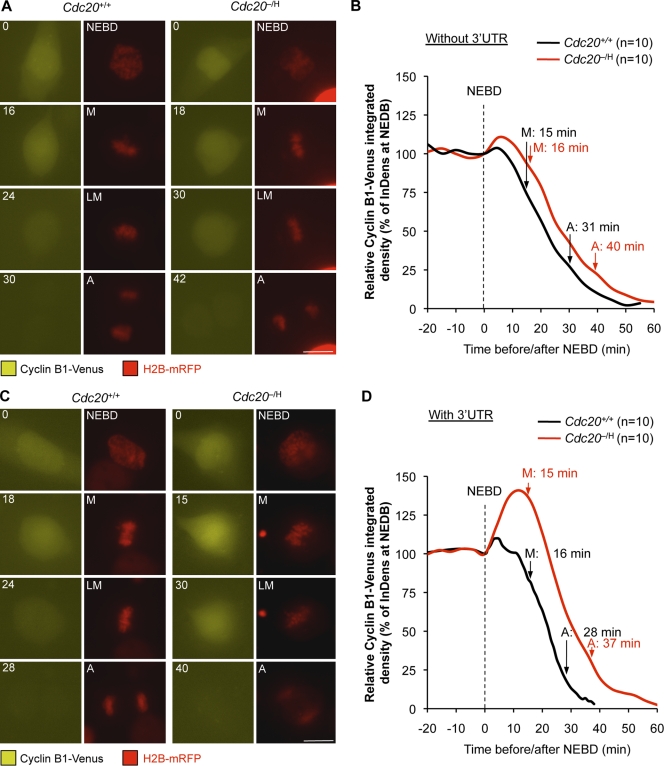
The 3′ untranslated region (UTR) of cyclin B1 regulates de novo synthesis in mitosis
To obtain additional evidence for de novo synthesis of cyclin B1 in mitosis, we followed cyclin B1–Venus protein levels in living Cdc20−/H and Cdc20+/+ MEFs using time-lapse microscopy. In Cdc20+/+ MEFs, cyclin B1–Venus levels were quite stable until ∼6 min after NEBD, after which they rapidly declined (Fig. 6, A and B). Surprisingly, Cdc20−/H MEFs did not show the expected dramatic incline in cyclin B1–Venus in the period after NEBD, although a small, statistically insignificant increase was noticeable. The onset of cyclin B1–Venus degradation, however, was noticeably delayed.
Figure 6.
Cyclin B1 synthesis in mitosis is mediated by the cyclin B1 3′UTR. (A) Time-lapse images of cyclin B1–Venus and H2B-mRFP expressing Cdc20+/+ and Cdc20−/H MEF progression through mitosis. The cyclin B1–Venus expression construct did not include the cyclin B1 3′UTR. (B) Cyclin B1–Venus fluorescence of Cdc20+/+ and Cdc20−/H MEFs was quantitated and plotted against the time before and after NEBD. (C and D) Same as in A and B, but in these experiments, the cyclin B1–Venus expression construct included the cyclin B1 3′UTR. M, metaphase; LM, late metaphase; A, anaphase. Bars, 10 µm.
We reasoned that our cyclin B1–Venus expression plasmid might lack critical cyclin B1 elements for expression of the protein in M phase. In frogs, the first few embryonic cell divisions after fertilization occur in the absence of gene transcription and are dependent on translation of selective oocyte-derived mRNAs, including cyclin B1 mRNA (Groisman et al., 2002; Richter, 2007). Selective translation of cyclin B1 and other transcripts is mediated by cytoplasmic polyadenylation elements (CPEs) in the 3′UTRs of these mRNAs, which bind CPE-binding protein (CPEB). As fertilized embryos enter M phase, CPEB is activated by aurora A phosphorylation at Ser174, which allows for poly(A) tail extension and translation initiation (Groisman et al., 2002; Richter, 2007). We wondered whether de novo synthesis of cyclin B1 during nonembryonic cell divisions might also be mediated by CPEB-dependent translation initiation. If so, the lack of cyclin B1 3′UTR sequences in our expression plasmid might explain why Cdc20−/H MEFs failed to show a marked increase in cyclin B1–Venus in mitosis.
To test this hypothesis, we cloned the mouse cyclin B1 3′UTR (Fig. S3 C) downstream of the cyclin B1–Venus coding region and repeated the aforementioned experiment. As illustrated in Fig. 6 (C and D), immediately after NEBD, cyclin B1–Venus started to accumulate profoundly in Cdc20−/H MEFs, reaching peak levels ∼12 min later. Cdc20+/+ MEFs also showed increased cyclin B1–Venus levels after NEBD, but this increase was very small compared with that of Cdc20−/H MEFs. Anaphase was preceded by a period of rapid cyclin B1–Venus proteolysis in both Cdc20−/H and Cdc20+/+ MEFs. However, the initiation of cyclin B1–Venus destruction was considerably delayed in Cdc20 hypomorphic cells. Importantly, when the cyclin B1 3′UTR was placed in the antisense orientation, Cdc20−/H MEFs failed to accumulate cyclin B1–Venus in mitosis (unpublished data). Collectively, these data strongly suggest that de novo synthesis of cyclin B1 during mitosis is driven by CPEB-dependent transcriptional control.
