Cep120, a protein involved in maintenance of neural progenitor cells, is required for centriole duplication in cycling cells and for centriole amplification in tracheal epithelial cells.
Abstract
Centrioles form the core of the centrosome in animal cells and function as basal bodies that nucleate and anchor cilia at the plasma membrane. In this paper, we report that Cep120 (Ccdc100), a protein previously shown to be involved in maintaining the neural progenitor pool in mouse brain, is associated with centriole structure and function. Cep120 is up-regulated sevenfold during differentiation of mouse tracheal epithelial cells (MTECs) and localizes to basal bodies. Cep120 localizes preferentially to the daughter centriole in cycling cells, and this asymmetry between mother and daughter centrioles is relieved coincident with new centriole assembly. Photobleaching recovery analysis identifies two pools of Cep120, differing in their halftime at the centriole. We find that Cep120 is required for centriole duplication in cycling cells, centriole amplification in MTECs, and centriole overduplication in S phase–arrested cells. We propose that Cep120 is required for centriole assembly and that the observed defect in neuronal migration might derive from a defect in this process.
Introduction
Centrioles are evolutionarily conserved, microtubule-based organelles that provide cells with diverse organization, motility, and sensory functions. Centrioles are the core components of the centrosome, the main microtubule-organizing center in animal cells. Another critical function of centrioles is to serve as basal bodies that nucleate the formation of cilia. There are two broad classes of cilia: (1) motile cilia, which move fluids over epithelial surfaces and provide the motive force for sperm; and (2) immotile primary cilia that have diverse roles in sensory perception, including the detection of light in the vertebrate eye, odorants in the nose, and flow in the kidney nephron (Pazour and Witman, 2003; Berbari et al., 2009). The axoneme of all cilia is composed of nine outer doublet microtubules extending directly from the microtubules of the basal body, which anchors the cilium just beneath the plasma membrane.
Defects in centrioles, centrosomes, and cilia can have serious phenotypic consequences for cells and organisms. For example, defects in maintaining centriole/centrosome number lead to an increased frequency of aberrant chromosome segregation and genetic instability and can ultimately drive tumorigenesis (for reviews see Sluder and Nordberg, 2004; Zyss and Gergely, 2009). Importantly, defects in centriole structure/function also impact cilia function. It has recently become appreciated that dysfunction of cilia leads to a set of human disease conditions, referred to as ciliopathies, including polycystic kidney disease, hydrocephalus, retinal degeneration, and Bardet-Biedl syndrome (Quarmby and Parker, 2005; Baker and Beales, 2009). Thus, there is an intimate relationship between centriole/basal body formation and proper cilia assembly and function. However, little is known about this synergy and its implications in human disease.
Despite their importance, we know little of the mechanism of centriole duplication, basal body maturation, and cilium initiation. This is partly because of the presence of only a single centrosome and cilium in most cell types, which makes certain experimental approaches (for example biochemical characterization) difficult. Application of a variety of experimental methods that circumvent this difficulty, for example comparative genomic (Avidor-Reiss et al., 2004; Li et al., 2004), proteomic (Keller et al., 2005; Pazour et al., 2005), and gene expression analysis (Ross et al., 2007), has identified several conserved basal body and ciliary components. We recently established a mouse tracheal epithelial cell (MTEC) culture system (Vladar and Stearns, 2007), which provides a unique opportunity to study centriole assembly and ciliogenesis in mammalian cells that produce hundreds of centrioles during differentiation, each one nucleating a motile cilium (Fig. S1 A). The cultured MTECs acquire cilia over the course of several days, similar to the timing of ciliogenesis during airway development and tracheal epithelium reformation in vivo after damage (Vladar and Stearns, 2007). To identify new components of the centriole/cilium assembly pathway, we have examined gene expression changes in differentiating MTECs and identified genes that are specifically up-regulated during the early stages of differentiation, when centrioles are formed (unpublished data). Here, we focus on Cep120 (centrosomal protein 120), which is up-regulated approximately sevenfold during the early stages of centriole assembly in MTECs.
Three lines of evidence suggest a role for Cep120 in centriole and/or centrosome function. The protein, originally named Ccdc100 (coiled-coil domain containing 100), was first identified in a proteomic screen of purified human centrosomes (Andersen et al., 2003). Subsequently, Xie et al. (2007) showed that Cep120 is highly expressed in mouse brain and localizes to centrosomes in neural progenitor cells during neocortical development. They determined that Cep120 interacts with transforming acidic coiled-coil proteins to regulate centrosome-associated microtubules in the neural progenitors. Silencing of Cep120 in the developing neocortex impaired interkinetic nuclear migration, an essential step in proper neocortical development, as well as neural progenitor self-renewal (Xie et al., 2007). Finally, a homologue of Cep120 (Uni2) in the ciliated alga, Chlamydomonas reinhardtii, was shown to be essential for basal body maturation and ciliogenesis, with uni2 mutants having structural defects in the transition zone at the distal ends of basal bodies (Piasecki et al., 2008).
Here, we report that Cep120 is associated with centrioles in mammalian cells, with a specific enrichment at daughter centrioles. This asymmetry between the mother and daughter centriole is relieved coincident with new centriole assembly. Depletion of Cep120 protein causes defects in centriole duplication in cycling cells and MTECs. Together, these data reveal a conserved role for Cep120 in centriole formation and suggest a molecular mechanism for the previously observed defect in neuronal migration.
Results
Cep120 localizes to centrioles in ciliated epithelial cells
To identify and characterize components of the centriole biogenesis and cilia assembly pathway, we previously established an MTEC culture system that recapitulates the differentiation of multiciliated cells (Fig. S1 A; Vladar and Stearns, 2007). Reasoning that gene expression changes that take place during differentiation would reveal important proteins, we have generated a dataset of genes that are specifically up-regulated during initiation of centriole and cilia assembly (unpublished data). We have identified CEP120 (Ccdc100) as one such up-regulated gene; the CEP120 transcript is up-regulated sevenfold early in the differentiation of ciliated epithelial cells. Cep120 is a protein of 988 amino acids, contains a C-terminal coiled-coil domain, and is conserved in organisms containing centrioles/basal bodies and cilia, ranging from humans to C. reinhardtii (Fig. S1 B).
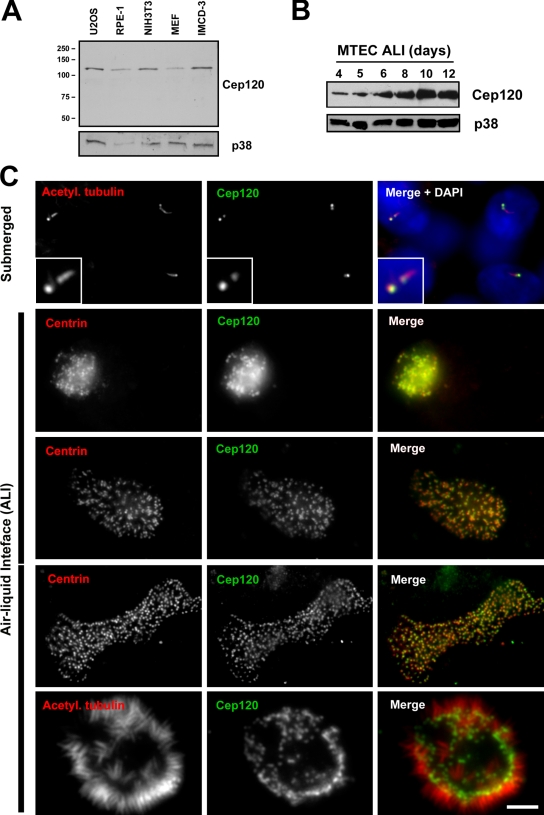
To characterize the expression and localization of Cep120, we used a polyclonal antibody directed against the C-terminal half of the protein (Xie et al., 2007). The antibody recognizes a protein of ∼120 kD on Western blots of lysates from mouse and human cell lines (Fig. 1 A). Western blots of MTEC lysates showed increasing amounts of Cep120 during differentiation (Fig. 1 B), which correlates with the up-regulation in RNA levels. Indirect immunofluorescence staining of MTEC cultures showed Cep120 localization to centrioles in proliferating cells before differentiation (Fig. 1 C, submerged). Interestingly, the Cep120 signal appeared to be brighter on one of the two centrioles in each cell. Centriole amplification begins ∼3–4 d after shifting to an air–liquid interface (ALI) in these cultures, as indicated by the appearance of dense clusters of centrin-positive spots (Fig. 1 C, second row). The Cep120 signal overlapped with the centrin signal at this early stage of centriole assembly. The fully assembled centrioles then disperse throughout the cytoplasm (Fig. 1 C, third row), dock at the apical surface (Fig. 1 C, fourth row), and initiate the formation of cilia (Fig. 1 C, fifth row). Cep120 remained associated with centrioles throughout the course of ciliogenesis (Fig. 1 C).
Figure 1.
