Both Cdc2 and PKC-mediated phosphorylation of EBP50 alter accessibility of its first PDZ domain when its second PDZ domain is occupied to regulate microvilli formation on epithelial cells.
Abstract
The mechanisms by which epithelial cells regulate the presence of microvilli on their apical surface are largely unknown. A potential regulator is EBP50/NHERF1 (ERM-binding phosphoprotein of 50 kD/Na+-H+ exchanger regulatory factor), a microvillar scaffolding protein with two PDZ domains followed by a C-terminal ezrin-binding domain. Using RNAi and expression of RNAi-resistant EBP50 mutants we systematically show that EBP50 is necessary for microvillar assembly and requires that EBP50 has both a functional first PDZ domain and an ezrin-binding site. Expression of mutants mimicking Cdc2 or PKC phosphorylation are nonfunctional in microvillar assembly. Biochemical analysis reveals that these mutants are defective in PDZ1 accessibility when PDZ2 is occupied, and can be rendered functional in vivo by additional mutation of PDZ2. EBP50 is not necessary for mitotic cell microvilli, and PKC activation causes a rearrangement of microvilli on cells due to phosphorylation-dependent loss of EBP50 function. Thus, EBP50 is a critical factor that regulates microvilli assembly and whose activity is regulated by signaling pathways and occupation of its PDZ2 domain.
Introduction
The vast majority of cells are, or have the potential to be, polarized. Polarity requires the ability to control the protein composition of the plasma membrane in time, location, and in response to signaling pathways. A well-studied example is epithelial cells that have compositionally distinct apical and basolateral membranes. In addition to composition, cells regulate their morphology, for example the apical surface of epithelial cells is usually studded with microvilli. Cell morphology, as well as aspects of endocytosis, exocytosis, and membrane signaling are integrated processes, yet relatively little is known about how this is achieved. Part of this integration is provided by the actin-based cytoskeleton that is intimately associated with the plasma membrane. We have been studying in detail the microfilaments that support microvilli on epithelial cells as a model system to uncover how these processes are controlled and integrated. Here we report on mechanisms that regulate the presence of microvilli on cells.
Epithelial cell microvilli consist of a core of bundled actin filaments tethered laterally to the plasma membrane in part by ezrin, a member of the ERM (ezrin/radixin/moesin) family of regulated membrane cytoskeleton linking proteins (Fehon et al., 2010). Ezrin can exist in a dormant-inactive form in which its C-terminal F-actin binding site as well as binding sites for membrane-associated proteins on its N-terminal FERM (4.1 ERM) domain are masked by their intramolecular association. Upon activation by phosphorylation, the FERM domain can bind the cytoplasmic tails of various transmembrane proteins, or to the PDZ (postsynaptic density 95/discs large/zona occludens-1) domain containing scaffolding protein EBP50 (ERM-binding phosphoprotein of 50 kD). The EBP50 protein family, also known as the NHERF (Na+-H+ exchanger regulatory factor) family (Shenolikar et al., 2004), includes E3KARP (NHERF2) and PDZK1 (NHERF3), and together represent the only known PDZ scaffolding proteins present in the apical aspect of polarized epithelial cells, and have been suggested to coordinate functions in the apical domain. In addition, a broader role for EBP50, and the importance of its apical localization, is emerging in tumorigenesis (Georgescu et al., 2008).
We initially identified EBP50 as a protein that binds active, but not dormant, ezrin (Reczek et al., 1997). It contains two PDZ domains followed by a tail whose last 38 residues associate with ezrin’s N-terminal FERM domain. EBP50 was independently discovered as a factor necessary for conferring protein kinase A (PKA) regulation on NHE3 (Na+-H+ exchanger 3), hence the alternative name NHERF1 (Weinman et al., 1995). It has subsequently been implicated in scaffolding and regulating membrane proteins such as CFTR (cystic fibrosis transmembrane conductance regulator), NaPi IIa cotransporter, and the β2-adrenergic receptor (Short et al., 1998; Cao et al., 1999; Hernando et al., 2002). In addition to binding integral membrane proteins, EBP50 has been shown to bind the RabGAP protein EPI64 (EBP50-PDZ interactor of 64 kD), the RhoGAP nadrin through its first PDZ domain (Reczek and Bretscher, 2001), and PDZK1 through either domain (LaLonde et al., 2010).