Cdc20 hypomorphism delays anaphase onset
The finding that the onset cyclin B1–Venus degradation was consistently delayed in Cdc20–/H MEFs suggested that the timing of mitosis was altered in these cells. However, it cannot be excluded that the cyclin B1–Venus overexpression in these experiments is at least in part responsible for the delay. To assess the true impact of Cdc20 hypomorphism on mitotic timing, we followed mRFP-H2B–positive Cdc20+/+ and Cdc20−/H MEFs through mitosis and calculated the duration of each mitotic stage. Cells showing aberrant chromosome segregation were excluded from the analysis, as altered mitotic timing in these cells might simply be caused by prolonged mitotic checkpoint activation. We found that prophase, defined in this study as the period between chromosome condensation and the start of NEBD, was significantly accelerated in Cdc20−/H MEFs, whereas prometaphase showed normal timing (Fig. 7 A). However, metaphase was significantly delayed when Cdc20 levels were low, with Cdc20+/+ MEFs executing metaphase on average in 11 min versus Cdc20−/H MEFs in 18 min. The duration of anaphase was normal, although Cdc20−/H MEFs had elevated cyclin B1 levels at this stage of mitosis. To examine whether the observed increase in cyclin B1 expression in Cdc20−/H metaphases might be linked to the delay in anaphase onset, we repeated the aforementioned experiment in the presence of CHX to prevent de novo cyclin B1 synthesis. Only cells that entered mitosis within 10 min after adding CHX to the culture medium were analyzed to preclude that alterations in mitotic timing might stem from protein biosynthesis deficiencies in G2 phase. Consistent with the notion that high cyclin B1 levels prolong metaphase, CHX restored normal metaphase timing in Cdc20−/H MEFs (Fig. 7 A). Notably, inhibition of protein biosynthesis had no significant impact on the timing of the various mitotic stages in Cdc20+/+ MEFs.
Figure 7.
Cdc20 hypomorphism specifically prolongs metaphase. (A) Analysis of mitosis timing in Cdc20+/+ and Cdc20−/H MEFs in the presence or absence of CHX (n = 10 to 15 cells/group). Error bars represent SEM. *, P < 0.0005 versus Cdc20+/+ (by Mann–Whitney test). P, prophase; PM, prometaphase; M, metaphase; A, anaphase. (B) Analysis of mitotic checkpoint activity of Cdc20+/+ (n = 64 cells) and Cdc20−/H MEFs (n = 68 cells) challenged with nocodazole (Noc) or taxol. Error bars represent SD. (C) Images of Cdc20+/+ MEFs in which Mad2 levels were normal or depleted by RNA interference. Prometaphases stained for Cdc20, centromeres, and DNA are shown. (D) Western blot showing that Mad2 was efficiently depleted. (E) Images of wild-type and BubR1−/− prometaphases stained for Cdc20, centromeres, and DNA. Bars, 10 µm.
Given that Cdc20 is the main target of the mitotic checkpoint, we wondered whether Cdc20 hypomorphism affects the activity of this checkpoint. To measure the checkpoint status, we used an established live cell imaging–based nocodazole challenge assay. In this assay, YFP-H2B–positive Cdc20+/+ and Cdc20−/H MEFs were exposed to nocodazole to activate the mitotic checkpoint, and the time between NEBD and DNA decondensation is measured (Baker et al., 2006; van Ree et al., 2010). Cells with an intact checkpoint arrest in prometaphase for a prolonged period of time before exiting mitosis without chromosome separation. The duration of this period was on average 4.8 h for our Cdc20+/+ MEFs (Fig. 7 B). Cdc20−/H MEFs exited mitosis slightly faster, but the difference with Cdc20+/+ MEFs was not statistically significant. Similar results were obtained when paclitaxel was used instead of nocodazole (Fig. 7 B). Thus, the mitotic checkpoint seems to function properly at low amounts of Cdc20 and is unlikely to be a source of chromosome missegregation in Cdc20−/H MEFs. In addition, Cdc20 abundantly accumulated at unattached kinetochores in Mad2-depleted Cdc20+/+ MEFs (Fig. 7, C and D), indicating that its kinetochore association is independent of mitotic checkpoint signaling. However, BubR1-deficient cells failed to enrich Cdc20 at kinetochores (Fig. 7 E), suggesting that BubR1 plays a key role in recruitment of Cdc20 to kinetochores.
Cdc20 hypomorphic mice are not tumor prone
Mutant mouse strains with aneuploidy are usually susceptible to tumor development, particularly to carcinogen-induced tumorigenesis (for reviews see Ricke et al., 2008; Holland and Cleveland, 2009; Schvartzman et al., 2010). To determine the extent to which Cdc20 mutant mice are prone to cancer, we treated the dorsal skin of Cdc20+/+, Cdc20+/−, Cdc20H/H, and Cdc20−/H pups with a low dose of the carcinogen 7, 12-dimethylbenz(a)anthracene (DMBA) at 3–5 d after birth (Dawlaty et al., 2008). Animals were sacrificed at 5 mo and screened for tumors. Consistent with the literature, this treatment protocol induced primarily lung tumors as well as an occasional skin tumor. We found that overall, lung, and skin tumor incidence rates of Cdc20+/−, Cdc20H/H, and Cdc20−/H animals were not significantly different from those of Cdc20+/+ mice (Fig. 8, A–C). The mean number of lung tumors was lower in Cdc20+/−, Cdc20H/H, and Cdc20−/H mice, but only the decrease in Cdc20+/− mice reached statistical significance (Fig. 8 D). The median number of lung tumors was not significantly different among the various mouse genotypes (Fig. 8 E).