Cep120 localizes to centrioles in MTECs. (A) Western blot of lysates from human (U2OS and RPE-1) and mouse (NIH3T3, IMCD-3, and MEF) cells. Samples were probed with a polyclonal antibody against Cep120 and MAPK p38 as a loading control. Numbers on the left indicate molecular mass of markers in kilodaltons. (B) Expression levels of Cep120 in MTECs. Cells were grown on Transwell permeable filter supports, and ALI was established 2 d after cells reached confluence. Cells were harvested on the indicated days, and lysates were analyzed by Western blotting using antibodies against Cep120 and p38. (C) MTEC cultures during differentiation (see Introduction). Cells were grown on Transwell filters, fixed, and labeled with antibodies against acetylated α-tubulin or centrin (red) and Cep120 (green). Insets are magnified images of the centrosome region. Bar, 10 µm.
Cep120 is enriched at the daughter centriole
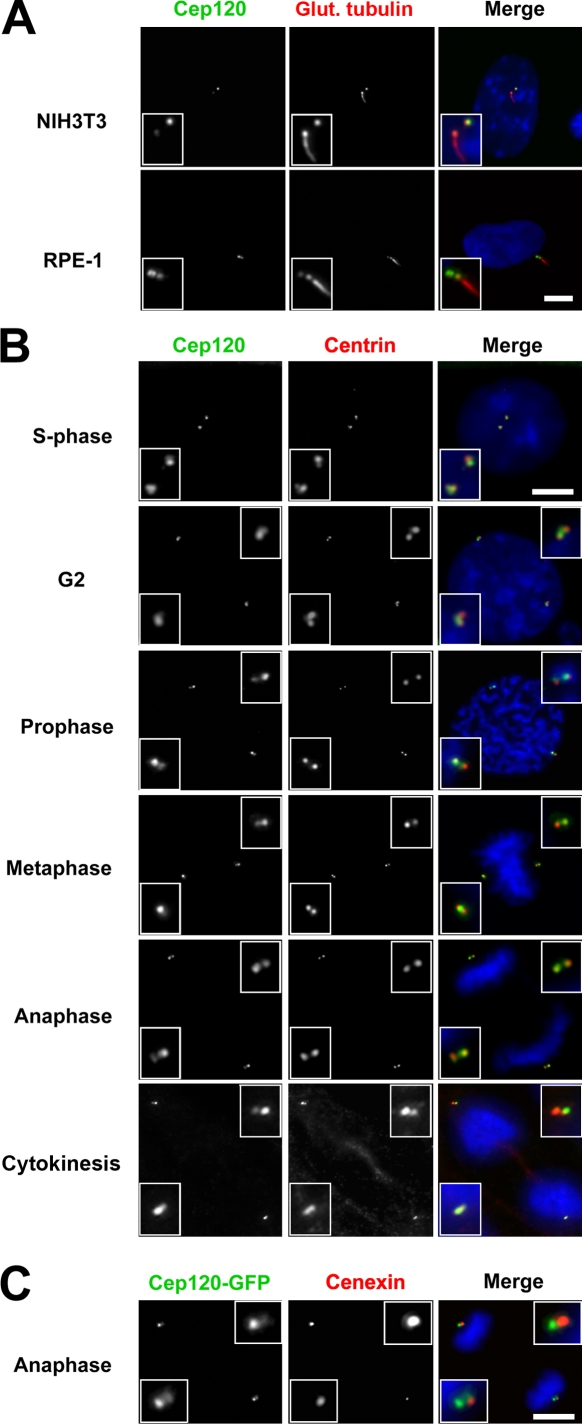
Cep120 was enriched on one of the two centrioles in G1-phase MTECs before differentiation, and the brighter signal was associated with the centriole lacking a primary cilium (Fig. 1 C, submerged). Following convention, we refer to the cilium-associated centriole as the mother centriole and the other as the daughter centriole. To confirm the enrichment of Cep120 on the daughter centriole, mouse NIH3T3 and human RPE-1 cells were serum starved to induce primary cilium formation and stained for Cep120 and glutamylated tubulin, a marker of both centrioles and cilia. As in MTECs, the Cep120 signal was brighter on the daughter centriole (Fig. 2 A). To determine whether this asymmetric localization pattern is maintained throughout the cell cycle, we examined NIH3T3 cells synchronized by release from serum starvation. In S phase, new daughter centrioles grow adjacent to both the original mother centriole (now the grandmother centriole) and the original daughter centriole (now the mother centriole). Centrin labeling identifies the new daughter centrioles as faint spots adjacent to the brighter grandmother and mother centrioles. Cep120 signal was brighter on these newly formed daughter centrioles (Fig. 2 B). The asymmetric distribution of Cep120 persisted through G2 and all stages of mitosis (Fig. 2 B). Because mitotic cells do not have a cilium, daughter centrioles were distinguished from mother and grandmother centrioles by staining with the mother centriole appendage protein cenexin/Odf2 (Lange and Gull, 1995). Double labeling with Cep120-GFP showed that, as in interphase, Cep120 is enriched at the daughter centriole at each spindle pole (Fig. 2 C). Consistent with these results, the asymmetric localization of Cep120-GFP was also observed by fluorescence imaging of a Cep120-GFP fusion protein in living cells (see Fig. 4 C).
Figure 2.
Cep120 is enriched at the daughter centriole. (A) NIH3T3 and RPE-1 cells were serum starved, fixed, and stained for Cep120 (green), glutamylated tubulin (red), and DNA (DAPI, blue). Note that Cep120 signal is greater on the daughter centriole. (B) NIH3T3 cells were released from serum starvation and followed over time as they progressed through the various cell cycle stages. Samples were fixed and stained for Cep120 (green), centrin (red), and DNA (blue). The asymmetric distribution of Cep120 is conserved through S phase, G2, and mitosis. (C) NIH3T3 cells were infected with lentivirus expressing Cep120-GFP. Cells were fixed and stained for GFP (green), cenexin (red), and DNA (blue). Cep120-GFP is enriched at the centrioles lacking cenexin, which corresponds to daughter centrioles at each pole. Insets are magnified images of the centrosome region. Bars, 10 µm.
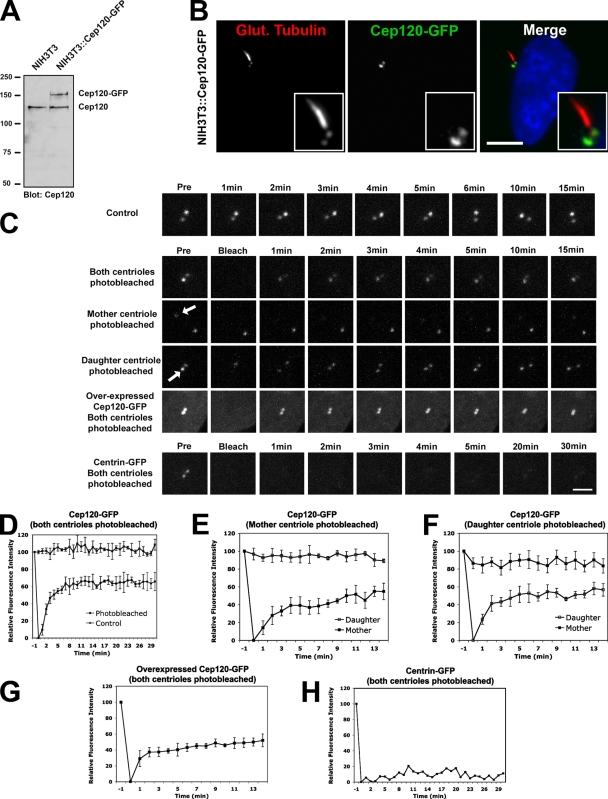
Figure 4.
FRAP analysis of Cep120 dynamics at centrioles. (A) Western blot of lysates from NIH3T3 and a Cep120-GFP–expressing NIH3T3 stable cell line (NIH3T3::Cep120-GFP) probed with anti-Cep120 antibody. Numbers on the left indicate molecular mass of markers in kilodaltons. (B) NIH3T3::Cep120-GFP cells stained for GFP (green) and glutamylated tubulin (red). Insets are magnified images of the centrosome region. Bar, 10 µm. (C) Photobleaching and recovery of Cep120 and centrin. The top and middle panels show NIH3T3::Cep120-GFP cells, and the bottom panels show NIH3T3::centrin-GFP cells. Control cells (top row) were not photobleached. In all other cases, the indicated centrioles were photobleached and then were assessed at the indicated times after photobleaching. Arrows indicate the centrioles that were photobleached. Bar, 1 µm. (D–H) For each time point before and after photobleaching, the relative fluorescence intensity of centrioles is shown. Results are a mean of three cells per bleaching experiment (one cell for centrin-GFP control). Data are means ± SD.