EBP50 is subject to phosphorylation by several kinases, and these modifications have been suggested to alter its binding activity and possibly its oligomeric state. The G protein–coupled kinase, GRK6A, phosphorylates EBP50 on serine 290 (Hall et al., 1999), and this is reported to increase its oligomerization (Lau and Hall, 2001). The phosphorylation of serines 162, 339, and 340 by PKC is also thought to promote oligomerization of EBP50 as well as increase its affinity for PDZ ligands (Raghuram et al., 2003; Fouassier et al., 2005; Li et al., 2007). During mitosis EBP50 is phosphorylated on serines 280 and 302 by the cyclin-dependent kinase Cdc2, and this phosphorylation is reported to inhibit oligomerization (He et al., 2001). Some of these studies suggesting regulation of oligomerization have used blot overlays (Fouassier et al., 2000; Shenolikar et al., 2001) and were not aware of the ability of the EBP50 tail to bind PDZ2 in an intramolecular manner (Morales et al., 2007; Cheng et al., 2009). However, recent work on wild-type EBP50 suggests that it exists as a monomer in solution (Li et al., 2007, 2009).
Mice lacking either ezrin or EBP50 have short aberrant microvilli on their epithelia, suggesting that the two proteins function together in microvillus structure or regulation (Morales et al., 2004; Saotome et al., 2004). Consistent with this, we have shown that siRNA knockdown of EBP50 in human choriocarcinoma JEG-3 cells, which are normally robustly covered in microvilli, results is a significant reduction of cells with microvilli (Hanono et al., 2006). Expression of an RNAi-resistant GFP-EBP50 construct maintains microvilli when endogenous EBP50 is knocked down (LaLonde et al., 2010). This provides an assay to explore properties of EBP50 that are necessary for maintaining microvilli. Here we use this in vivo system, coupled with biophysical studies and biochemical characterization of EBP50 variants, to show that both its function in microvilli and biochemical scaffolding properties are regulated by phosphorylation. Our biochemical analysis of purified wild-type and mutant EBP50 reveal that it is an elongated monomer in solution with no tendency to oligomerize regardless of its phosphorylation state.
Results
EBP50 is required for microvillar assembly in JEG-3 cells
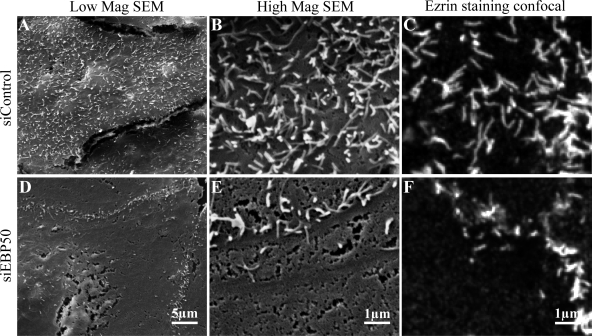
Microvilli are dynamic structures with life cycles averaging around 12 min and involving assembly, steady-state length, and retraction phases (Gorelik et al., 2003; unpublished data). Depletion of EBP50 by siRNA in human choriocarcinoma JEG-3 cells causes a significant loss of cells displaying apical microvilli as visualized by ezrin and actin staining (Hanono et al., 2006). To determine if this was due to a defect in a particular phase of the microvillar life cycle, we examined JEG-3 cells treated with either control siRNA–targeting luciferase or siRNA against EBP50 (siEBP50) by scanning electron microscopy (Fig. 1). In siEBP50-treated cells the apical surface is smooth and shows no evidence of small microvillar protrusions, suggesting that loss of EBP50 prevents the assembly phase of microvilli (Fig. 1, D and E). Comparison of ezrin staining by confocal microscopy at the same final magnification shows that ezrin localization is as good as scanning electron microscopy at identifying surface microvilli (Fig. 1, C and F). Therefore, to explore in more depth the possibility that EBP50 loss might reduce both the number and lifetime of microvilli, we prepared cells expressing ezrin-GFP treated with control siRNA or siEBP50 and imaged them in an environmental chamber for 20–30 min, taking confocal stacks every 30 s (Videos 1 and 2, respectively). Control cells showed numerous dynamic microvilli, whereas the siEBP50 treated cells were essentially featureless. Occasionally small punctuate ezrin-GFP structures formed only to disappear very quickly, suggesting that EBP50 is required to assemble microvilli. As noted previously, microvilli above junctions are often resistant to siEBP50 treatment (Fig. 1 and Video 2; Hanono et al., 2006). Examination of these structures by both scanning electron microscopy and confocal microscopy of ezrin staining (Fig. 1, A–F) again showed that the latter technique accurately reflects the presence of microvilli on these cells.
