The augmin protein complex nucleates noncentrosomal microtubules during anaphase to promote completion of cell division.
Abstract
The central spindle forms between segregating chromosomes during anaphase and is required for cytokinesis. Although anaphase-specific bundling and stabilization of interpolar microtubules (MTs) contribute to formation of the central spindle, it remains largely unknown how these MTs are prepared. Using live imaging of MT plus ends and an MT depolymerization and regrowth assay, we show that de novo MT generation in the interchromosomal region during anaphase is important for central spindle formation in human cells. Generation of interchromosomal MTs and subsequent formation of the central spindle occur independently of preanaphase MTs or centrosomal MT nucleation but require augmin, a protein complex implicated in nucleation of noncentrosomal MTs during preanaphase. MTs generated in a hepatoma up-regulated protein (HURP)–dependent manner during anaphase also contribute to central spindle formation redundantly with preanaphase MTs. Based on these results, a new model for central spindle assembly is proposed.
Introduction
Since the very early stages of mitosis research, the central spindle (also known as interzonal fibers) has been the hallmark of dividing animal cells. It consists of two masses of parallel microtubule (MT) bundles with opposite directionalities that interdigitate in the center of the interchromosomal region (McDonald et al., 1979). The center of this interzonal MT structure provides a scaffold for signaling molecules required for the initiation, progression, and completion of cytokinesis (Straight and Field, 2000; Ruchaud et al., 2007; Glotzer, 2009). The robust central spindle is assembled in anaphase through anaphase-specific stabilization and bundling of interpolar MTs in which the activation/deactivation of several MT-associated proteins occurs (Saxton and McIntosh, 1987; Mollinari et al., 2002; Canman et al., 2003; Inoue et al., 2004; Mishima et al., 2004; Cheerambathur et al., 2007). However, it is unclear when and how these interpolar MTs are prepared for central spindle formation. An established source of interpolar MTs in anaphase is the MTs constituting the preanaphase spindle. Although this is often emphasized in the drawings describing central spindle MTs, it cannot be directly estimated how much of a portion of the preformed MTs actually contributes to central spindle formation. Another possible source is those MTs that are generated de novo during anaphase. Several lines of evidence have suggested that cells have the potential to nucleate MT de novo in postanaphase (Canman et al., 2000; Rosa et al., 2006; Glotzer, 2009) and that γ-tubulin–dependent MT nucleation in postanaphase is required for completion of cytokinesis (Julian et al., 1993). However, the physiological importance, precise locations and timings, or molecular basis for controlling of de novo MT generation in anaphase has been unclear.
Augmin is a protein complex that is localized at the spindle and is required for centrosome-independent, MT-dependent MT generation through the recruitment of γ-tubulin to spindle MTs during preanaphase (Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009). Previously, we found that augmin depletion in HeLa cells resulted in severe defects not only in metaphase spindle MT generation but also in anaphase central spindle formation; consequently, the cleavage furrow that was in the process of invagination regressed, and cytokinesis could not be completed (Uehara et al., 2009). This observation raised a possibility that augmin might also actively generate noncentrosomal MTs during anaphase.
In this study, to understand how central spindle MTs are formed, we performed an anaphase-specific MT depolymerization/regrowth assay and live imaging of MT plus ends in human tissue culture cells. Our data indicate that, in contrast to the conventional view, de novo MT generation near the segregating chromosomes during anaphase makes a major contribution to central spindle MT formation and that this process requires augmin but not centrosomes or preanaphase MTs.
Results and discussion
Augmin-dependent de novo MT generation in the interchromosomal region during anaphase
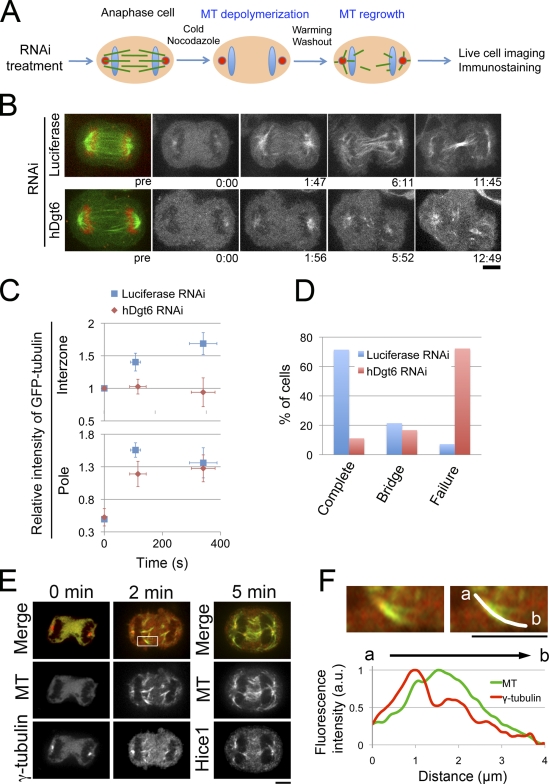