Figure 8.
Cdc20 hypomorphic mice are not cancer prone despite aneuploidy. (A–C) Overall (A), skin (B), and lung (C) tumor incidence of DMBA-treated Cdc20+/+, Cdc20+/−, Cdc20H/H, and Cdc20−/H mice. Error bars represent 95% confidence intervals using the modified Wald method for proportions. There were no statistical differences between Cdc20 mutant and wild-type mice (by Fisher’s exact χ2 test). (D and E) Mean (D) and median (E) number of DMBA-induced lung tumors in mice of the indicated genotypes. Error bars in D represent SEM, and error bars in E represent nonparametric two-sided 95% confidence intervals for the median. *, P = 0.0289 versus Cdc20+/+ (by unpaired t test). (F) Kaplan–Meier overall survival curves of Cdc20+/+ and Cdc20−/H mice. We note that Cdc20−/H mice remained indistinguishable from Cdc20+/+ and developed no obvious pathologies. (G) Kaplan–Meier tumor-free survival curves of mice that are presented in F. Animals that died without tumors were censored from the data.
To determine whether Cdc20 hypomorphic mice are prone to spontaneous tumors, Cdc20+/+ and Cdc20−/H mice were monitored for signs of ill health for nearly 3 yr. Survival curves of Cdc20+/+ and Cdc20−/H mice were overlapping, indicating that Cdc20 hypomorphism has no impact on lifespan (Fig. 8 F). Tumor-free survival curves were also superimposed, underscoring the conclusion from our tumor bioassay with DMBA that mice with low amounts of Cdc20 are not tumor prone (Fig. 8 G). Also, the spectrum of spontaneous tumors in Cdc20−/H mice was similar to that of Cdc20+/+ mice (Fig. S4).
Neuronal development is normal in Cdc20 hypomorphic mice
Cdc20 knockdown experiments in postmitotic rat neurons suggest that centrosome-associated Cdc20 controls the formation of primary, secondary, and tertiary dendritic branches (Kim et al., 2009). These findings prompted us to screen Cdc20−/H pups for defects in dendrite morphogenesis. Primary cerebellar granule cells harvested from postnatal day 6 (P6) Cdc20+/+ and Cdc20−/H mice were cultured and dendrite branches visualized by immunostaining for MAP2. Although the overall dendrite length of Cdc20-depleted rat neurons is ∼2-3-fold reduced, no reduction in length was observed for Cdc20−/H neurons (Fig. S5, A and B). As judged by immunofluorescence staining for Cdc20 and centrosomes, Cdc20−/H neurons showed no obvious reduction in centrosome-bound Cdc20 (Fig. S5 C), although overall Cdc20 levels were reduced (Fig. S5 D). Cdc20−/H MEFs also accumulated normal amounts of Cdc20 at centrosomes in interphase (unpublished data). Thus, it seems that Cdc20 preferentially targets to centrosomes, which might explain why dendrite morphogenesis was unperturbed in our model.
Cdc20 has also been implicated in presynaptic differentiation (Yang et al., 2009), a process required for formation of synapses and neuronal circuits during brain development. Cdc20 presumably induces presynaptic differentiation by ordering the destruction of neuroD2, a neuronal transcription factor that inhibits formation of presynapsis by repressing complexin II gene transcription. To probe Cdc20−/H mice for defects in presynaptic differentiation, we measured neuroD2 levels in cerebellar lysates of P22 Cdc20+/+ and Cdc20−/H mice by Western blot analysis. Unlike cerebellum of Cdc20-depleted rats (Yang et al., 2009), cerebellum of Cdc20−/H mice showed no elevated neuroD2 levels (Fig. S5 D). Furthermore, staining of brain sections of P26 Cdc20+/+ and Cdc20−/H mice for synapsin I revealed that Cdc20 hypomorphism had no impact on the amount of synapses in the granular and molecular layers of the cerebellum (Fig. S5, E and F). Furthermore, Cdc20−/H mice showed no evidence of neurological dysfunction in two behavioral tests, one screening for memory deficits (Fig. S5 G) and the other for motor deficits (Fig. S5 H). When taken together with the observation that Cdc20−/H mice have normal health and life spans, the aforementioned data imply that Cdc20 levels in Cdc20−/H mice are above the physiologically critical threshold for development of overt neuropathology.
Discussion
To address the physiological role of Cdc20, we used a hypomorphic approach in mice. We were successful in generating viable mice that express 15–25% of normal Cdc20 protein levels. Using these mice and cells thereof, we have yielded important new insights into the functions of Cdc20 in vivo. Our experiments unveil a hitherto unrecognized requirement for Cdc20 in efficient kinetochore–MT attachment and show that partial loss of Cdc20 function leads to inaccurate chromosome separation and near diploid aneuploidies. Furthermore, our data show that cyclin B1 is de novo synthesized in mitosis via CPEB-dependent translation, causing accumulation of cyclin B1 in prometaphase and, most likely, delayed anaphase onset in Cdc20 hypomorphic cells. Remarkably, Cdc20 insufficiency failed to promote tumorigenesis, which separates Cdc20 from other chromosomal instability genes that cause aneuploidy when defective. Cdc20-insufficient mice showed no evidence of abnormal brain development or neuropathology, which is unexpected given the recently ascribed effects of APC/CCdc20 ubiquitin signaling on dendrite morphogenesis and presynaptic differentiation.