Figure 1.
Depletion of EBP50 by siRNA causes a defect in microvilli formation. JEG-3 cells were treated with siRNA against luciferase as a control (A–C) or against EBP50 (D–F) and examined by scanning electron microscopy (SEM; A, B, D, and E) or confocal microscopy by ezrin staining (C and F). Low and high magnification SEM images were acquired at 4,000 and 20,000x, respectively. Control cells showed numerous microvilli, whereas siEBP50 cells showed a dramatic reduction in microvilli by both SEM and ezrin staining. Confocal images were processed to be shown at the same scale as the high magnification SEM micrographs.
EBP50 requires an ezrin binding site and a functional PDZ1 domain for microvilli assembly
Loss of microvilli in siEBP50-treated cells is reversed by expression of an RNAi-resistant GFP-EBP50 (LaLonde et al., 2010). We exploited this system to systematically determine the elements of EBP50 necessary to maintain microvilli in EBP50 knockdown cells. We first made and characterized mutations in EBP50’s tail specifically defective in binding the ezrin FERM domain, and then tested their function in vivo.
Structural analyses of the tail of EBP50 bound to the radixin (Terawaki et al., 2006) or moesin (Finnerty et al., 2004) FERM domain show that the C-terminal 12 amino acids form an α-helix that associates with the FERM domain (Fig. 2 A). Structure-based mutations to reduce this interaction were made in an expression construct of MBP (maltose-binding protein) N-terminally fused to the C-terminal 39 residues of EBP50 (Fig. 2 A). The ability of immobilized ezrin FERM domain to retain soluble EBP50 tail fusion constructs was then assessed (Fig. 2 B). Deletions of either the C-terminal 17 residues (EBP50-320-341), or the four terminal residues of EBP50 (ΔFSNL), or even just the terminal leucine (ΔL), abolished the binding to the ezrin FERM domain. Other critical residues in the α-helix necessary for interacting with the FERM domain are methionine 346, tryptophan 348, and phenylalanine 355 (Terawaki et al., 2006), and both the F355R and M346A/W348A mutations also abolished interaction with the ezrin FERM domain. Surprisingly, the F355A mutation only had a modest negative effect on binding ezrin FERM under these conditions. Additionally, EBP50 can form an intramolecular association between its C-terminal −FSNL sequence and its second PDZ domain (Morales et al., 2007). Addition of a single alanine to the C terminus abolishes the ability of the tail to bind its own PDZ2 domain, but this had little effect on the EBP50 tail–ezrin FERM interaction.
Figure 2.
Mutations in the tail of EBP50 perturb binding to ezrin. (A) Schematic of MBP-EBP50 tail (320–358) constructs, with the α-helical region that interacts with the ERM FERM domain indicated. Mutations or truncations were made in the tail of EBP50 that alter or remove key residues necessary for interaction with ezrin based on known structures. The additional alanine shown in red was added in the EBP50-LA construct to inhibit the tail–PDZ2 interaction. (B) Ezrin FERM coupled to CNBr-agarose was used to recover soluble MBP-EBP50 tails with indicated mutations/truncations. Gels were stained for total protein with IRDye.