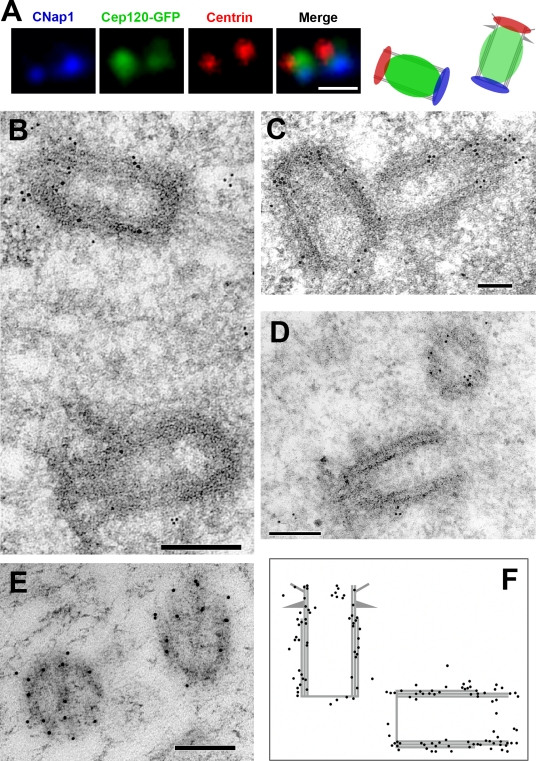
To refine the centriolar localization of Cep120, we costained RPE-1 cells expressing GFP-tagged Cep120 with antibodies against centrin (which marks the distal end of the centriole barrel) and C-Nap-1 (which stains the proximal end of centrioles; Fry et al., 1998). Cep120-GFP localized between these two markers, suggesting that Cep120 is distributed along the length of the centriole barrel (Fig. 3 A). We then used immunoelectron microscopy to test this directly. NIH3T3 cells were processed for transmission electron microscopy and incubated with anti-Cep120 antibodies followed by gold-conjugated secondary antibodies. Cep120-specific labeling was found along the barrel of the centrioles (Fig. 3, B–F), with little labeling in the centriole lumen (Fig. 3 F). In some cases, there was more Cep120 labeling associated with the daughter centriole (identified by the lack of appendages; Fig. 3 B), although this was not consistently observed.
Figure 3.
Cep120 is associated with the outer wall of centrioles. (A) Immunofluorescence localization of Cep120 relative to centriole markers. G1-phase RPE-1 cells expressing Cep120-GFP were fixed and stained for centrin to mark the distal end of centrioles (red), C-Nap-1 to mark the proximal end of centrioles (blue), and Cep120-GFP (green). The panel on the right is a representation of the observed spatial distribution. (B–E) Immunoelectron microscopy localization of Cep120 on centrioles. NIH3T3 cells were fixed and processed for transmission electron microscopy and incubated with anti-Cep120 antibodies followed by 10-nm gold-conjugated secondary antibodies. (F) Schematic of observed gold particle distribution (130 particles). Bars: (A) 0.5 µm; (B, C, and E) 100 nm; (D) 200 nm.
Cep120 dynamics at centrioles
Transport of proteins along microtubules is a common method of targeting them to the centrosome (Zimmerman and Doxsey, 2000). To test whether Cep120 association with centrioles is dependent on microtubules, NIH3T3 cells were treated with nocodazole to depolymerize microtubules. Cells were then fixed and stained for Cep120, α-tubulin, and centrin. Cep120 localization to centrioles was unaffected by the absence of microtubules (Fig. S1 C). Next, the relative amounts of cytoplasmic versus centriole-bound Cep120 were determined by fractionation and Western blot analysis of lysates from IMCD-3 cells. The majority of Cep120 protein was found in the centrosome-containing insoluble fraction (Fig. S1 D). There was very little Cep120 in the soluble cytoplasmic fraction; however, the protein could be extracted from centrosomes under high salt conditions (Fig. S1 D).
The dynamics of Cep120 association with the centrioles were investigated using FRAP. We generated an NIH3T3 cell line that stably expresses a Cep120-GFP fusion protein at near-endogenous levels (Fig. 4 A). Both immunofluorescence staining of these cells with anti-GFP antibody and live cell fluorescence of GFP showed the same daughter centriole enrichment of Cep120-GFP as with the endogenous protein (Fig. 4, B and C). In this cell line, both centrioles of an interphase cell were photobleached and assayed for fluorescence recovery by 4D confocal microscopy. Considering both centrioles together, ∼60% of the Cep120-GFP fluorescence recovered rapidly (halftime of ∼2 min), but the remaining 40% of the original signal did not recover during the observation period (Fig. 4, C and D; and Video 1). As a control, we examined the dynamics of centrin-GFP, a protein that is stably incorporated into centrioles (Paoletti et al., 1996). As expected, most of the centrin-GFP signal at centrioles did not recover after photobleaching (Fig. 4, C and H; and Video 2).
We next photobleached either the mother or daughter centriole alone in the Cep120-GFP cell line and measured the fluorescence levels at both centrioles over time. The observed recovery dynamics were similar for both centrioles, with a total fluorescence recovery of ∼60% at both mother and daughter centrioles (Fig. 4, C, E, and F; and Video 3). There are two possible reasons why recovery of Cep120-GFP reaches only 60% of initial levels: (1) because most Cep120 is associated with the centrioles (Fig. S1 D), there could be an insufficient amount of cytoplasmic Cep120 to replace the bleached pool at the centrioles; and (2) there could be two distinct pools of Cep120 at the centriole, one that is dynamic and exchangeable and another that is stably incorporated and not exchangeable. To distinguish between these two possibilities, we performed FRAP on NIH3T3 cells that overexpress Cep120-GFP so that the cytoplasmic pool of Cep120-GFP is presumably not limiting. The dynamics and total fluorescence recovery (∼60%) of Cep120-GFP in these cells were similar to the previous experiments (Fig. 4, C and G; and Video 4). This suggests that there are two distinct populations of Cep120 at centrioles, one pool that is dynamic and another that is stably incorporated. We also noted that the asymmetric distribution of Cep120 on mother and daughter centrioles was maintained in the overexpressing cells (unpublished data), suggesting that the available binding sites rather than the amount of Cep120 are limiting.
A C-terminal coiled-coil domain of Cep120 directs centriolar localization
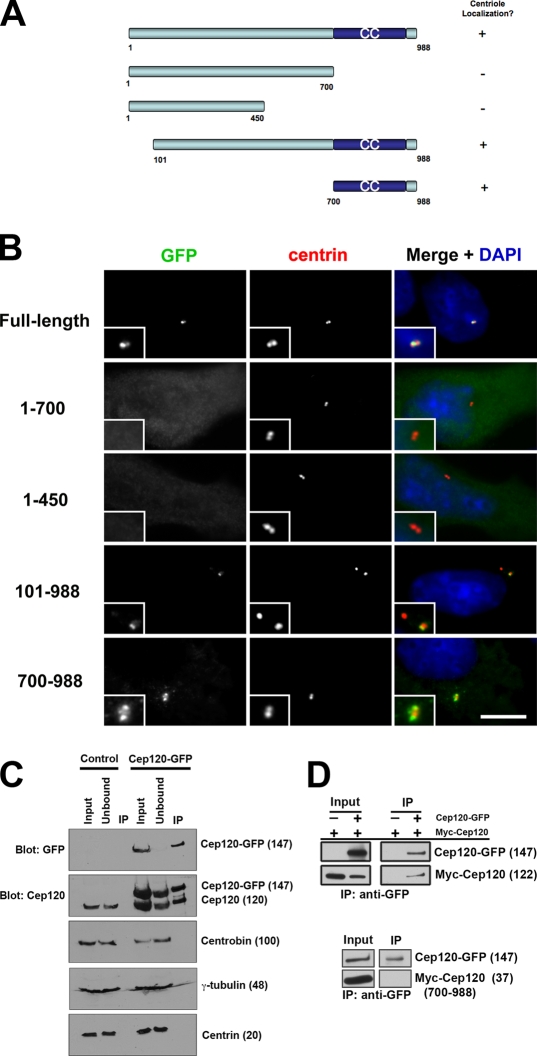
To identify functionally important domains of Cep120, we generated a series of deletion mutants (Fig. 5 A). These mutant forms of Cep120 were expressed in NIH3T3 cells as GFP fusion proteins, and their localization was determined by immunofluorescence. A C-terminal fragment of Cep120 containing a predicted coiled-coil domain (amino acids 700–988) is both necessary and sufficient for localization to centrioles (Fig. 5 B). Fragments lacking this region failed to localize to centrioles and displayed a diffuse cytoplasmic distribution. Similar results were obtained using the myc epitope to mark the expressed fragments (unpublished data).
Figure 5.
Cep120 forms a complex at centrioles. (A) Schematic of Cep120 deletion mutant constructs. Numbers correspond to amino acid position. CC, coiled-coil. (B) Localization of Cep120 deletion proteins. NIH3T3 cells transfected with the indicated Cep120 deletion–GFP fusion constructs and stained for GFP (green), centrin (red), and DNA (blue). Insets are magnified images of the centrosome region. Bar, 10 µm. (C and D) Self-interaction of Cep120. (C) Cep120-GFP was immunoprecipitated with anti-GFP antibodies from NIH3T3::Cep120-GFP cells or control NIH3T3 cells, and samples were probed with the indicated antibodies. (D) Cep120-GFP was coexpressed with myc-Cep120 or myc-Cep120 (700–988) in reticulocyte lysate, immunoprecipitated with anti-GFP antibodies, and probed with anti-GFP and anti-myc antibodies. Numbers in parentheses indicate molecular mass in kilodaltons.