Cdc20 and chromosome–MT attachment
A surprising finding was that cells with low amounts of Cdc20 are prone to chromosome misalignment. Chromosomes that fail to achieve biorientation consist of chromosome pairs attached to MTs coming from one spindle pole. Although these MT attachments are unstable, the bioriented chromosomes of Cdc20 hypomorphic cells are stably attached to spindle MTs and are under normal tension, suggesting that Cdc20 insufficiency reduces the efficiency of MT-capturing process rather than the stability of the newly established MT–kinetochore interaction. Because Cdc20 is a kinetochore-associated protein, it is conceivable that it exerts its influence on MT–kinetochore attachment from this location. If so, it is unlikely to do so through a mechanism that involves kinetochore assembly because various kinetochore-associated proteins with key roles in the attachment process, such as Bub1, BubR1, and CENP-E, are normally recruited to kinetochores of Cdc20 hypomorphic cells. We find that recruitment of Cdc20 to kinetochores is dependent on its binding partner BubR1, raising the possibility that Cdc20 might exert its effect on MT–kinetochore attachment through BubR1. However, BubR1 seems to be more important in stabilizing MT–kinetochore attachments rather than their initial MT capture (Lampson and Kapoor, 2005), arguing against this idea.
Interestingly, kinetochore-associated Cdc20 levels peak in prometaphase and drop significantly after biorientation in metaphase (Fig. 2, G and H; Kallio et al., 2002). However, Cdc20 levels remain high at chromosomes that fail to align in the metaphase plate, raising the possibility that Cdc20 might play an active role in mitotic checkpoint signaling. Cdc20 fails to accumulate at high levels at kinetochores of misaligned chromosomes in hypomorphic cells, which might result in weakened checkpoint signaling and thus cause premature anaphase onset, chromosome missegregation, and aneuploidy. Although attractive, this scenario seems unlikely because like normal cells, Cdc20 hypomorphic cells undergo prolonged mitotic arrest in the presence of nocodazole, suggesting that their mitotic checkpoint is properly activated and maintained when Cdc20 levels are low. Moreover, if a checkpoint defect was to underlie chromosome misalignment, one would expect MG132 treatment to ameliorate the attachment phenotype rather than to aggravate it.
It is equally possible that the attachment phenotype of Cdc20 hypomorphic cells are caused by impaired APC/CCdc20 activity in the mitotic cytosol. As a result of inadequate APC/CCdc20 activity, Cdc20 hypomorphic cells retain high levels of cyclin B1 in metaphase, allowing cyclin B1–Cdk1 activity to persist at a time that it should be rapidly declining. This leads to aberrant phosphorylation of substrates, as suggested by increased immunofluorescence of phosphorylated Cdk substrates in Cdc20 hypomorphic metaphases, which may include regulators of MT–kinetochore attachment, thereby perhaps altering their function. Consistent with this model, we find that chromosome misalignment is the primary chromosome segregation defect of mutant MEFs that ectopically express cyclin B1 (unpublished data).
De novo cyclin B1 synthesis in mitosis
Cells with low amounts of Cdc20 show a remarkable increase in cyclin B1 levels that starts after NEBD and peaks in metaphase. Until now, de novo synthesis of cyclin B1 protein in M phase was thought to be limited to the first few divisions of frog embryogenesis. During M phase of these embryonic divisions, CPEB is known to regulate poly(A) tail extension of oocyte-derived cyclin B1 transcripts, thereby inducing translational activation (Groisman et al., 2002). This embryonic mechanism seems to be conserved in MEFs, as we find that de novo synthesis of cyclin B1 in mitotic Cdc20 hypomorphic MEFs is dependent on CPEB-binding sequences in the 3′UTR of cyclin B1 mRNA. We find that wild-type MEFs and HeLa cells promptly raise cyclin B1 protein levels in a translation-dependent fashion in response to nocodazole, suggesting that cyclin B1 synthesis in mitosis is not just a feature of Cdc20 hypomorphic cells. This notion is further supported by the observation that normal cells in metaphase exhibit reduced cyclin B1 levels after a brief period of translation inhibition.
Future studies will be needed to understand the biological significance of the newfound cyclin B1 synthesis in mitosis. However, one attractive model is that it serves to counteract cyclin B1 proteolysis during prometaphase. As more and more kinetochores become attached to spindle MTs and mitotic checkpoint signaling starts to decline, APC/CCdc20 activity may gradually increase and drive cyclin B1 proteolysis in prometaphase. Perhaps, de novo synthesis of cyclin B1 in mitosis is particularly important in situations where attachment errors that do not invoke the mitotic checkpoint would persist in late metaphase to provide sufficient time to correct such errors before anaphase onset and avoid chromosome missegregation. Importantly, protein synthesis during mitosis is not restricted to cyclin B1, as demonstrated by a recent study showing that Cdc20 continues to be synthesized after mitosis onset as a means to counteract APC/C-mediated Cdc20 degradation in the presence of active mitotic checkpoint signaling (Nilsson et al., 2008). A recent study reported that Cdc20 transcripts undergo CPEB-regulated poly(A) tail elongation in mitotic HeLa cells (Novoa et al., 2010), suggesting that synthesis of cyclin B1 and Cdc20 during mitosis is orchestrated through a common mechanism. Mad2, securin, and cyclin A2 protein levels were not impacted by inhibition of translation during mitosis (Fig. 5 C), which is consistent with a lack of CPE elements in transcripts encoding for these proteins.