To visualize anaphase-specific MT generation and test the involvement of augmin in this process, we developed a single cell–based, anaphase-specific MT depolymerization/regrowth assay in HeLa cells (Fig. 1 A). In this assay, anaphase cells were treated with cold medium containing nocodazole, and almost all of the MTs were first depolymerized. Live imaging of GFP-tubulin in the control cells for 20 min showed that MTs were generated after drug washout, as expected. Interestingly, confocal imaging revealed that MTs were regenerated not only in the polar regions but also in the interchromosomal regions (Fig. 1 B). In the very early phase of MT regrowth, typically within 2 min, a burst of MT nucleation occurred from the centrosomes while MTs were also nucleated in the interchromosomal region (Fig. 1 B, 1:47). However, from 2 to 6 min, intensity of centrosomal MTs stopped increasing, whereas MTs in the interchromosomal region kept increasing and became more clearly visible (Fig. 1 C, blue). These interzonal MTs were organized into a highly bundled MT structure (Fig. 1 B, 6:11). This suggests that de novo MT nucleation in this region contributes to central spindle formation. However, in augmin-depleted cells (generated using RNAi directed toward the Dgt6 subunit), MT regrowth was severely impaired in the interchromosomal region but remained largely intact in the polar region (Fig. 1, B and C). As a result, augmin-depleted cells failed to form the central spindle in this assay (compare 5:52 [augmin] and 6:11 [control]). We checked the completion of cytokinesis for each cell traced during MT regrowth (Fig. 1 D). Cytokinesis was completed (the cells divided completely) in 10 of 14 control cells; of the remaining cells, three still contained an intercellular bridge (probably in the process of abscission), and one failed to complete cytokinesis and showed furrow regression. In contrast, cytokinesis was completed in only 2 of 18 augmin-depleted cells; of the remaining cells, three still contained a bridge and 13 failed to complete cytokinesis and showed furrow regression. It is likely that the RNAi efficiency was lower in those two cells that succeeded in cytokinesis despite the hDgt6 siRNA treatment because they had more central spindle MTs than other cells that failed cytokinesis (unpublished data). These results indicate that de novo MT generation does occur in the interchromosomal region in an augmin-dependent manner during anaphase. Moreover, the generated MTs were sufficient for completing cytokinesis in MT-depolymerized/regrown cells.
Figure 1.
Functional central spindle formation by augmin-dependent de novo MT generation in anaphase. (A) Schematic representation of the MT depolymerization/regrowth assay in anaphase cells. (B) GFP-tubulin–expressing cells treated with control (luciferase) or hDgt6 siRNA in the assay. The initial time point of observation is represented as 0:00 (min:s), which is ∼60 s after nocodazole was washed out. (C) Relative fluorescence intensity of GFP-tubulin in the interzone and the polar region of control (n = 14) and augmin-depleted cells (n = 18) in the assay. Means ± SEM (y axis) or SD (x axis) of single cells from six independent experiments are shown. (D) Completion or failure of cytokinesis after the assay in control (n = 14) and augmin-depleted cells (n = 18). Means of single cells from six independent experiments are shown. Bridge indicates the percentage of cells that possess an intercellular bridge at the end of the observation period. (E) Immunostaining of MT (green), γ-tubulin (red at 0 and 2 min), and Hice1 (red at 5 min) in anaphase cells during the MT regrowth phase. MT regrowth began at time 0. (F, top) A magnified image of the region indicated in the box in E. (bottom) MT and γ-tubulin intensities along an MT bundle. Chromosomes are located at the left. Bars, 5 µm.
We next performed a population-based MT depolymerization/regrowth assay in which MT depolymerization/regrowth treatment was followed by cell fixation and immunostaining at various time points. Consistent with results from the single-cell assay, newly nucleated MTs appeared in the interchromosomal region 2 min after treatment, and these gradually grew to form an ordered MT bundle structure (5 min). We observed that most MTs in the interzone were located close to the chromosomes. Moreover, γ-tubulin (Fig. 1, E and F) and Hice1 (an augmin subunit; Fig. 1 E) were concentrated at the chromosome-proximal ends of these MTs (Fig. S1 A [quantification]). These results suggest that the majority of the minus ends of these MTs were located proximal to the chromosomes.