A simple explanation for the ability of the putative coiled-coil domain to direct localization to the centriole would be that it is able to interact with endogenous Cep120. We first tested for self-interaction between full-length Cep120 molecules in vivo. Cep120-GFP was immunoprecipitated from the stably expressing NIH3T3 cells (Fig. 4 A). Endogenous Cep120 coimmunoprecipitated with Cep120-GFP from these cells (Fig. 5 C), and this interaction was maintained even under higher stringency buffer conditions (0.6 M NaCl; not depicted). This suggests that Cep120 either interacts with itself or is part of a larger complex with at least two molecules of Cep120. We also probed these precipitates with antibodies against other centriolar proteins, including centrin, γ-tubulin, and centrobin, but failed to identify any that coimmunoprecipitate with Cep120-GFP (Fig. 5 C). To test whether Cep120 is able to interact with itself, full-length Cep120-myc and Cep120-GFP were expressed together in reticulocyte lysate, which lacks most centrosomal proteins. Cep120-myc efficiently coimmunoprecipitated with Cep120-GFP, consistent with self-interaction (Fig. 5 D). Interestingly, the putative coiled-coil domain was not able to interact with full-length Cep120 in this experiment (Fig. 5 D), suggesting that this domain directs centriolar localization by interaction with some other binding partner.
Cep120 is required for centriole assembly
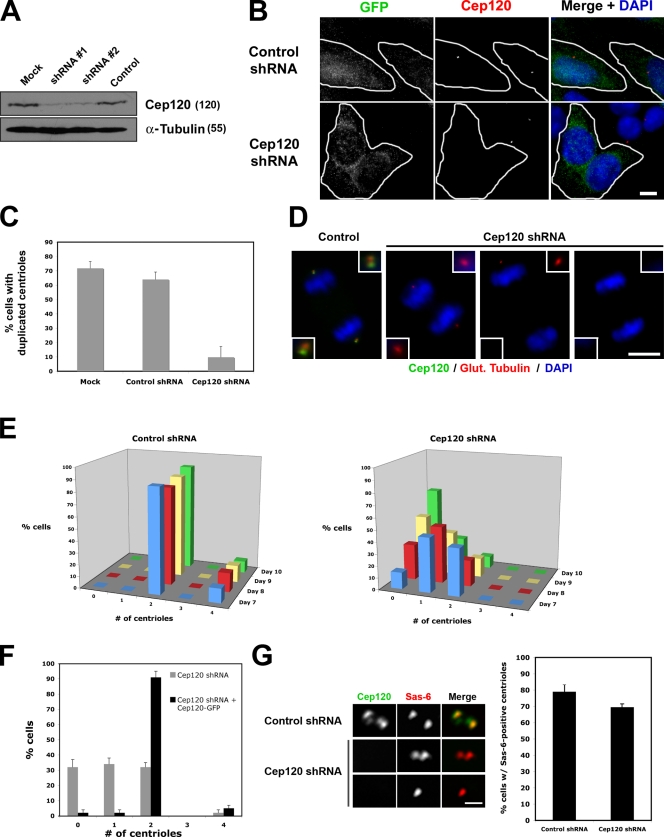
To determine the function of Cep120, we used RNA interference to deplete the protein from cells. Primary mouse embryonic fibroblasts (MEFs) were infected with a lentivirus expressing GFP to identify infected cells and short hairpin RNA (shRNA) targeting Cep120. Maximal depletion of Cep120 was observed 7 d after infection by Western blotting (Fig. 6 A) and by indirect immunofluorescence (Fig. 6 B). Although depletion of some centrosomal proteins can lead to cell cycle arrest in G1 (Mikule et al., 2007), infected cells apparently continued to proliferate throughout the peak of Cep120 depletion. This lack of an effect on cell cycle progression was confirmed in two ways. First, the fraction of cells entering S phase 7 d after infection was assessed by EdU (5-ethynyl-2′-deoxyuridine) labeling. 66% of cells expressing control shRNA incorporated the label EdU into nuclear DNA over a 16-h pulse compared with 61% of cells expressing Cep120 shRNA (n = 400 for each sample), confirming that cells proceed through S phase in the absence of Cep120 (Fig. S1 E). Second, the fraction of cells dividing over time was determined by time-lapse microscopy of live cells. Imaging of cells 7 d after infection (20 cells per sample) was performed at 10-min intervals for 36 h. Cells expressing Cep120 shRNA and control shRNA divided at similar rates (17/20 and 19/20 cells divided at least once, respectively).
Figure 6.
Cep120 is required for centriole duplication. (A) Cep120 depletion from MEF cells by lentiviral expression of shRNAs. Lysates were probed for Cep120 and α-tubulin. Numbers in parentheses indicate molecular mass in kilodaltons. (B) Loss of Cep120 in MEFs infected with lentivirus expressing control or Cep120 shRNA and GFP to identify infected cells (outlined in white). Bar, 10 µM. (C) Centriole duplication in S phase–arrested MEFs after treatment with the indicated shRNAs. Results shown are the mean of three independent experiments (n = 600 for each sample). (D) Representative images showing reduction of centriole number at spindle poles of mitotic MEFs depleted of Cep120. Insets are magnified images of the centrosome region. (E) Quantification of centriole number over time in control and Cep120-depleted MEFs; centrioles were counted beginning on day 7 after infection. n = 400 for each time point. (F) The loss-of-centriole phenotype in Cep120-depleted human RPE-1 cells is rescued by expression of mouse Cep120. n = 400 for each time point. (G) Sas-6 recruitment to centrioles is unaffected by loss of Cep120. S phase–arrested MEFs were stained for Cep120 (green) and Sas-6 (red). n = 200 for each sample. (E–G) Results shown are a mean of two independent experiments. Data are means ± SD. Bar, 0.5 µm.
We tested whether Cep120 was required for centriole duplication by depleting cells of Cep120 and counting centrioles. MEFs were mock infected or infected with lentivirus expressing shRNA targeting Cep120 or a control shRNA. On day 7 after infection, cells were accumulated in S phase by treatment with thymidine to maximize the fraction of cells that would have duplicated centrioles and then were fixed and stained for GFP (to identify infected cells), centrin (to count centrioles), and Cep120. In cultures that were mock infected or infected with control shRNA lentivirus, most cells had duplicated centrioles (72 and 64% of cells with duplicated centrioles, respectively; Fig. 6 C). In contrast, only 10% of Cep120-depleted cells had duplicated centrioles (Fig. 6 C), consistent with a defect in centriole duplication.
We noted that many Cep120-depleted cells had either a single centriole or no centrioles, as determined by staining for several centriolar proteins (centrin, acetylated or glutamylated tubulin, Cep135, ninein, centrobin, and cenexin). We hypothesized that this was caused by successive cell divisions without centriole duplication, resulting in a progressive dilution of centriole number (Fig. 6 D). To test this, centrioles in control and Cep120-depleted cells were counted over time. Cells expressing control shRNA had either two or four centrioles, consistent with a typical cell cycle distribution (Fig. 6 E, left). A significant fraction of Cep120-depleted cells had either one or zero centrioles at day 7 after infection (Fig. 6 E, right), and this fraction increased over the next 3 d (Fig. 6 E, right), consistent with the hypothesis. This phenotype is a result of the loss of Cep120 rather than a nonspecific effect of RNAi, as it can be complemented in human RPE-1 cells depleted of endogenous Cep120 by expression of mouse Cep120-GFP immune to the shRNA (Fig. 6 F).
We next addressed whether Cep120 is required for initiation of centriole assembly or a later step. The Sas-6 protein is recruited to the site of new centriole assembly early in the process and is required for centriole duplication (Leidel et al., 2005). In mammalian cells, Sas-6 is absent from the mother and daughter centrioles in G1 and is recruited during new daughter centriole assembly in G1/S (Strnad et al., 2007). To test whether Sas-6 could be recruited to centrioles in the absence of Cep120, MEFs were depleted of Cep120 and treated to enrich for S-phase cells with duplicated centrioles, as described in Fig. 6 C. In S phase–arrested cultures that were infected with control shRNA, the majority of cells had two Sas-6 foci (79%; Fig. 6 G), representing the sites of new centriole assembly. Interestingly, most Cep120-depleted cells also contained Sas-6 foci (70%; Fig. 6 G). Some cells had only one Sas-6 focus, which was never observed in control cells. Based on the aforementioned results, these likely represent cells that had experienced a failure of centriole duplication in the previous cycle and, therefore, had only one centriole (Fig. 6 G, bottom row). This suggests that Cep120 acts downstream of Sas-6 recruitment in the centriole assembly pathway.
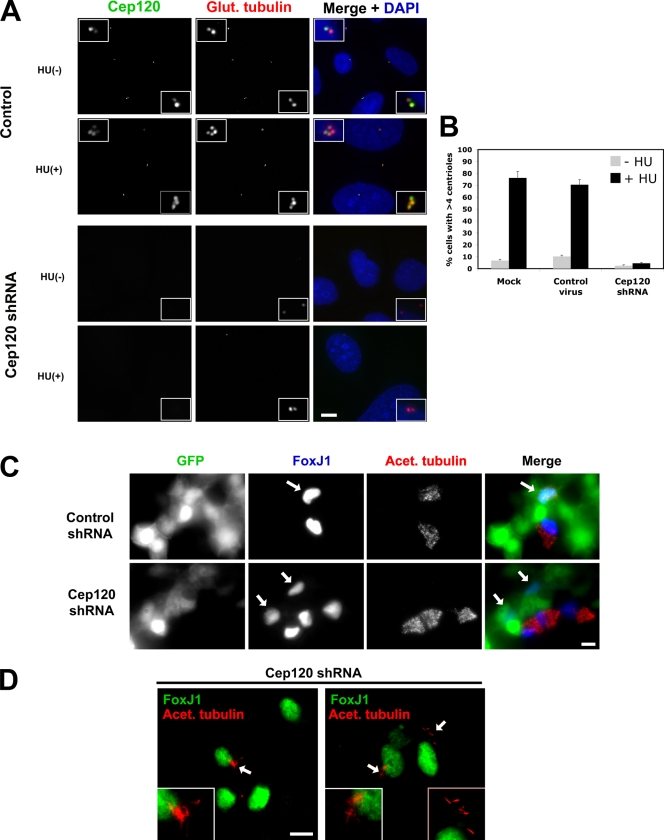
Human U2OS cells overduplicate centrioles when arrested in S phase (Meraldi et al., 1999), and we used this phenotype to further examine the effect of Cep120 depletion on centriole amplification. U2OS cells infected with lentivirus expressing control shRNA or shRNA targeting Cep120 were treated with hydroxyurea to block DNA replication on day 7 after infection and incubated for an additional 48 h. Whereas a high percentage (70–80%) of control cells had extra centrioles indicative of S-phase arrest overduplication, Cep120 depletion inhibited the formation of extra centrioles (Fig. 7, A and B).