Cdc20 depletion and tumorigenesis
Experiments of mutant mice that are prone to chromosome missegregation have provided compelling evidence for the hypothesis that aneuploidy is causally implicated in tumor development. Most mouse models of aneuploidy are predisposed to spontaneous tumors, whereas those that are not are typically prone to carcinogen-induced tumors. Also prone to various spontaneous tumors are mutant mice that suffer from APC/CCdc20 hyperactivity caused by a Cdc20 mutation that prevents Mad2 binding (Li et al., 2009). Our finding that Cdc20 hypomorphic mice were neither prone to spontaneous nor carcinogen-induced tumors is very surprising in light of this evidence. One could argue that the aneuploidy rates of Cdc20 hypomorphic mice might be too low, but this explanation is unlikely because chromosome counts on splenocytes and MEFs suggest that aneuploidy rates of several of the cancer-prone strains are in the same range as those of our Cdc20 hypomorphic mice. It is reasonable to assume that aneuploidy rates of splenocytes and MEFs are indicative of aneuploidies in other tissues and cell types, as indicated by chromosome counts on Cdc20 hypomorphic and wild-type lung fibroblasts. A recent study demonstrated that inhibition of Cdc20 to the extent that it blocks anaphase onset promotes programmed cell death of cancer cells in a mitotic checkpoint–independent fashion (Huang et al., 2009). Although anaphase onset is not blocked in Cdc20 hypomorphic mice, it is significantly delayed. We speculate that this delay might slightly sensitize cells that are at risk for neoplastic transformation to apoptosis, thereby perhaps counteracting the tumor-promoting effect that aneuploidy of Cdc20 hypomorphic mice might have. Thus, Cdc20 hypomorphism might induce both tumor-suppressive and tumor-promoting effects that largely neutralize each other.
Materials and methods
Generation of Cdc20 mutant mice
The gene targeting procedure used to generate the hypomorphic Cdc20 allele was as previously described (Dawlaty and van Deursen, 2006). Correctly targeted ES cell clones were injected into blastocysts, and Cdc20+/H offspring were obtained from the resulting chimeras through standard procedures (Babu et al., 2003). The Cdc20− allele used in our experiments was derived from gene trap ES clone XE368 (purchased from BayGenomics). DMBA tumor bioassays were performed as previously described (Babu et al., 2003). All mice used were on a 129 × C57BL/6 mixed genetic background. Cohorts of Cdc20+/+ and Cdc20–/H mice were monitored three times weekly for general health, well being, and tumor formation. At biopsy, spontaneous tumors were collected and processed for standard histopathology. All mouse experiments were conducted after approval of the Mayo Clinic Committee on Animal Care and Use. All mice were housed in a pathogen-free barrier environment. Mouse protocols were reviewed and approved by the institutional animal care and use committee.
Generation and culture of MEFs and chromosome counts
Cdc20 wild-type and mutant MEFs were generated and cultured as previously described (Baker et al., 2004). MEFs were frozen at passage 2 (P2) or 3 and used for experimentation between P4 and P6. At least three independently generated MEF lines per genotype were used. Chromosome counts on mouse splenocytes, MEFs, and primary lung fibroblasts were as previously described (Babu et al., 2003; Baker et al., 2009). In karyotyping experiments in which MG132 was used, cells were cultured for 2 h in the presence of 10 µM of the drug. mMad2-shRNAmir clone V2MM_6980 was used to knockdown Mad2 in wild-type MEFs as described previously (Malureanu et al., 2009). BubR1−/− MEFs were prepared through Cre-mediated recombination as previously described (Malureanu et al., 2009).
Western blot analysis, indirect immunofluorescence, and confocal microscopy
Western blot analysis was performed as described previously (Kasper et al., 1999). Tissue extracts were prepared in PBS containing 0.1% NP-40, 10% glycerol, and complete protease inhibitor cocktail (Roche) using a gentleMACS dissociator (program m_impTumor_03 was run twice; Miltenyi Biotech). Extracts were centrifuged at 20,000 g for 15 min at 4°C, and supernatants were collected for electrophoresis. To analyze de novo synthesis of cyclin B1 by Western blotting, mitotic HeLa cells were harvested from asynchronous cultures (4 × 15–cm dishes) by shake off. Harvested cells were pelleted and cultured in medium containing 100 ng/ml nocodazole or 100 ng/ml nocodazole plus 1 µg/ml CHX for 15 min at 37°C.