Contribution of hepatoma up-regulated protein (HURP) to the de novo MT formation in the interzone
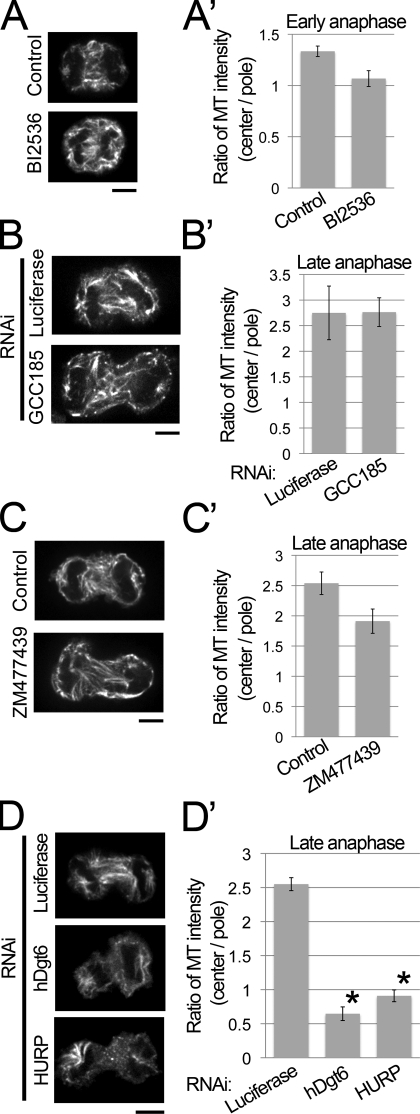
In preanaphase, augmin is not essential for the initial nucleation of MTs but is necessary for MT amplification (Goshima et al., 2008). This may also be the case in anaphase because we observed a small but detectable amount of MTs at the beginning of MT regrowth in our assays in augmin-depleted cells (Fig. 1 B and Fig. S1 B). We addressed the question of how chromosome-proximal MTs are initially formed in anaphase. First, we tested the contribution of centrosomal MTs, which might be released and translocated to the interchromosomal region (Belmont et al., 1990). Inhibition of the Plk1 kinase activity by the inhibitor BI2536 severely blocked recruitment of γ-tubulin to the centrosomes (Fig. S2 A), resulting in the loss of functional centrosomes, as reported in previous studies (Lénárt et al., 2007; Torosantucci et al., 2008). However, when we performed the population-based MT depolymerization/regrowth assay in the presence of the same concentration of BI2536, MTs were still formed in the interchromosomal region in early anaphase (Fig. 2, A and A′; and Fig. S1 C [definition of early and late anaphase]). Quantification showed that in the presence of BI2536, regrowth of interchromosomal MTs occurred nearly to the same extent as in control cells (80% of control; >32 cells analyzed; Fig. 2 A′), whereas inhibition of augmin by RNAi targeting hDgt6 resulted in much larger reduction of interchromosomal MTs in early anaphase (39% of control; >11 cells analyzed; Fig. S1 D). BI2536 blocked initiation of furrow ingression (Petronczki et al., 2007), hindering us from investigating its effect on MT generation in late anaphase. Therefore, we performed the MT regrowth assay using cells depleted of the centriole duplication factor Plk4. In such cells, the lack of centriole duplication occasionally results in the formation of monastral bipolar spindles in which the centrosome is present at only one of two poles (Fig. S2, B and C; Bettencourt-Dias et al., 2005; Habedanck et al., 2005). Monastral bipolar spindles were also found in anaphase after Plk4 knockdown (Fig. S2 D), which facilitated the comparison of interzonal MT regrowth in the acentrosomal and centrosomal halves of the spindle. We found that intensity of regrown interzonal MTs was only slightly lower in acentrosomal half-spindle than centrosomal one (Fig. S2 D), suggesting that the contribution of centrosomes to regrowth of interzonal MTs is minor. These results suggest that functional centrosomes are not a prerequisite for central spindle formation.
Figure 2.
HURP and augmin are required for de novo interzonal MT generation in the absence of preexisting MTs. (A–D) Immunostaining of MT in control, BI2536-treated (A), GCC185-depleted (B), ZM477439-treated (C), augmin- or HURP-depleted (D) cells fixed at 5 min after the MT depolymerization/regrowth treatment. Bars, 5 µm. (A’–D’) The ratio of central and polar MT intensities of anaphase cells in A–D. Means ± SEM of three independent experiments are shown (>16 cells were analyzed for each treatment). The ratio in both the augmin- and HURP-depleted cells but not in the other cells was significantly lower (augmin and HURP [asterisks], P < 0.001; other cells, P > 0.04; by t test) than that in the control cells.
Another possible MT-nucleating organelle that might contribute to central spindle formation is the Golgi apparatus, which is located around the spindle poles and chromosomes during anaphase (Altan-Bonnet et al., 2006). However, interzonal MTs reappeared normally in cells depleted of GCC185, a protein essential for the Golgi-mediated MT generation (Efimov et al., 2007), suggesting that MT generation from the Golgi is probably not required for central spindle formation (Fig. 2, B and B′; and Fig. S2, F–H).