Figure 7.
Cep120 is required for centriole amplification. (A) Block of centriole overduplication in Cep120-depleted cells. U2OS cells were infected with lentivirus expressing shRNA targeting Cep120 or a control shRNA, arrested in S phase by hydroxyurea (HU) treatment for 48 h, and stained for Cep120 (green), glutamylated tubulin (red), and DNA (blue). (B) Quantification of centriole overduplication in Cep120 shRNA, control shRNA, or mock-infected U2OS cells. Results shown are a mean of two independent experiments (n = 400 for each sample). (C) Block of centriole and cilia assembly in Cep120-depleted MTECs. MTECs were infected with Cep120 shRNA or control shRNA lentivirus 2 d before establishing ALI and then were stained at ALI+8 for GFP to mark infected cells (green), FoxJ1 to identify cells committed to the ciliated cell fate (blue), and acetylated tubulin (red). Arrows indicate cells infected with shRNA-expressing lentivirus. (D) Examples of cilia formation in MTECs with partial centriole duplication defects. Cells were stained for FoxJ1 (green) and acetylated tubulin (red; arrows mark regions magnified in insets). Insets are magnified images of the centrosome region. Data are means ± SD. Bars, 10 µm.
Finally, we tested the role of Cep120 in centriole formation during ciliogenesis in MTECs. Centriole formation in these multiciliated cells differs from canonical centriole duplication in four ways: (1) cells go from having two centrioles to several hundred centrioles; (2) a mother centriole simultaneously nucleates more than one daughter centriole; (3) noncentriolar structures (deuterosomes) facilitate the simultaneous nucleation of multiple centrioles; and (4) centrioles are generated in terminally differentiated, nondividing cells. Cep120 is up-regulated during centriole biogenesis in differentiating tracheal epithelial cells (Fig. 1 B). MTECs were harvested and cultured to confluence, infected with shRNA-expressing lentivirus 2 d before establishing ALI (ALI-2), and then assayed on days ALI+4–ALI+8. Cells were examined by immunofluorescence with antibodies against GFP (to mark infected cells), FoxJ1 (to identify cells committed to the ciliated cell fate), and acetylated tubulin (to mark cilia and basal bodies). MTEC cultures expressing control shRNA assembled centrioles and cilia (Fig. 7 C) as previously described (Vladar and Stearns, 2007). In contrast, 87% of MTECs expressing Cep120 shRNA failed to assemble centrioles and, thus, cilia (Fig. 7 C). Quantification of the FoxJ1 levels in control and Cep120-depleted cells (n = 20) revealed no difference in expression, indicating that the centriole assembly defect was not caused by a block in differentiation (Fig. 7 C and not depicted). In the remaining 13% of cells depleted for Cep120, the cells assembled a few centrioles, and these centrioles were competent to form cilia (Fig. 7 D). Consistent with our results on cycling cells, the role of Cep120 in these differentiated cells appears to be in the initial assembly step of centrioles rather than the function of these centrioles as basal bodies.
Cep120 asymmetry is relieved during centriole duplication and requires daughter centriole assembly
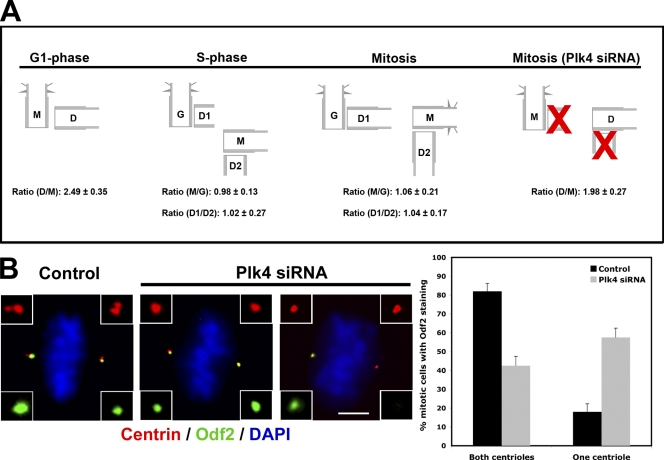
Cep120 is enriched on the daughter centriole in G1, and this asymmetric localization pattern is maintained throughout the cell cycle (Fig. 2 A). Importantly, Cep120 signal was brighter on the newly formed daughter centrioles, indicating that Cep120 asymmetry must be newly established at some point after the mother and daughter centrioles transitioned to grandmother and mother, respectively, and began duplication (Fig. 2 B). We sought to determine more precisely when this switch occurs. Asynchronous U2OS cells were fixed and stained with antibodies against Cep120 and cenexin/Odf2, which marks the mother centriole in G1 and both grandmother and mother centrioles during mitosis (Lange and Gull, 1995). The combination of the two markers allowed us to identify the relative ages of centrioles in G1, S, and mitosis. In G1 cells, the amount of Cep120 on daughter centrioles was ∼2.5-fold that on mother centrioles (Fig. 8 A). However, in S phase, the amount of Cep120 on these centrioles, now grandmother and mother, was approximately equal (Fig. 8 A). The amount of Cep120 on the two daughter centrioles within a cell was also approximately equal. Thus, the switch from a G1 daughter centriole with high Cep120 to a mother centriole with low Cep120 occurs in early S phase, coincident with centriole duplication. These relative ratios of Cep120 were maintained through mitosis (Fig. 8 A).
Figure 8.
Centriole maturation requires daughter centriole assembly. (A) Schematic showing relative Cep120 levels on centrioles at stages of the cell cycle. The Cep120 fluorescence intensity at daughter (D, D1, and D2), mother (M), and grandmother (G) centrioles in U2OS cells was quantified at different cell cycle stages and presented as the indicated ratios. Mitotic U2OS cells with single centrioles at each pole were generated by depletion of Plk4. Results shown are a mean of two independent experiments (n = 50 for each cell cycle stage). (B) Maturation of the daughter centriole is dependent on the presence of a procentriole. (left) U2OS cells were transfected with control or Plk4 siRNA and stained for centrin (red), cenexin/Odf2 (green), and DNA (blue). (right) Mitotic cells with two centrioles (control siRNA) or single centrioles (Plk4 siRNA) at each spindle pole were scored for the presence or absence of cenexin/Odf2. Results shown are a mean of two independent experiments (n = 200). Insets are magnified images of the centrosome region. Data are means ± SD. Bar, 10 µm.
Next, we asked whether this switch from asymmetric to symmetric distribution of Cep120 on grandmother and mother centrioles was caused by passage of cells through S phase or whether it depended on the assembly of daughter centrioles. To test this, daughter centriole formation was blocked by siRNA-mediated depletion of Plk4 in U2OS cells (Habedanck et al., 2005). Failure of daughter centriole formation results in cells having only two centrioles in the ensuing mitosis, the original mother and daughter, one at each spindle pole (Habedanck et al., 2005). Quantification of Cep120 ratios in such cells revealed that the asymmetric distribution on mother and daughter was maintained (Fig. 8 A). This suggests that daughter centriole assembly itself, and not simply passage through S phase, is required for breaking Cep120 asymmetry.
Conversion of a daughter centriole with high Cep120 to a mother centriole with low Cep120 is a form of centriole maturation in the cell cycle; the other commonly considered centriole maturation event is the acquisition by that centriole of centriolar appendage proteins, which occurs at G2/M (for review see Hoyer-Fender, 2010). Because Cep120 level conversion required daughter centriole assembly, we asked whether the acquisition of appendage proteins might share this requirement. To test this, U2OS cells were depleted of Plk4 and then stained for the centriole appendage protein cenexin/Odf2, which accumulates on the new mother centriole during G2/M (Lange and Gull, 1995). In ∼80% of control mitotic cells, both the mother and grandmother centrioles were labeled with cenexin/Odf2 (Fig. 8 B). In contrast, more than half of Plk4-depleted mitotic cells had only a single cenexin/Odf2-positive centriole (Fig. 8 B), presumably representing cases in which the centriole that would be the new mother, had duplication occurred, failed to acquire this appendage protein. Thus, both reduction of Cep120 and acquisition of appendages fail on a mother centriole lacking an associated daughter by virtue of Plk4 depletion.