Indirect immunofluorescence was performed as described previously (Kasper et al., 1999; Taylor et al., 2001). Standard fixations for immunostainings were with 3% paraformaldehyde for 12 min at RT. For kinetochore localization experiments, cells were fixed with 1% paraformaldehyde for 5 min at RT. For localization of Cdc20 at centrosomes, cerebellar granule cell cultures were fixed with ice-cold methanol for 10 min at −20°C. CHX and nocodazole treatments were performed at 1 µg/ml and 100 ng/ml, respectively. A laser-scanning microscope (LSM 510 v3.2SP2; Carl Zeiss, Inc.) with Axiovert 100M (Carl Zeiss, Inc.) with a c-Apochromat 100× oil immersion objective was used to analyze immunostained cells and to capture representative images. To analyze endogenous cyclin B1 levels, Cdc20+/+ and Cdc20−/H MEFs were initially immunostained with two independent mouse monoclonal antibodies against cyclin B1: GNS-1 (mouse anti–human cyclin B1; Santa Cruz Biotechnology, Inc.) and GNS-11 (mouse anti–human cyclin B1; BD). By immunostaining of wild-type MEFs in which cyclin B1 was depleted with cyclin B1–shRNAmir lentivirus, we confirmed that immunofluorescence signals from both these antibodies were specific for cyclin B1 protein. To measure endogenous cyclin A2 levels, MEFs were immunostained with rabbit anti–cyclin A2 (Santa Cruz Biotechnology, Inc.). For quantitation of cyclin B1 and cyclin A2 levels, we used ImageJ software (National Institutes of Health). Confocal images were converted to 8-bit grayscale. Cell edges were traced using the freehand tool, and the mean pixel intensity within the marked area was calculated. The integrated density (in arbitrary units), defined as the mean pixel intensity multiplied by the area, was used as a measure for cyclin B1 and cyclin A2 levels. For quantification of synapsin I signals, ImageJ software was used as follows. Confocal images were converted to 8-bit grayscale, and a “black and white” threshold of 0 to 22 or 0 to 45 was set up. Images were transformed to binary, and the “find edges” function was applied. The counts from “analyze particles” for 0–25 size were recorded for each field. Each field had a size of 93 × 93 µm. Random fields were manually counted to confirm the automated counting.
Antibodies
The following antibodies were used for immunofluorescence and Western blotting: mouse anti–human cyclin B1 (Santa Cruz Biotechnology, Inc.) and mouse anti–human cyclin B1 (BD), rabbit anti–cyclin A2 (Santa Cruz Biotechnology, Inc.), rabbit anti-Cdc20 (Santa Cruz Biotechnology, Inc.), in-house rabbit anti-Mad2, mouse anti–aurora B (BD), mouse anti–β-actin (Sigma-Aldrich), mouse anti–α-tubulin (Sigma-Aldrich), rabbit anti–human BubR1 (Baker et al., 2004), human anti-centromere antibody (Antibodies, Inc.), mouse anti–human securin (Novus Biologicals), mouse anti–histone H3 phosphorylated at Ser10 (Millipore), sheep anti–hamster MCAK (provided by L. Wordeman, University of Washington School of Medicine, Seattle, WA; Andrews et al., 2004), rabbit anti–mouse Bub1 (Jeganathan et al., 2007), rabbit anti–CENP-E (provided by D. Cleveland, Ludwig Institute for Cancer Research, La Jolla, CA), in-house rabbit anti-mMad1 (510–998), rabbit anti–phospho-Ser Cdks substrate antibody (Cell Signaling), mouse anti–centrin 2 (provided by J. Salisbury, Mayo Clinic, Rochester, MN), rabbit anti-MAP2 (Millipore), rabbit anti-neuroD2 (Millipore), and mouse anti–synapsin I (Millipore).
Deconvolution microscopy
Microscopy was performed using a fluorescence microscope (IX70; Olympus) fitted with FITC, CY5, rhodamine, and DAPI filters and a digital camera (CoolSNAP HQ; Photometrics). Images were acquired using Resolve 3D as z stacks with 0.2-µm intervals using a U Plan Apo 100×/1.35 NA oil immersion objective (Olympus). Images were deconvolved using SoftWoRx software (version 3.5.1; Applied Precision). When suitable kinetochore pairs were identified, the interkinetochore stretch was determined using the “distance measure” function in SoftWoRx. To determine interkinetochore distances of chromosomes in the absence of MT tension, cells were cultured in medium containing 10 µg/ml nocodazole for 1 h. Analysis of kinetochores with cold stable–stable MT attachments were as described previously (Lampson and Kapoor, 2005). ACA and α-tubulin immunostaining were used to visualize kinetochores and MTs in deconvolved image z stacks. A kinetochore was scored as attached when an MT bundle ended at the kinetochore. Kinetochores for which the attachment status could not be determined with high fidelity because of MT crowding were excluded. At least 40 kinetochores were scored per genotype.
Plasmids
pVenus-N1/Hs-CcnB1 was constructed by PCR amplifying Hs-CcnB1 cDNA from pCMX-Hs-CcnB1-GFP (provided by J. Pines, Gurdon Institute, Cambridge, England, UK; [forward primer] 5′-CTATACAAGCTTGCCACCATGGCGCTCCGAGTCACCAGG-3′ and [reverse primer] 5′-CTATACGGATCCGTCACCTTGCCACAGCCTTGGC-3′) and cloning it into HindIII–BamHI cut pVenus-N1 (pEYFP-N1 [Takara Bio Inc.], in which EYFP was replaced with Venus). pVenus-N1/Hs-CcnB1/Ms-3′UTR was generated by PCR amplifying the mouse cyclin B1 3′UTR sequence from pBluescript SK-Ms-CcnB1 (forward primer, 5′-CGGGGTACCCCGGCGGCCGCCTCCAATAGACTGCTGCTACATCTGCAG-3′; and reverse primer, 5′-CGGGGTACCCCGGCGGCCGCAATTGTGAAAGCTTTCCACCAATAAATTTTA-3′; Chapman and Wolgemuth, 1993) and ligating the polished PCR fragment into the EcoRV site of pVenus-N1/Hs-CcnB1. Insertion of the 3′UTR sequence in the sense orientation yielded pVenus-N1/Hs-CcnB1/Ms-3′UTR, and insertion in the antisense orientation yielded pVenus-N1/Hs-CcnB1/Ms-3′UTR-AS.