The EBP50 tail mutations were introduced into an RNAi-resistant GFP-EBP50 construct and expressed in siEBP50-treated JEG-3 cells (Fig. 3 and Fig. S1 A). Expression levels of each GFP-EBP50 construct were similar to normal endogenous EBP50 levels, and in siEBP50-treated cells were three- to fourfold higher than the residual endogenous protein (Fig. S1 A). About 68% of JEG-3 cells normally show abundant microvilli on their apical surface, and when EBP50 levels are reduced 70% by siRNA treatment the number of cells with microvilli is reduced to ∼23% as assessed by ezrin and F-actin localization (Fig. 3, A and B; Fig. S1 A). As we have noted earlier (Hanono et al., 2006), and as can be seen in Fig. 1, D–F and Fig. 3 B, microvilli immediately above cell–cell junctions are especially resistant to EBP50 knockdown and these are not considered when counting cells lacking microvilli. Expression of RNAi-resistant GFP-EBP50 restored microvilli in ∼50% of the cells, whereas expression of a control plasmid expressing a GFP protein targeted to the nucleus (GFP-Nuc) did not. Expression of all constructs harboring mutations that abolish the EBP50–ezrin interaction failed to restore microvilli (Fig. 3, A and B and equivalent expression levels: Fig. S1 A). Surprisingly, the EBP50-F355A mutant, which is still able to bind the ezrin FERM, was unable to restore microvilli perhaps for some other reason, as it was unable to incorporate into microvilli even in the presence of endogenous EBP50 (unpublished data). Interestingly, inhibition of the EBP50 intramolecular interaction in the EBP50-LA mutant was fully competent to form microvilli. Thus, the interaction of EBP50 with ezrin is critical for the presence of microvilli on JEG-3 cells.
Figure 3.
The EBP50–ezrin interaction is critical for microvilli. GFP-tagged RNAi-resistant mutants of EBP50 were expressed in EBP50 siRNA (siEBP50)–treated JEG-3 cells and their abilities to maintain microvilli were assayed. (A) Results from scoring cells for the presence of microvilli; error bars indicate mean ± SD. (B) Representative maximum projection images of the apical surface from the microvillar rescue assay. Top row was not treated with siEBP50, and shows control cells transfected to express the GFP-Nuc control plasmid. Cells were stained for ezrin (red) and actin (blue) and transfected cells identified as those expressing the GFP-tagged construct (green). Microvilli were visualized on the apical surface of cells by ezrin and phalloidin staining. Bar, 10 µm.
To investigate the roles that EBP50’s PDZ domains play in microvilli maintenance, the conserved GYGF motif in the beginning of each PDZ domain was mutated to GYAA to specifically abolish binding to PDZ ligands (Doyle et al., 1996). The single and double PDZ mutants were expressed in cells treated with siEBP50 at levels 3–6 times higher than residual endogenous levels, and their ability to maintain microvilli was assayed as before (Fig. 4 and expression levels: Fig. S1 B). A truncated form of EBP50 containing only the last 39 amino acids of the tail, which is able to bind ezrin but lacks PDZ domains (Fig. 2), was unable to restore microvilli (Fig. 4 A) despite the fact that it localizes to microvilli in nonsiRNA-treated cells (unpublished data), suggesting that EBP50 isn’t merely binding to ERMs to keep them in their open active conformation. Constructs with both PDZ1 and PDZ2 mutated, or just PDZ1 mutated showed a loss of microvilli similar to the levels of the EBP50 tail alone (Fig. 4, A and B). Interestingly, cells expressing EBP50 with mutation of PDZ2 alone were able to form microvilli at levels indistinguishable from wild type. To determine if defects in PDZ binding were also causing defects during the assembly stage of microvilli, cells coexpressing ezrin-GFP and either RNAi-resistant TagRFPT-EBP50 wild-type or with both PDZ1 and 2 mutated were treated with siEBP50 and imaged for 7 min every 15 s (Videos 3 and 4, respectively). Cells expressing wild-type EBP50 showed numerous microvilli at various stages in the microvillar life cycle, whereas cells expressing the EBP50 PDZ1/2 mutant showed no evidence of microvilli assembly. Collectively, these results indicate that EBP50’s ability to bind PDZ1 but not PDZ2 ligands is critical for the assembly of microvilli.