Next, we tested the involvement of the chromosome-dependent MT nucleation pathway in central spindle formation. Aurora B is a subunit of the chromosomal passenger complex that is required for chromosome-based MT generation in preanaphase cells and for the completion of cytokinesis in postanaphase cells (Giet and Glover, 2001; Tulu et al., 2006; Kelly et al., 2007; Ruchaud et al., 2007; Fuller et al., 2008). However, in the presence of aurora B–specific inhibitor ZM477439, de novo MT generation in the MT regrowth phase was not significantly affected (Fig. 2, C and C′). Two lines of evidence indicate that ZM477439 efficiently inhibited aurora B activity in this experiment: (1) under identical experimental conditions, chromosome-proximal MT generation was severely impaired in preanaphase cells (Fig. S2 I; Sampath et al., 2004; Tulu et al., 2006), and (2) phosphorylation of vimentin (Ser72) at the cell equator, which is an anaphase-specific substrate of aurora B (Goto et al., 2003), was severely inhibited in the presence of ZM477439 (Fig. S2 J). These results strongly suggest that aurora B is not essential for de novo MT generation in anaphase. Another pathway from chromosomes to MT nucleation is the RanGTP-dependent pathway (Bastiaens et al., 2006; Kalab and Heald, 2008), and HURP is a downstream factor of RanGTP in preanaphase (Koffa et al., 2006; Silljé et al., 2006; Wong and Fang, 2006). We found that in the MT regrowth phase, HURP knockdown cells showed a severe defect in MT generation in the interzone, similar to that observed in the case of augmin (Fig. 2, D and D′; and Fig. S1, E and F). Next, we tested whether HURP is responsible for the chromosome-proximal MTs in RNAi cells for augmin by simultaneously depleting augmin and HURP. Double knockdown of UCHL5IP (an augmin subunit) and HURP resulted in reduction of initial nucleation of interchromosomal MTs in the early phase of MT regrowth (Fig. S1, G and H). We concluded that HURP-mediated chromatin-based MT nucleation is involved in the de novo generation of interzonal MTs in MT-depolymerized/regrown anaphase cells. The augmin–γ-tubulin complex might bind to HURP-nucleating MTs and nucleate the new MTs.
Augmin-dependent de novo MT generation is also critical in unperturbed anaphase
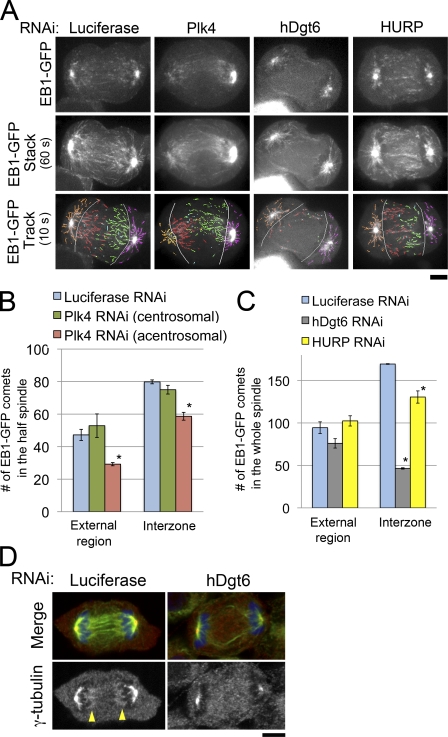
We next tested the involvement of centrosomes, augmin, and HURP in MT generation in unperturbed anaphase (i.e., without MT depolymerization treatment) using EB1-GFP, a marker for growing MT plus ends (Mahoney et al., 2006). In control anaphase cells, EB1-GFP comets were observed in both spindle external and interchromosomal regions (Fig. 3 A and Video 1). In the interchromosomal regions, many EB1-GFP comets moved across the cell equatorial plane, suggesting that these MTs participate in constructing interdigitating antiparallel MT bundles in the spindle interzone (Fig. 3 A, 60-s stacks; and Video 1). The comets in the spindle external region mainly arose from the centrosome (a bright dot at each pole) and moved radially (Fig. 3 A, orange and magenta), whereas the majority of the interzonal comets moved from the chromosome-proximal area to the interior region parallel to the long axis of the cell (Fig. 3 A, red and green). This suggests that most of the MTs in the interchromosomal region are generated at sites distinct from the centrosomes. To confirm this observation, we next examined EB1-GFP in monastral anaphase cells created by Plk4 RNAi (Fig. 3, A and B; and Fig. S2 E). We found that the distribution of EB1-GFP in the interzone was only slightly asymmetric; the number of interzonal comets migrating from the acentrosomal to centrosomal side (Fig. 3 A, red) was 22% less than that in the opposite direction (nine cells analyzed; Fig. 3 A, green). This suggests that the majority (∼75%) of interzonal MTs are generated independent of the centrosomes. In contrast, in augmin-depleted anaphase cells in which γ-tubulin accumulation in the chromosome-proximal region was specifically impaired (Fig. 3 D), the number of comets decreased dramatically in the interzone (>70% reduction; nine cells analyzed) but decreased only slightly in the external region (Fig. 3, A and C; and Video 1). HURP depletion also reduced the number of comets in the interzone but to a lesser extent (23% reduction; nine cells analyzed) than augmin depletion (Fig. 3, A and C; and Video 1). This indicates that the HURP-dependent pathway also contributes to de novo interzonal MT generation in unperturbed anaphase cells, but the contribution is smaller than that in the case in which MTs have been depolymerized once. In contrast, augmin function was critical both in the presence and absence of preformed MTs.