Live cell imaging
For chromosome missegregation analysis, chromosome movements of H2B-YFP or H2B-mRFP–positive MEFs progressing through an unchallenged mitosis were followed at interframe intervals of 3 min as previously described (Jeganathan et al., 2007). Destruction of securin-EYFP and cyclin B1–Venus was monitored as follows. H2B-mRFP expressing Cdc20+/+ and Cdc20−/H MEFs were nucleofected with 5 mg pVenus-N1/Hs-CcnB1, pVenus-N1/Hs-CcnB1/Ms-3′UTR, pVenus-N1/Hs-CcnB1/Ms-3′UTR-AS, or 2 mg pEYFP-N1-securin (provided by J. Pines) using a Nucleofector II (Lonza). Nucleofections were performed on 2 × 106 cells in MEF2 buffer (VPD-1005; Lonza). Nucleofected MEFs were seeded into 35-mm glass-bottom dishes and analyzed 6–8 h later. G2 phase cells were marked, and images were captured every 3 min. Quantification of fluorescence levels was performed using ImageJ software as previously described (Wolthuis et al., 2008). In brief, a line was drawn around the edge of the cell, and the mean pixel intensity was calculated. For the background subtraction purposes, an open region was selected near the cell, and the mean pixel intensity was calculated. The total cell fluorescence was calculated by (mean cell fluorescence) − (mean background fluorescence) × cell area.
Nocodazole and taxol challenge assays and mitotic timing experiments were performed as previously described (Malureanu et al., 2009). For mitotic timing experiments, H2B-mRFP–positive MEFs progressing through an unchallenged mitosis were followed at interframe intervals of 3 min, and the durations of prophase, prometaphase, metaphase, and anaphase were determined (Dawlaty et al., 2008). Note that cells with chromosome alignment and segregation errors were excluded from the analysis. AxioVision software (version 10.6) and PowerPoint (for Mac; Microsoft) were used for image processing, and Prism software (for Mac; version 4.0a; GraphPad Software, Inc.) was used for statistical analysis. At least three independent clones per genotype were used in the aforementioned experiments unless otherwise noted.
Analysis of dendrite morphogenesis
Cerebellar granule cell cultures were prepared from cerebella of 6-d-old Cdc20+/+ and Cdc20−/H pups as described previously (Galvin et al., 2008). In brief, the whole cerebellum was collected and dissociated. Suspended cells were plated on coverslips coated with 500 µg/ml poly-d-lysine. Plated cells were cultured overnight in neurobasal A medium (Invitrogen) supplemented with B-27 and glutamax I. After 24 h, medium was refreshed. After 5 d in culture, granule cells were fixed for immunohistochemistry. For quantification of total dendrite length of individual neurons, NeuronJ plug in (version 1.4.1; National Institutes of Health) for ImageJ was used. Images were converted to 8-bit grayscale, and tracings of all primary, secondary, and tertiary dendrites for individual neurons were measured and recorded.
Object recognition and rotarod tests
A scent-based object recognition test developed by Buenz et al. (2009) was used to test memory. In brief, a single mouse was placed in a 30 × 30–cm acrylic box containing wood shavings. After 5 min, the animal was removed and held in a normal cage. Two berry-scented candles were placed in the acrylic box for the training session. The mouse was returned to the box and videotaped for the next 10 min to assess the number of interrogations of each object. The mouse was relocated in a holding cage. One of the two berry-scented candles was replaced with an apple-scented candle for the testing session. After 5 min, the mouse was returned to the box and videotaped for 10 min. The discrimination index was determined by dividing the number of interrogations of the novel object by the number of investigations of the control object. Normal recognition memory produces a discrimination index of about one for the training session and a discrimination index considerably greater than one for the testing session, which is consistent with greater interrogation of the novel object. Rotarod tests were performed as previously described (Brooks and Dunnett, 2009).
Online supplemental material
Fig. S1 shows that core mitotic checkpoint proteins properly target to kinetochores in early mitosis when Cdc20 levels are low. Fig. S2 shows that securin does not accumulate in Cdc20 hypomorphic MEFs. Fig. S3 shows that synthesis of cyclin B1 during mitosis is transcription independent and provides a map of CPE consensus sequences in the 3′UTR of cyclin B1. Fig. S4 shows that the spontaneous tumor incidence and the tumor spectrum of Cdc20 hypomorphic mice are normal. Fig. S5 shows that dendrite morphogenesis and presynaptic differentiation are not impaired in Cdc20 hypomorphic mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003090/DC1.
Acknowledgments
We thank W. Zhou, M. Li, and D. Norris for technical assistance, E. Thrushina, B. Sauer, and C. Howe for help in performing motor and memory tests, R. Lennon for consultation on statistics, and D. Katzmann for assistance with deconvolution microscopy. We thank P. Galardy, M. Hamada, and R. Ricke for comments on the manuscript.