Figure 4.
PDZ1 but not PDZ2 binding is required for microvilli. (A) Quantification of cells scored for the presence of microvilli; error bars indicate mean ± SD. (B) Representative images of the apical surface from the microvillar rescue assay. Cells were stained for ezrin (red) and actin (blue) and transfected cells identified as those expressing the GFP-tagged construct (green). Bar, 10 µm.
We have shown that overexpressing a construct of EPI64 that cannot bind the first PDZ domain of EBP50 causes a loss of microvilli (Hanono et al., 2006). To investigate whether the PDZ1 mutant of EBP50 is unable to assemble microvilli because it can no longer bind to EPI64, we generated a chimera of EPI64 fused to the last 60 residues of EBP50 that directly links EPI64 to ezrin, effectively bypassing EBP50. When this chimera was expressed in siEBP50-treated cells it was unable to restore microvilli (Fig. S2). In cells that still had a few remaining microvilli the chimera localized exclusively to the base of microvilli (Fig. S2 B), which is where endogenous EPI64 has been shown to localize (Hanono et al., 2006). Therefore, an inability to bind EPI64 is unlikely to be the only reason mutation of PDZ1 prevents EBP50 from forming microvilli.
Cell cycle–regulated phosphorylation of EBP50 inhibits its function in interphase, but not mitotic, cell microvilli
As introduced earlier, EBP50 is subject to phosphorylation on multiple sites by several different kinases, so we set out to evaluate the role of these modifications by each kinase in microvilli formation. The kinase GRK6A phosphorylates rabbit EBP50 on serine 289 (serine 290 in human EBP50) and is responsible for 75% of the constitutive levels of EBP50 phosphorylation (Hall et al., 1999). We generated mutations in human EBP50 in both serine 290 and the adjacent serine 291 to either alanine or aspartate to mimic nonphosphorylated and phosphorylated forms of EBP50, respectively. These phospho-deficient and -mimetic mutants of GFP-EBP50 were expressed in cells depleted of endogenous EBP50 and scored for their abilities to form microvilli (Fig. S3). Interestingly, both the phospho-deficient and -mimetic mutants of EBP50 were able to maintain microvilli in siEBP50-treated cells (Fig. S3, A and B), indicating that phosphorylation of EBP50 by GRK6A has no effect on microvilli assembly or maintenance.