Figure 3.
Augmin, but not centrosomes or HURP, is critical for interzonal MT generation in unperturbed anaphase cells. (A) Single time frame images (top), 60-s stacked images (middle), and 10-s tracings (bottom) of EB1-GFP in RNAi-treated cells. Absence of the centrosome at the left pole of the monastral bipolar spindle of the Plk4-depleted cell was confirmed by z-axis confocal sectioning of the whole cell (Fig. S2 E). The following comets were distinguished: interzonal comets moving from left to right (acentrosomal to centrosomal side in Plk4-depleted cells; red), those moving in the opposite direction (green), comets moving in the external region on the left side (acentrosomal side in Plk-depleted cells; orange), or those on the opposite side (magenta). Light blue, interzonal comets with unknown directionalities; gray lines, borders between the external region and interzone. (B) Number of comets observed in each region of the half-spindle in control (blue) or Plk4-depleted cells (centrosomal or acentrosomal halves shown in green or red, respectively). Asterisks indicate statistically significant difference from the control (P < 0.008 by t test). The numbers of interzonal comets with unknown directionalities were 9.7 ± 2.3 (SEM) and 9.7 ± 3.5 in control and Plk4-depleted cells, respectively (not depicted). (C) The number of comets observed in each region of the whole spindle in control (blue), hDgt6-depleted (gray), and HURP-depleted cells (yellow) is shown. Means ± SEM of three independent experiments were shown (nine cells were analyzed for each region). Asterisks indicate statistically significant difference from control (P < 0.005 by t test). An identical dataset was used for luciferase RNAi cells in B and C. (D) Immunostaining of MTs (green), γ-tubulin (red), and chromosomes (blue) in control and hDgt6-depleted cells in anaphase. In the control cell, γ-tubulin was concentrated at both ends of the central spindle (arrowheads); however, such concentration was not observed in the augmin-depleted cell. Bars, 5 µm.
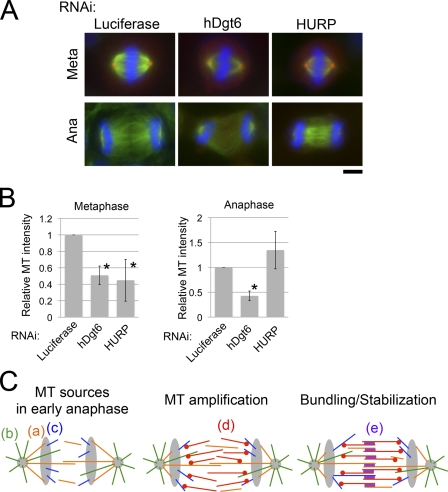
Finally, we compared the amounts of MT in preanaphase and anaphase spindles after augmin and HURP knockdowns, both of which resulted in a reduction in MT intensity in the metaphase spindle, including the kinetochore fibers (Fig. 4, A and B; Silljé et al., 2006; Wong and Fang, 2006; Goshima et al., 2008; Uehara et al., 2009). In anaphase, the amount of MTs in the interzone was dramatically reduced in augmin-depleted cells, but the amount in HURP-depleted cells was similar to that in control cells (Fig. 4, A and B). These results, together with those from the MT depolymerization/regrowth assay and EB1-GFP imaging, strongly suggest that in addition to preexisting MTs, augmin-mediated de novo MT generation in anaphase is a critical determinant of the interzonal MT number in anaphase.
Figure 4.
Physiological importance of de novo interzonal MT generation in anaphase. (A) Immunostaining of MT (green), γ-tubulin (red), and chromosomes (blue) in control, augmin-depleted, or HURP-depleted cells in metaphase (meta) or anaphase (ana). Bar, 5 µm. (B) MT intensities in the metaphase spindle or interzone in anaphase. Means ± SD of three independent experiments are shown (>18 cells were analyzed for each treatment). Depletion of hDgt6 and HURP resulted in significant reduction in MT amounts in metaphase (*, P < 0.01), whereas only the former caused significant reduction in anaphase interzonal MTs (*, P < 0.0003 for hDgt6-depleted anaphase cells; P > 0.09 for HURP-depleted anaphase cells). (C) A model for the central spindle formation process. In anaphase, MTs that are formed in preanaphase (a; orange), those constantly nucleated from centrosomes (b; green), and those nucleated in the interzone near the chromosomes in an HURP-dependent manner (c; blue) are available as MT sources. (d; red) These MTs are used as the templates for MT amplification mediated by augmin and γ-TuRC. (e; purple) All of the interzonal MTs formed by processes a–d are stabilized and bundled by antiparallel MT-bundling proteins, resulting in robust central spindle formation.