Discussion
We identified CEP120 as a gene up-regulated during ciliogenesis, which had previously been shown to function in interkinetic nuclear migration in neural progenitors (Xie et al., 2007) and was identified in a proteomic analysis of purified human centrosomes (Andersen et al., 2003). The putative orthologue of Cep120 in the ciliated alga C. reinhardtii (Uni2) is required for basal body maturation and ciliogenesis (Piasecki et al., 2008). The Uni2 protein was shown to localize to both probasal bodies and mature basal bodies in C. reinhardtii (Piasecki et al., 2008, Piasecki and Silflow, 2009), and uni2 mutants display structural defects in the transition zone at the distal end of the basal bodies. However, basal body assembly itself is not impaired in uni2 mutants, and it is not clear whether Uni2 and Cep120 are truly functional orthologues. We have shown that Cep120 is associated with centrioles in the context of both centrosomes and basal bodies, that Cep120 is enriched at daughter centrioles in cycling cells, and that Cep120 is required for centriole formation in all contexts tested. Here, we consider the implications of these results for centriole duplication and relate this molecular function to the observed phenotypic defect in neural differentiation.
The two centrioles in a G1 animal cell differ in age: the younger (daughter) formed in the previous cell cycle, and the older (mother) formed at least one cell cycle prior. The mother centriole has associated appendage structures, which the daughter only acquires after becoming a mother after centriole duplication. Several proteins are specific to the mother centriole and thought to be part of these appendages, including Odf2/cenexin (Lange and Gull, 1995), Cep164 (Graser et al., 2007), centriolin (Gromley et al., 2003), ninein (Ou et al., 2002), and ε-tubulin (Chang and Stearns, 2000). In contrast, few proteins are known to be preferentially associated with the daughter centriole. Some proteins, such as Sas-4 (Leidel and Gönczy, 2003) and Sas-6 (Leidel et al., 2005), are recruited to daughter centrioles early in duplication, but this enrichment is lost shortly thereafter.
The one previously described example of a daughter centriole–associated protein is centrobin; Zou et al. (2005) identified centrobin as a centriole-associated protein that asymmetrically localizes to the daughter centriole. siRNA of centrobin inhibited centriole duplication and resulted in centrosomes with one or no centriole, demonstrating that centrobin is required for centriole duplication. Also, inhibition of centriole duplication by centrobin depletion led to impaired cytokinesis (Zou et al., 2005). Subsequently, it was reported that depletion of centrobin caused significant reduction of microtubule-organizing activity, cell shrinkage, defects in spindle assembly, and abnormal nuclear morphology (Jeong et al., 2007). Our finding that Cep120 is also enriched at daughter centrioles led us to test whether it plays a similar role in cells as centrobin.
Depletion of Cep120 inhibited centriole assembly in three similar, yet distinct, pathways: (1) centriole duplication in the G1/S phase of cycling cells; (2) centriole overduplication in S phase–arrested cells; and (3) centriole amplification in ciliated epithelial cells. These data strongly support our hypothesis that Cep120 is essential for centriole assembly and also show that centriole duplication in cycling cells and centriole amplification in multiciliated cells share common mechanisms despite their morphological differences. During the preparation of this work, Cep120 was identified in a high-throughput RNAi screen for genes having a centriole duplication defect (Hutchins et al., 2010), strongly supporting our findings. Unlike centrobin, however, we did not find defects in microtubule organization or cytokinesis in cells lacking Cep120. Furthermore, although depletion of Cep120 led to loss of centrioles, the ability of cells to organize a microtubule cytoskeleton was not grossly affected (unpublished data). Finally, loss of Cep120 function did not perturb cell cycle progression or cell division (Fig. 6 D and Fig. S1 F). Thus, Cep120 appears to play a specific role in the assembly of centrioles but not other centrosome-associated functions. The observation that Sas-6 recruitment still occurs in the absence of Cep120 suggests that Cep120 might be required either for a specific post–Sas-6 event in centriole assembly or might be required for stabilizing structures that form after Sas-6 recruitment.
We found that the majority of Cep120 is associated with centrioles, suggesting that Cep120 might be stably incorporated into the structure of the centrioles. We tested this by performing FRAP experiments on cells stably expressing Cep120-GFP. Surprisingly, FRAP analysis showed that Cep120 at centrioles exchanges fairly rapidly, indicating that the association with the centriole is dynamic. The recovery dynamics were the same when both centrioles were bleached, when the mother and daughter centrioles were bleached separately, and even in the presence of excess cytoplasmic Cep120-GFP. This behavior is distinct from that of Nudel (Guo et al., 2006) and β-catenin (Bahmanyar et al., 2008), which exhibit almost complete recovery at the centrosome after photobleaching, and from centrin, which shows almost no FRAP (Pearson et al., 2009; Prosser and Fry, 2009). We posit that there are two populations of Cep120 at centrioles, one pool that is dynamic and another that is stably incorporated. One possibility is that some of the Cep120 may be incorporated into the centriolar structure, whereas the rest may perform a more transient role, perhaps functioning as a scaffold for assembly. As centrioles complete assembly and subsequently mature into mother centrioles, a portion of Cep120 at those centrioles is lost, accounting for the dynamic population and for the daughter centriole enrichment.
Although the mechanism of centriole biogenesis in mammalian cells is poorly understood, many proteins have been implicated in the process (Jaspersen and Stearns, 2008). Some of these proteins, such as the kinase Plk4 and the structural protein Sas-6, are not only essential for assembly of new centrioles but are also regarded as “master regulators” of the process because overexpression of either can stimulate the formation of centrioles (Habedanck et al., 2005; Strnad et al., 2007). We found that overexpression of Cep120 in a variety of cell types did not lead to overduplication of centrioles (unpublished data). Similarly, overexpression of the C-terminal coiled-coil domain, which is both necessary and sufficient for its centriolar targeting, did not cause any defects in centriole number.
A previous study examined Cep120 in neural progenitor cells during neocortical development in the mouse brain (Xie et al., 2007). The investigators determined that Cep120 is involved in regulating centrosome-associated microtubules in the neural progenitors. Silencing of Cep120 in the developing neocortex impaired interkinetic nuclear migration, an essential step in proper neocortical development, as well as neural progenitor self-renewal. These results were interpreted as revealing a critical role for Cep120 in interkinetic nuclear migration by regulating microtubules coupling the centrosome and the nucleus (Xie et al., 2007). Although we did not observe a defect in microtubule organization after Cep120 depletion in the fibroblast and epithelial cell lines in which we conducted our experiments, this could be because of differences in cell type. The neural progenitor cells are specialized in that they undergo a microtubule-dependent nuclear migration step during their differentiation, which clearly involves centrosome and microtubule function, although exactly how is not clear. Thus, microtubule dynamics during the movement of these cells might be more sensitive to loss of Cep120 or, perhaps, to loss of centrioles. Alternatively, the involvement of centrosomal proteins in neocortical neurogenesis might be more general, as most of the identified causative genes for autosomal-recessive primary microcephaly (a disorder likely caused by abnormal neurogenesis) encode centrosomal proteins (Hung et al., 2000; Bond et al., 2005; Kouprina et al., 2005; Zhong et al., 2005).
Previous studies on the mechanisms by which centrosomal proteins influence neurogenesis have focused on their roles in mitotic spindle assembly and orientation (Feng and Walsh, 2004; Fish et al., 2006). The findings by Xie et al., (2007) suggest that sustaining interkinetic nuclear migration may also be an important mechanism by which microtubule-regulating proteins maintain the neural progenitor pool during neocortical development. In the case of Cep120, this might occur by the previously described interaction with transforming acidic coiled-coil proteins (Xie et al., 2007), but our results suggest as an alternative that the root cause of the impaired interkinetic nuclear migration phenotype may be loss of centrioles in the absence of Cep120. It is tempting to speculate that loss of other core centriolar proteins would lead to similar phenotypes in the developing neocortex and that the main defect in diseases such as microcephaly may be caused by loss of centrioles.
Finally, our results reveal something of the logic of the centriole maturation cycle. Both reduction in Cep120 level at mother centrioles and acquisition of appendages by mother centrioles were dependent on formation of a daughter centriole. This suggests that the cellular machinery responsible for these maturation events might use the presence of an adjacent daughter centriole as the cue for their initiation, thus limiting them to mother centrioles. We note the interesting possibility that it is the activity of Plk4 itself, which was depleted to prevent daughter centriole formation, that is required for these events, rather than daughter centriole formation by itself. Further work will test the relationship between these two maturation events and the mechanism by which mother and daughter centrioles are distinguished.
Materials and methods
Cell culture and media
Cultured mammalian cells used in this study, including NIH3T3, Hek293T, RPE-1, U2OS, and MEFs, were grown in DME with 10% FBS (Invitrogen). IMCD-3 cells were grown in DME/F12 media with 10% FBS. MTEC cultures were established as previously described (You et al., 2002; Vladar and Stearns, 2007). C57BL/6J mice were sacrificed at 2–4 mo of age, and trachea were excised, opened longitudinally to expose the lumen, and placed in 1.5 mg/ml pronase E in F12K nutrient mixture (Invitrogen) at 4°C overnight. Tracheal epithelial cells were dislodged by gentle agitation and collected in F12K with 10% FBS. After centrifugation, cells were treated with 0.5 mg/ml DNase I for 5 min on ice and centrifuged at 4°C for 10 min at 400 g. Cells were resuspended in DME/F12 with 10% FBS and plated in a tissue culture dish for 3 h at 37°C with 5% CO2 to adhere contaminating fibroblasts. Nonadhered cells were collected, concentrated by centrifugation, resuspended in an appropriate volume of MTEC-Plus medium (as described in You et al., 2002), and seeded onto Transwell-Clear permeable filter supports (Corning). ALI was established 2 d after cells reached confluence by feeding MTECs serum-free medium (You et al., 2002) only in the lower chamber. Cells were cultured at 37°C with 5% CO2, and media were replaced every 2 d. All chemicals were obtained from Sigma-Aldrich unless otherwise indicated. All media were supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin, and 0.25 mg/ml Fungizone (all obtained from Invitrogen).