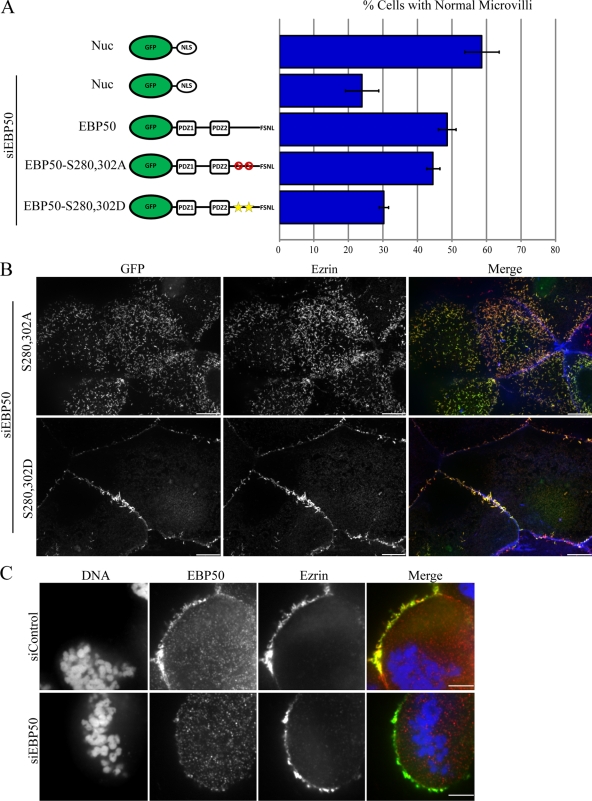
During the early stages of mitosis in HeLa cells, Cdc2 phosphorylates EBP50 on serine residues 280 and 302 (He et al., 2001). To investigate potential regulation by Cdc2, we generated phospho-mimetic and -deficient mutants of these serine residues and determined their ability to maintain microvilli in siEBP50-treated cells (Fig. 5, A and B and expression levels: Fig. S1 B). The phospho-deficient mutant was able to maintain microvilli at levels indistinguishable from wild type, whereas the phospho-mimetic mutant was unable to restore microvilli (Fig. 5, A and B). To determine if EBP50 phosphorylation by Cdc2 also causes a defect during the formation of microvilli, we coexpressed ezrin-GFP and a TagRFPT-tagged version of the Cdc2 site phospho-mimetic EBP50 in cells treated with siEBP50 and found that similar to cells depleted of EBP50 or expressing a PDZ1 mutant, microvilli were unable to form (Video 5). Thus, phosphorylation of EBP50 by Cdc2 inhibits its role in microvillar assembly in interphase cells.
Figure 5.
Phosphorylation by Cdc2 inhibits EBP50’s role in forming microvilli in interphase but not mitotic cells. (A) Results from scoring JEG-3 cells for the presence of microvilli; error bars indicate mean ± SD. (B) Representative images from the microvillar rescue assay. Cells were stained for ezrin (red) and actin (blue) and transfected cells identified as those expressing the GFP-tagged construct (green). Bar, 10 µm. (C) JEG-3 cells transfected with siRNA targeting EBP50 (siEBP50) or luciferase (siControl) were arrested in 50 ng/ml nocodazole overnight, fixed and stained for endogenous EBP50 (red), ezrin (green), and DNA (blue). Note the loss of EBP50 from microvilli of the cell cortex in siEBP50-treated cells, whereas ezrin localization remains unchanged. Bar, 5 µm.
Because mitotic cells are generally covered in small microvilli (Sanger and Sanger, 1980), we explored whether EBP50 becomes phosphorylated during mitosis in JEG-3 cells, and whether EBP50 is needed for mitotic cell microvilli. To examine if EBP50 becomes phosphorylated in mitosis, JEG-3 cells were arrested at prometaphase after incubation with 50 ng/ml nocodazole for 18 h, and then allowed to progress through mitosis by washout with full media. As is seen in HeLa cells, upon nocodazole arrest EBP50 becomes highly phosphorylated and remains heavily phosphorylated until 2–3 h after nocodazole washout (Fig. S4 A), and this phosphorylation can be largely abolished by inhibition of Cdc2 with 2 µM roscovitine (Fig. S4 B; He et al., 2001). During mitosis JEG-3 cells are covered with dense short microvilli rich in ezrin and EBP50 (Fig. 5 C). Strikingly, upon siEBP50 treatment microvilli are unaffected, maintaining an enrichment in ezrin despite a loss of EBP50 staining and expression (Fig. 5 C and Fig. S4 C). These results indicate that EBP50 is heavily phosphorylated during mitosis by Cdc2 but this phosphorylation, although inhibitory in interphase cells, has no observable effect in mitotic cells, indicating that mitotic microvilli are structurally and perhaps functionally distinct from those in interphase.