A model for the formation of central spindle MTs
Based upon the results of this study and previous ones, we suggest a model for the formation of central spindle MTs in human cells (Fig. 4 C). At the onset of anaphase, there are three major sources of MT in cells: preexisting MTs that are carried over from metaphase (Fig. 4 C, a), MTs that are newly generated at centrosomes (Fig. 4 C, b), or those that are formed around the chromosomes in an HURP-dependent manner (Fig. 4 C, c). The central spindle formation defect observed after HURP RNAi in the MT depolymerization/regrowth assay but not in unperturbed anaphase indicate that the HURP-dependent process and preexisting MTs are redundant (Fig. S3, A and B). Our data also show that sufficient amounts of MTs for constructing a functional central spindle cannot be obtained solely from these sources and that further amplification of MTs in the interzone is necessary. We propose that MTs formed by the aforementioned three mechanisms provide templates for augmin-dependent MT amplification (Goshima et al., 2008; Uehara et al., 2009) that occurs specifically in the interchromosomal region (Fig. 4 C, d). The augmin-dependent mechanism would generate enough amounts of MTs for compensating loss of one of the MT sources (this might explain an earlier observation in which robust central spindles are formed in chromosome- and centrosome-depleted insect spermatocytes that still possess preexisting MTs; Alsop and Zhang, 2003; Bucciarelli et al., 2003). A simple comparison of the interzonal EB1-GFP comet numbers (Fig. 3 C) and interzonal MT-staining intensities (Fig. 4 B) of augmin-depleted, HURP-depleted, and control cells suggests that 60–70% of the total central spindle MTs is generated via augmin-dependent de novo MT formation that occurs after anaphase onset, although accurate quantification of the contribution would require anaphase-specific inhibition of augmin. This model would also be largely consistent with previous electron microscopy observations in PtK1 cells in early anaphase in which the minus ends of central spindle MTs were distributed in the interzone away from the centrosome (Mastronarde et al., 1993). We assume that the γ-tubulin ring complex (γ-TuRC) is also an essential contributor to this process because we observed similar central spindle formation defects after the knockdown of a γ-TuRC subunit (Fig. S3, C and D). Moreover, an augmin mutant that failed to bind γ-TuRC did not rescue cytokinesis failure (Uehara et al., 2009). However, our model does not depend on the exact nature of the molecular activity of augmin–γ-TuRC, which has not been determined biochemically. For example, the roles of augmin and γ-TuRC in the stabilization of nucleated MTs rather than in nucleation per se cannot be ruled out (Goshima et al., 2008); however, this activity would also lead to augmentation of anaphase MTs. Amplified MTs, together with MTs from other sources, would be stabilized and bundled by the actions of several MT-associated proteins, including kinesin-6 motor proteins, thus forming a robust central spindle (Fig. 4 C, e; Mollinari et al., 2002; Mishima et al., 2004; Glotzer, 2009).
In a recent study involving whole genome sequencing of cancer cells obtained from breast cancer patients, mutations were most commonly found in the hDgt3/HAUS3 subunit of augmin (Shah et al., 2009). The mechanism underlying cell transformation in these patients might involve errors in central spindle formation and cytokinesis, which lead to tetraploidy and anueploidy.
Materials and methods
Cell culture, RNAi, and cell synchronization
HeLa cells were cultured as described previously (Goshima et al., 2008). siRNA transfection was performed using Lipofectamine RNAiMAX (Invitrogen). The siRNA sequences used in this study are shown in Table S1. To test the effects of hDgt6 or NEDD1 depletion on the anaphase MT structure, cells were observed 2 d after treatment with 20 or 100 µM siRNA that targeted each of the genes. Longer incubation (e.g., 3 d) severely affected mitotic progression, preventing the examination of anaphase cells. Cells were synchronized at anaphase as described previously (Petronczki et al., 2007) but with slight modifications. Cells were treated with 100 ng/ml nocodazole for 4 h, and this was followed by shaking to collect the mitotic cells from the culture dish. The cells were washed twice with DME and treated with 10 µM MG132 for 2 h on glass-bottom dishes coated with concanavalin A. The cells arrested in metaphase were washed twice with DME and incubated for 40 min; these cells entered anaphase during this time.
Reagents
To generate the anti-Hice1 or anti-UCHL5IP antibody, the C-terminal fragment of Hice1 (132–410 aa) or full-length UCHL5IP was cloned into pDEST17 (Invitrogen), respectively, bacterially expressed, and purified for rabbit immunization. Specificity of the obtained antisera was confirmed by immunoblotting (Fig. S1, H and I). The anti-HURP (Silljé et al., 2006), anti–phospho-vimentin (Yasui et al., 2001; Goto et al., 2003), or anti-GCC185 antibody (Efimov et al., 2007) was provided by E. Nigg (University of Basel, Basel, Switzerland), M. Inagaki (Aichi Cancer Center Research Institute, Nagoya, Japan), or I. Kaverina (Vanderbilt University Medical Center, Nashville, TN), respectively. Anti-NEDD1 antibody (clone 7D10) or anti-GM130 (clone 35) was purchased from Abnova or BD, respectively. The expression vector encoding EB1-GFP or Lifeact-mCherry was provided by K. Kaibuchi (Nagoya University, Nagoya, Japan) or S. Kojima (Gakushuin University, Tokyo, Japan), respectively.