Plasmids and transfections
A full-length cDNA clone of mouse Cep120 was obtained from Thermo Fisher Scientific (clone 6328829). The Cep120 ORF (2,967 bp encoding 988 amino acids) was amplified by PCR and cloned into vector pCS2+ at unique FseI and AscI sites in the multiple cloning site (gift of G. Fang, Genentech, San Francisco, CA). This vector contains a C-terminally located EGFP gene, resulting in a Cep120-GFP fusion protein. Alternatively, the full-length Cep120 fragment was cloned into a pCS2+ vector containing an N-terminal 5× myc tag. For generation of truncated versions of Cep120, PCR amplification of the desired Cep120 cDNA sequences was performed to yield fragments that encode amino acids 1–495, 1–700, 101–988, and 700–988. The PCR products were cloned into the aforementioned vectors and verified by sequencing. Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Lentivirus production and cell infection
HIV-derived recombinant lentivirus expressing shRNA targeting Cep120 (shRNA#1 nucleotides 845–863 and shRNA#2 nucleotides 2,589–2,607 of Cep120 cDNA) was made using the pLentiLox 3.7 lentiviral transfer vector that also expresses GFP from a CMV promoter to mark infected cells (Rubinson et al., 2003). Lentivirus expressing GFP-tagged Cep120 was made using the lentiviral transfer vector pRRL.sin-18.PPT.PGK.GFP.pre (Follenzi et al., 2000) by inserting the Cep120 ORF in frame with the GFP cassette at the MroI site. This yields a C-terminally tagged Cep120 fusion protein. All constructs were verified by sequencing. Recombinant lentivirus was produced by cotransfection of Hek293T cells with the appropriate transfer and lentiviral helper plasmids (pCMVDR8.74 packaging vector and pMD2.VSVG envelope vector; Dull et al., 1998) using the calcium phosphate coprecipitation method. The medium was exchanged 18 h after transfection, and lentiviral supernatant was harvested after another 48 h. The lentiviral supernatants were concentrated 100- to 500-fold by ultracentrifugation at 20°C for 180 min at 50,000 g.
All lentiviral constructs were titered on NIH3T3 cells. For infection of cultured mammalian cells, the cells were seeded onto 24-well tissue culture plates the day before infection. The medium was removed on the next day and replaced by medium containing lentivirus at an MOI of 1. Virus-containing medium was removed 24 h after infection. If necessary, another round of infection was performed. Cells were assayed from 2 to 12 d after initial infection.
To infect MTECs, cells were grown to confluence on filters, the medium was removed, and cells were rinsed twice with PBS. Epithelial tight junctions were disrupted by treatment with 12 mM EGTA in 10 mM Hepes, pH 7.4, at 37°C for 20 min. Cells were rinsed twice with PBS. Fresh medium was added to the bottom of the dish, and a mix of lentivirus (at an MOI of 1) and medium was placed on top of the cells. The plate was sealed with parafilm and centrifuged at 32°C for 80 min at 1,500 g. After centrifugation, the plate was unsealed and placed at 37°C. Virus-containing medium was removed 24 h after infection. ALI was established 2 d later. Control infections used virus made from transfer vectors without the transgene or short hairpin construct of interest.
Immunofluorescence and immunoelectron microscopy
For indirect immunofluorescence, cultured cells or MTECs were rinsed twice with PBS and fixed in either 100% ice-cold methanol at −20°C for 10 min or with 4% paraformaldehyde in PBS at room temperature for 10 min, depending on antigen. After fixation, cells were rinsed twice with PBS. For MTEC samples, filters were excised from their plastic supports and cut into quarters to provide multiple equivalent samples for parallel staining. Cells were briefly extracted with 0.2% Triton X-100 in PBS and blocked for 1 h at room temperature with 3% BSA (Sigma-Aldrich) in PBS. Samples were incubated with primary antibodies for 1 h at room temperature or 4°C overnight. Primary antibodies used in this study were rabbit anti-Cep120 (1:2,000), mouse anti-acetylated tubulin (clone 6-11b-1; 1:5,000; Sigma-Aldrich), mouse anti-centrin (clone 20H5; 1:2,000; gift from J. Salisbury, Mayo Clinic, Rochester, NY), mouse anti-glutamylated tubulin (GT335; 1:2,000; gift from C. Janke, Centre de Recherches de Biochimie Macromoléculaire, Montpellier, France), rabbit anti-cenexin (1:1,000; a gift from K. Lee, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD; Soung et al., 2006), rabbit anti–C-Nap-1 (1:100), mouse anti-GFP (clone 3E6; 1:2,000; Invitrogen), mouse anti-FoxJ1 (1:3,000; gift from S. Brody, Washington University School of Medicine, St. Louis, MO), and mouse anti–Sas-6 (1:200; Santa Cruz Biotechnology, Inc.). Alexa Fluor dye–conjugated secondary antibodies (Invitrogen) were used at a dilution of 1:500 at room temperature for 1 h. Samples were mounted using Mowiol mounting medium containing N-propyl gallate (Sigma-Aldrich). Images were captured using Openlab 4.0.4 (PerkinElmer) controlling a microscope (Axiovert 200M; Carl Zeiss, Inc.).
For immunoelectron microscopy, NIH3T3 cells grown on Aclar strips were rinsed with PBS, fixed with 4% paraformaldehyde for 10 min, treated with 1 mg/ml sodium borohydride in PBS for 10 min, and then extracted with 0.5% Triton X-100 in PBS for 2 min. Samples were dehydrated in a series of ethanol (30, 50, 70, and 95%) and then infiltrated and embedded with LR White resin (Ted Pella, Inc.). Gold sections were cut with a diamond knife, rehydrated with PBS for 10 min, blocked with 3% BSA in PBS for 30 min, and then incubated with rabbit anti-Cep120 (1:50) overnight at 4°C. Samples were rinsed three times with PBS and then incubated with 10-nm gold-conjugated anti–rabbit secondary antibodies for 2 h at room temperature. Cells were rinsed three times with PBS, postfixed with 2% glutaraldehyde for 30 min at room temperature, and then stained with uranyl acetate and lead citrate. Grids were examined on an electron microscope (TEM 1230; JEOL).
Centriole duplication and cell cycle analysis
Cells were arrested in G0 by incubation with DME containing 0.5% FBS for 24–48 h. For S-phase arrest, cells were incubated in growth media containing 2 mM thymidine for 12 h and then were incubated in fresh media for 12 h followed by media containing 2 mM thymidine for 12 h. For localization of Cep120 in the absence of microtubules, cells grown on coverslips were incubated with media containing 0.1 µg/ml nocodazole at 37°C for 1–4 h and then were fixed immediately with 100% ice-cold methanol.
To analyze centriole duplication in the absence of Cep120, cells were infected with lentivirus expressing control shRNA or shRNA targeting Cep120. Loss of Cep120 protein by Western blotting and immunofluorescence was typically seen ∼7 d after infection. On day 7, cells were arrested in S phase by incubation with media containing 2 mM thymidine for 16 h and then were fixed with 100% ice-cold methanol. For the overduplication experiments, U2OS cells infected with lentivirus expressing control shRNA or shRNA targeting Cep120 were incubated with 4 mM hydroxyurea-containing growth media on day 7 after infection for an additional 24–48 h before fixation with 100% ice-cold methanol.
To determine whether cells lacking Cep120 progress through the cell cycle, cells were infected with lentivirus expressing control shRNA or shRNA targeting Cep120. 7 d after infection, cells were incubated for 16 h with growth media containing 1 µM EdU, a modified nucleoside that is incorporated during DNA synthesis (Click-iT EdU cell proliferation assay kit; Invitrogen). Cells were fixed with 100% ice-cold methanol and processed per the manufacturer’s instructions. Alternatively, infected cells were grown in coverglass-bottom dishes (Laboratory-Tek II Chamber; Nalge Nunc International), mounted on a microscope (Diaphot; Nikon) equipped with a humidified chamber, and maintained at 37°C and 5% CO2. Time-lapse images were captured at 10-min intervals for 24–36 h.