This work was supported by grants from the National Cancer Institute (CA96985 and CA91956).
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- CENP-E
- centromere protein E
- CHX
- cycloheximide
- CPE
- cytoplasmic polyadenylation element
- CPEB
- CPE-binding protein
- DMBA
- 12-dimethylbenz(a)anthracene
- ES
- embryonic stem
- MEF
- mouse embryonic fibroblast
- MT
- microtubule
- PMSCS
- premature sister chromatid separation
- UTR
- untranslated region
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 10.1083/jcb.200211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Malureanu L., Perez-Terzic C., Terzic A., van Deursen J.M. 2006. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172:529–540 10.1083/jcb.200507081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jin F., Jeganathan K.B., van Deursen J.M. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 16:475–486 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S.P., Dunnett S.B. 2009. Tests to assess motor phenotype in mice: a user’s guide. Nat. Rev. Neurosci. 10:519–529 10.1038/nrn2652 [DOI] [PubMed] [Google Scholar]
- Buenz E.J., Sauer B.M., Lafrance-Corey R.G., Deb C., Denic A., German C.L., Howe C.L. 2009. Apoptosis of hippocampal pyramidal neurons is virus independent in a mouse model of acute neurovirulent picornavirus infection. Am. J. Pathol. 175:668–684 10.2353/ajpath.2009.081126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D.L., Wolgemuth D.J. 1993. Isolation of the murine cyclin B2 cDNA and characterization of the lineage and temporal specificity of expression of the B1 and B2 cyclins during oogenesis, spermatogenesis and early embryogenesis. Development. 118:229–240 [DOI] [PubMed] [Google Scholar]
- Dawlaty M.M., van Deursen J.M. 2006. Gene targeting methods for studying nuclear transport factors in mice. Methods. 39:370–378 10.1016/j.ymeth.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin K.E., Ye H., Erstad D.J., Feddersen R., Wetmore C. 2008. Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells. 26:1027–1036 10.1634/stemcells.2007-0879 [DOI] [PubMed] [Google Scholar]
- Geley S., Kramer E., Gieffers C., Gannon J., Peters J.M., Hunt T. 2001. Anaphase-promoting complex/cyclosome–dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153:137–148 10.1083/jcb.153.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I., Jung M.Y., Sarkissian M., Cao Q., Richter J.D. 2002. Translational control of the embryonic cell cycle. Cell. 109:473–483 10.1016/S0092-8674(02)00733-X [DOI] [PubMed] [Google Scholar]
- Hames R.S., Wattam S.L., Yamano H., Bacchieri R., Fry A.M. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20:7117–7127 10.1093/emboj/20.24.7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10:478–487 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.C., Shi J., Orth J.D., Mitchison T.J. 2009. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 16:347–358 10.1016/j.ccr.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M.J., Beardmore V.A., Weinstein J., Gorbsky G.J. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158:841–847 10.1083/jcb.200201135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper L.H., Brindle P.K., Schnabel C.A., Pritchard C.E., Cleary M.L., van Deursen J.M. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.H., Puram S.V., Bilimoria P.M., Ikeuchi Y., Keough S., Wong M., Rowitch D., Bonni A. 2009. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 136:322–336 10.1016/j.cell.2008.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98 10.1038/ncb1208 [DOI] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- Li M., York J.P., Zhang P. 2007. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell. Biol. 27:3481–3488 10.1128/MCB.02088-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Shin Y.H., Hou L., Huang X., Wei Z., Klann E., Zhang P. 2008. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 10:1083–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang X., Wei Z., York J.P., Zhang P. 2009. Loss of spindle assembly checkpoint–mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 185:983–994 10.1083/jcb.200904020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74:595–648 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Gallego J., Ferreira P.G., Mendez R. 2010. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell Biol. 12:447–456 10.1038/ncb2046 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Peters J.M., Tedeschi A., Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089–3114 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- Pines J. 2006. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 16:55–63 10.1016/j.tcb.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Richter J.D. 2007. CPEB: a life in translation. Trends Biochem. Sci. 32:279–285 10.1016/j.tibs.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Ricke R.M., van Ree J.H., van Deursen J.M. 2008. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 24:457–466 10.1016/j.tig.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.M., Sotillo R., Benezra R. 2010. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 10:102–115 10.1038/nrc2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciortino S., Gurtner A., Manni I., Fontemaggi G., Dey A., Sacchi A., Ozato K., Piaggio G. 2001. The cyclin B1 gene is actively transcribed during mitosis in HeLa cells. EMBO Rep. 2:1018–1023 10.1093/embo-reports/kve223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., Benezra R. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 11:9–23 10.1016/j.ccr.2006.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395 [DOI] [PubMed] [Google Scholar]
- van Ree J.H., Jeganathan K.B., Malureanu L., van Deursen J.M. 2010. Overexpression of the E2 ubiquitin–conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 188:83–100 10.1083/jcb.200906147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. 2008. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell. 30:290–302 10.1016/j.molcel.2008.02.027 [DOI] [PubMed] [Google Scholar]
- Yang Y., Kim A.H., Yamada T., Wu B., Bilimoria P.M., Ikeuchi Y., de la Iglesia N., Shen J., Bonni A. 2009. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 326:575–578 10.1126/science.1177087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. 2006. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 173:153–157 10.1083/jcb.200601172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 27:3–16 10.1016/j.molcel.2007.06.009 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]