MT depolymerization/regrowth assay
Cells that just entered anaphase were treated with ice-cold DME containing 40 ng/ml (single-cell assay) or 100 ng/ml (population assay) nocodazole and incubated on ice for 20 min. A higher concentration of nocodazole was used for MT depolymerization in the population assay to attenuate MT nucleation from centrosomes and clearly visualize MT nucleation in the chromosome-proximal region. In the experiments shown in Fig. S1 G and Fig. S2 (F and G), cold nocodazole incubation was extended to 50 min, which was found to slow down initial phase of MT regrowth and enable us to investigate the MT nucleation process in more detail. When BI2536 was used in the assay, the incubation time was increased to 50 min to completely depolymerize MTs. This was because the MTs were stabilized by this drug for some unknown reason. Nocodazole was washed out with prewarmed DME, and the regrowth of MTs was observed by live cell imaging or immunostaining. During the cold nocodazole treatment, neither ingression nor regression of the cleavage furrow occurred, and F-actin (visualized by Lifeact-mCherry; Riedl et al., 2008) remained enriched at the equatorial cortex, albeit with an ∼50% reduced level (Fig. S1 J). Upon warm up of cells, the cleavage furrows resumed ingression, which enabled us to check completion of cytokinesis in these cells. The appearance of MTs around the poles at time 0 in Fig. 1 B was because of the time gap between washing out and image acquisition (∼1 min was required to locate the cell under the microscope). In the single-cell–based assay, HeLa cells expressing GFP-tubulin and histone H2B–mCherry were cultured on a coverslip in which grids were printed to identify the position of each cell. When BI2536 or ZM477439 cells were used in the population assay, they were treated with the drugs after synchronization at anaphase. When RNAi-treated cells were used for the population assay, asynchronous cultures were used.
Immunostaining and live cell imaging
Fixation with methanol (Hice1) or PFA (others) was performed according to a previously described procedure (Goshima et al., 2008). For live and fixed cell imaging, a microscope (TE2000; Nikon) equipped with a 100× 1.4 NA Plan Apochromatic oil immersion objective lens (Nikon), a confocal unit (CSU-X; Yokogawa), and an electron multiplying charge-coupled device camera (for confocal imaging; ImagEM; Hamamatsu Photonics) or charge-coupled device camera (for epifluorescence imaging; MicroMax; Roper Industries) were used. Image acquisition was controlled using the µManager software.
RT-PCR
RNA extraction and cDNA synthesis were performed using RNeasy mini kit (QIAGEN) and Primescript II (Takara Bio Inc.), respectively. Primers 5′-AGGATCATTTGCTGGTGTCTACAG-3′ and 5′-GAAGGATGTTTCAATTGGCAATGTATTTTC-3′ (Bettencourt-Dias et al., 2005) were used for amplifying the Plk4 cDNA fragment, and 5′-GCCGGGACCTGACTGACTAC-3′ and 5′-TCCTTAATGTCACGCACGATTTC-3′ were used for β-actin.
Image analysis
For quantification in Fig. 1 C, we measured mean fluorescence intensities of manually selected interzonal and polar regions, subtracted background (measured in the cytoplasm), and multiplied them by the areas of these regions to obtain total fluorescence intensity. Fluorescence intensity of each region at each time point was normalized by that of the interzonal region at time point 0 because it varied from cell to cell as a result of the difference in GFP-tubulin expression levels. For quantification in Fig. 2, total MT intensities in the interzone and polar regions were measured using ImageJ (National Institutes of Health), and the ratio was obtained by dividing the intensity in the interzone by sum of that in two polar regions. Quantification of total fluorescence intensities in the whole spindle region in metaphase or interzone in anaphase in Fig. 4 B was performed in the same way as Fig. 1 C, and the intensities were normalized to that of control cells for comparison among different experiments.
Online supplemental material
Fig. S1 shows additional data on the MT depolymerization/regrowth assay. Fig. S2 shows the characterization of BI2536-, Plk4 RNAi–, GCC185 RNAi–, or ZM477439-treated cells. Fig. S3 shows a schematic model for redundancy in the process of central spindle MT formation and impaired central spindle formation in NEDD1-depleted cells. Video 1 shows live imaging of EB1-GFP in RNAi-treated anaphase cells. Table S1 shows sequences of the siRNAs used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004150/DC1.
Acknowledgments
We are grateful to Drs. Hiroshi Hanafusa, Masaki Inagaki, Kozo Kaibuchi, Irina Kaverina, Shin-ichiro Kojima, Erich Nigg, and Takashi Watanabe for providing reagents, Kunihiro Matsumoto for the microscopy setup, Masanori Mishima for critical comments on the manuscript, and Tomoko Kamasaki and Akiko Tomida for technical assistance.