Cell extracts and immunoprecipitation
To obtain whole-cell extracts for Western blotting, cells were washed in PBS and lysed by the addition of SDS sample buffer. For immunoprecipitation of Cep120-GFP, NIH3T3 cells stably expressing Cep120-GFP at near-endogenous levels (see FRAP analysis) were washed in PBS and lysed in Hepes buffer (50 mM Hepes, pH 7.5, 1 mM MgCl2, 1 mM EGTA, and 150–600 mM NaCl) containing protease inhibitors and 0.5% Triton X-100 for 30 min on ice. After centrifugation for 15 min at 16,000 g at 4°C, cleared lysates were obtained. Extracts were incubated with goat anti-GFP antibodies (Rockland) for 1–16 h at 4°C. Protein G beads (Sigma-Aldrich) were added, and the mixture was incubated for an additional hour at 4°C. The beads were pelleted and washed five times with Hepes buffer with 0.5% Triton X-100. Samples were prepared for SDS-PAGE by boiling in sample buffer. Antibodies used for Western blotting were rabbit anti-Cep120 (1:10,000), mouse anti–γ-tubulin (1:10,000; Sigma-Aldrich), rabbit anti-p38 kinase (1:5,000; Santa Cruz Biotechnology, Inc.), goat anti-GFP (1:5,000; Rockland), mouse anti-centrin (clone 20H5; 1:10,000), and rabbit anti-centrobin (1:100; gift from K. Rhee, Seoul National University, Seoul, South Korea).
FRAP analysis
NIH3T3 cells stably expressing Cep120-GFP were generated by infecting cells with lentivirus containing a Cep120-GFP transfer vector. Enrichment for cells expressing Cep120-GFP was achieved by FACS sorting. Western blotting and immunofluorescence microscopy confirmed that these cells express Cep120-GFP at a level comparable to endogenous levels. NIH3T3 cells stably expressing centrin-GFP were generated previously in our laboratory (Anderson and Stearns, 2009).
Photobleaching experiments were performed on a confocal laser scanner (LSM 510; Carl Zeiss, Inc.) using a 100× oil objective (NA 1.4) and a scan zoom of 5. Cells were cultured in coverglass-bottom dishes (Laboratory-Tek II Chamber) and maintained on the stage at 5% CO2 and 37°C. Before photobleaching, a series of digital optical sections consisting of 14 sections in the Z axis (total of 6 µm) was captured, and then a region of interest encompassing the centrosome was selected and photobleached with seven iterations at 100% laser power (488-nm argon laser). Image stacks were subsequently captured immediately after photobleaching and at 1-min intervals for 15–30 min. For each time point, 3D image reconstructions were produced, and the relative fluorescent intensity was calculated and plotted against time.
Online supplemental material
Fig. S1 shows data relevant to the Cep120 domain structure, localization, biochemical properties, and depletion phenotype. Video 1 shows photobleaching of Cep120-GFP (both centrioles). Video 2 shows photobleaching of centrin-GFP (both centrioles). Video 3 shows photobleaching of Cep120-GFP (mother versus daughter centrioles). Video 4 shows photobleaching of Cep120-GFP (both centrioles, with overexpressed Cep120-GFP). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003009/DC1.
Acknowledgments
We thank Jon Mulholland, John Perrino, and Lydia Joubert of the Stanford Cell Sciences Imaging Facility for help with light and electron microscopy. We also thank Eszter Vladar for help with laboratory methods.
M.R. Mahjoub was supported by a Stanford University Dean’s Fellowship. This work was supported by National Institutes of Health grant GM52022 to T. Stearns.
Footnotes
Abbreviations used in this paper:
- ALI
- air–liquid interface
- EdU
- 5-ethynyl-2′-deoxyuridine
- MEF
- mouse embryonic fibroblast
- MTEC
- mouse tracheal epithelial cell
- shRNA
- short hairpin RNA
References
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Anderson C.T., Stearns T. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19:1498–1502 10.1016/j.cub.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A.M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C.S. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Bahmanyar S., Kaplan D.D., Deluca J.G., Giddings T.H., Jr, O’Toole E.T., Winey M., Salmon E.D., Casey P.J., Nelson W.J., Barth A.I. 2008. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22:91–105 10.1101/gad.1596308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K., Beales P.L. 2009. Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 151C:281–295 10.1002/ajmg.c.30231 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., O’Connor A.K., Haycraft C.J., Yoder B.K. 2009. The primary cilium as a complex signaling center. Curr. Biol. 19:R526–R535 10.1016/j.cub.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Lizarraga S., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Chang P., Stearns T. 2000. Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2:30–35 10.1038/71350 [DOI] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Walsh C.A. 2004. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 44:279–293 10.1016/j.neuron.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Fish J.L., Kosodo Y., Enard W., Pääbo S., Huttner W.B. 2006. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA. 103:10438–10443 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A., Ailles L.E., Bakovic S., Geuna M., Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217–222 10.1038/76095 [DOI] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle–regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. 2006. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 17:680–689 10.1091/mbc.E05-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S. 2010. Centriole maturation and transformation to basal body. Semin. Cell Dev. Biol. 21:142–147 10.1016/j.semcdb.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Hung L.Y., Tang C.J., Tang T.K. 2000. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol. Cell. Biol. 20:7813–7825 10.1128/MCB.20.20.7813-7825.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J.R., Toyoda Y., Hegemann B., Poser I., Hériché J.K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J., et al. 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 328:593–599 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Stearns T. 2008. Exploring the pole: an EMBO conference on centrosomes and spindle pole bodies. Nat. Cell Biol. 10:1375–1378 10.1038/ncb1208-1375 [DOI] [PubMed] [Google Scholar]
- Jeong Y., Lee J., Kim K., Yoo J.C., Rhee K. 2007. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 120:2106–2116 10.1242/jcs.03458 [DOI] [PubMed] [Google Scholar]
- Keller L.C., Romijn E.P., Zamora I., Yates J.R., III, Marshall W.F. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098 10.1016/j.cub.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Kouprina N., Pavlicek A., Collins N.K., Nakano M., Noskov V.N., Ohzeki J., Mochida G.H., Risinger J.I., Goldsmith P., Gunsior M., et al. 2005. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum. Mol. Genet. 14:2155–2165 10.1093/hmg/ddi220 [DOI] [PubMed] [Google Scholar]
- Lange B.M., Gull K. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gönczy P. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Li J.B., Gerdes J.M., Haycraft C.J., Fan Y., Teslovich T.M., May-Simera H., Li H., Blacque O.E., Li L., Leitch C.C., et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 117:541–552 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., Nigg E.A. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88–93 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160–170 10.1038/ncb1529 [DOI] [PubMed] [Google Scholar]
- Ou Y.Y., Mack G.J., Zhang M., Rattner J.B. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115:1825–1835 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J.L., Bornens M. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089–3102 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Witman G.B. 2003. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15:105–110 10.1016/S0955-0674(02)00012-1 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Giddings T.H., Jr, Winey M. 2009. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol. Biol. Cell. 20:904–914 10.1091/mbc.E08-08-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki B.P., LaVoie M., Tam L.W., Lefebvre P.A., Silflow C.D. 2008. The Uni2 phosphoprotein is a cell cycle regulated component of the basal body maturation pathway in Chlamydomonas reinhardtii. Mol. Biol. Cell. 19:262–273 10.1091/mbc.E07-08-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki B.P., Silflow C.D. 2009. The UNI1 and UNI2 genes function in the transition of triplet to doublet microtubules between the centriole and cilium in Chlamydomonas. Mol. Biol. Cell. 20:368–378 10.1091/mbc.E08-09-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S.L., Fry A.M. 2009. Fluorescence imaging of the centrosome cycle in mammalian cells. Methods Mol. Biol. 545:165–183 10.1007/978-1-60327-993-2_10 [DOI] [PubMed] [Google Scholar]
- Quarmby L.M., Parker J.D. 2005. Cilia and the cell cycle? J. Cell Biol. 169:707–710 10.1083/jcb.200503053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A.J., Dailey L.A., Brighton L.E., Devlin R.B. 2007. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 37:169–185 10.1165/rcmb.2006-0466OC [DOI] [PubMed] [Google Scholar]
- Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Zhang M., Ihrig M.M., McManus M.T., et al. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401–406 10.1038/ng1117 [DOI] [PubMed] [Google Scholar]
- Sluder G., Nordberg J.J. 2004. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 16:49–54 10.1016/j.ceb.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Soung N.K., Kang Y.H., Kim K., Kamijo K., Yoon H., Seong Y.S., Kuo Y.L., Miki T., Kim S.R., Kuriyama R., et al. 2006. Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol. Cell. Biol. 26:8316–8335 10.1128/MCB.00671-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E.K., Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Moy L.Y., Sanada K., Zhou Y., Buchman J.J., Tsai L.H. 2007. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 56:79–93 10.1016/j.neuron.2007.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Richer E.J., Huang T., Brody S.L. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L1315–L1321 [DOI] [PubMed] [Google Scholar]
- Zhong X., Liu L., Zhao A., Pfeifer G.P., Xu X. 2005. The abnormal spindle-like, microcephaly-associated (ASPM) gene encodes a centrosomal protein. Cell Cycle. 4:1227–1229 [DOI] [PubMed] [Google Scholar]
- Zimmerman W., Doxsey S.J. 2000. Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic. 1:927–934 10.1034/j.1600-0854.2000.011202.x [DOI] [PubMed] [Google Scholar]
- Zou C., Li J., Bai Y., Gunning W.T., Wazer D.E., Band V., Gao Q. 2005. Centrobin: a novel daughter centriole–associated protein that is required for centriole duplication. J. Cell Biol. 171:437–445 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyss D., Gergely F. 2009. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 19:334–346 10.1016/j.tcb.2009.04.001 [DOI] [PubMed] [Google Scholar]