This study was supported by Special Coordination Funds for Promoting Science and Technology, the Inoue Foundation, the Uehara Foundation, the Naito Foundation, the Asahi-glass Foundation, a Human Frontier Science Program grant (to G. Goshima), Grant-in-Aid for Scientific Research (Ministry of Education, Culture, Sports, Science and Technology; to G. Goshima and R. Uehara), and a Sasagawa Scientific Research grant (to R. Uehara).
Footnotes
Abbreviations used in this paper:
- γ-TuRC
- γ-tubulin ring complex
- HURP
- hepatoma up-regulated protein
- MT
- microtubule
References
- Alsop G.B., Zhang D. 2003. Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J. Cell Biol. 162:383–390 10.1083/jcb.200301073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N., Sougrat R., Liu W., Snapp E.L., Ward T., Lippincott-Schwartz J. 2006. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell. 17:990–1005 10.1091/mbc.E05-02-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens P., Caudron M., Niethammer P., Karsenti E. 2006. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 16:125–134 10.1016/j.tcb.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Belmont L.D., Hyman A.A., Sawin K.E., Mitchison T.J. 1990. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 62:579–589 10.1016/0092-8674(90)90022-7 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bucciarelli E., Giansanti M.G., Bonaccorsi S., Gatti M. 2003. Spindle assembly and cytokinesis in the absence of chromosomes during Drosophila male meiosis. J. Cell Biol. 160:993–999 10.1083/jcb.200211029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman J.C., Hoffman D.B., Salmon E.D. 2000. The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr. Biol. 10:611–614 10.1016/S0960-9822(00)00490-5 [DOI] [PubMed] [Google Scholar]
- Canman J.C., Cameron L.A., Maddox P.S., Straight A., Tirnauer J.S., Mitchison T.J., Fang G., Kapoor T.M., Salmon E.D. 2003. Determining the position of the cell division plane. Nature. 424:1074–1078 10.1038/nature01860 [DOI] [PubMed] [Google Scholar]
- Cheerambathur D.K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., Scholey J.M. 2007. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 177:995–1004 10.1083/jcb.200611113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 12:917–930 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Yasui Y., Kawajiri A., Nigg E.A., Terada Y., Tatsuka M., Nagata K., Inagaki M. 2003. Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 278:8526–8530 10.1074/jbc.M210892200 [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Inoue Y.H., Savoian M.S., Suzuki T., Máthé E., Yamamoto M.T., Glover D.M. 2004. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166:49–60 10.1083/jcb.200402052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian M., Tollon Y., Lajoie-Mazenc I., Moisand A., Mazarguil H., Puget A., Wright M. 1993. gamma-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J. Cell Sci. 105:145–156 [DOI] [PubMed] [Google Scholar]
- Kalab P., Heald R. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121:1577–1586 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., Funabiki H. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M.D., Casanova C.M., Santarella R., Köcher T., Wilm M., Mattaj I.W. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16:743–754 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Lawo S., Bashkurov M., Mullin M., Ferreria M.G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J.R., Hegemann B., et al. 2009. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19:816–826 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.M. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Mahoney N.M., Goshima G., Douglass A.D., Vale R.D. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569 10.1016/j.cub.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N., McDonald K.L., Ding R., McIntosh J.R. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489 10.1083/jcb.123.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K.L., Edwards M.K., McIntosh J.R. 1979. Cross-sectional structure of the central mitotic spindle of Diatoma vulgare. Evidence for specific interactions between antiparallel microtubules. J. Cell Biol. 83:443–461 10.1083/jcb.83.2.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E.A., Glotzer M. 2004. Cell cycle regulation of central spindle assembly. Nature. 430:908–913 10.1038/nature02767 [DOI] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.P., Jiang W., Schoehn G., Hunter T., Margolis R.L. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J.M. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 12:713–725 10.1016/j.devcel.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J., Canovas P., Islam A., Altieri D.C., Doxsey S.J. 2006. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell. 17:1483–1493 10.1091/mbc.E05-08-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Sampath S.C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Saxton W.M., McIntosh J.R. 1987. Interzone microtubule behavior in late anaphase and telophase spindles. J. Cell Biol. 105:875–886 10.1083/jcb.105.2.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S.P., Morin R.D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J., et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 461:809–813 10.1038/nature08489 [DOI] [PubMed] [Google Scholar]
- Silljé H.H., Nagel S., Körner R., Nigg E.A. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Field C.M. 2000. Microtubules, membranes and cytokinesis. Curr. Biol. 10:R760–R770 10.1016/S0960-9822(00)00746-6 [DOI] [PubMed] [Google Scholar]
- Torosantucci L., De Luca M., Guarguaglini G., Lavia P., Degrassi F. 2008. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol. Biol. Cell. 19:1873–1882 10.1091/mbc.E07-10-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., Wadsworth P. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541 10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., Goshima G. 2009. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 106:6998–7003 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Fang G. 2006. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173:879–891 10.1083/jcb.200511132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Goto H., Matsui S., Manser E., Lim L., Nagata Ki, Inagaki M. 2001. Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene. 20:2868–2876 10.1038/sj.onc.1204407 [DOI] [PubMed] [Google Scholar]