Phosphorylation of EBP50 by PKC regulates its role in microvilli and contributes to microvillar organization
PKC phosphorylates EBP50 on serines 162 (Raghuram et al., 2003), 339, and 340 (Fouassier et al., 2005), and this has been reported to result in an increased affinity for PDZ-binding ligands (Li et al., 2007). To determine if PKC plays any role in regulating EBP50 in microvillar formation, phospho-mimetic and -deficient point mutants were created at these serine residues and the control or mutant GFP-EBP50 constructs were expressed in JEG-3 cells treated with siEBP50, and their ability to preserve microvilli was assayed as before (Fig. 6 and expression levels: Fig. S1 B). Interestingly, both the phospho-deficient and -mimetic mutants were unable to maintain microvilli (Fig. 6, A and B). To determine if EBP50 phosphorylation by PKC also causes a defect during the formation of microvilli, we coexpressed ezrin-GFP and a TagRFPT-tagged version of the PKC site phospho-mimetic EBP50 in cells treated with siEBP50 and found that similar to cells depleted of EBP50 or expressing a PDZ1 mutant or Cdc2 site phospho-mimetic mutant, microvilli were unable to form (Video 6). Strikingly however, coexpression of both the phospho-mimetic and -deficient mutants was able to restore microvilli at levels indistinguishable from wild type (Fig. 6). Collectively, these results suggest that either the phosphorylation state of EBP50 mediated by PKC must cycle or that once phosphorylated by PKC, EBP50 engages in different interactions than its non-PKC–phosphorylated form and that all of these interactions are required for microvilli formation. In a small fraction of the cells coexpressing both the phospho-mimetic and -deficient mutants we noticed a clustering of microvilli, suggesting that PKC phosphorylation of EBP50 may alter the arrangement of microvilli along the apical surface (Fig. 6 B, arrows).
Figure 6.
Phosphorylation by PKC regulates EBP50’s ability to maintain microvilli. (A) Results from scoring JEG-3 cells for the presence of microvilli; error bars indicate mean ± SD. (B) Representative images from the microvillar rescue assay. Cells were stained for ezrin (red) and actin (blue) and transfected cells identified as those expressing the GFP-tagged construct (green). Bar, 10 µm.
To further explore the possible role of PKC in microvillar regulation, JEG-3 cells were serum starved overnight and then treated with 1 µM PMA for 30 min to activate PKC. Strikingly, treatment with PMA caused some cells to develop either dorsal ruffles or clusters of microvilli reminiscent of those we observed with coexpression of the PKC site phospho-mimetic and -deficient EBP50 mutants (Fig. 7 A). These microvilli clusters were enriched in both EBP50 and ezrin and were centered around a tight knot of actin in the terminal web region at the base of the microvilli. To determine if these microvilli clusters were a result of PKC phosphorylation of EBP50, we overexpressed wild-type GFP-EBP50 or either the PKC phospho-mimetic or -deficient mutant of EBP50 in cells treated with PMA (Fig. 7 B). Both the wild-type and PKC site phospho-mimetic constructs showed microvilli clusters in ∼20% of cells, whereas the phospho-deficient mutant almost completely blocked microvilli clustering in PMA-treated cells (Fig. 7 C). The clustering of microvilli seen with the expression of the PKC phospho-mimetic mutant only became pronounced with PMA treatment, indicating that in addition to EBP50, some other factor(s) need to be phosphorylated to induce microvilli clustering. Thus, phosphorylation of EBP50 by PKC, as well as other factors, leads to the rearrangement of microvilli into apical clusters.
Figure 7.
PKC activation and phosphorylation of EBP50 promotes microvillar rearrangement. (A) JEG-3 cells were serum starved overnight, treated with 1 µM PMA for 30 min, then fixed and stained for endogenous EBP50 (red), ezrin (green), and actin (blue). Bar, 5 µm. (B) GFP-EBP50 wild-type and PKC phospho-mimetic (S162,339,340D) or -deficient (S162,339,340A) mutants were overexpressed in cells treated with PMA, stained for ezrin (red) and actin (blue), and their apical surface phenotype was scored. Bar, 10 µm. (C) Results from scoring JEG-3 cells for their presence of microvilli clusters; error bars indicate mean ± SD.
EBP50 is an elongated monomer regardless of its phosphorylation state
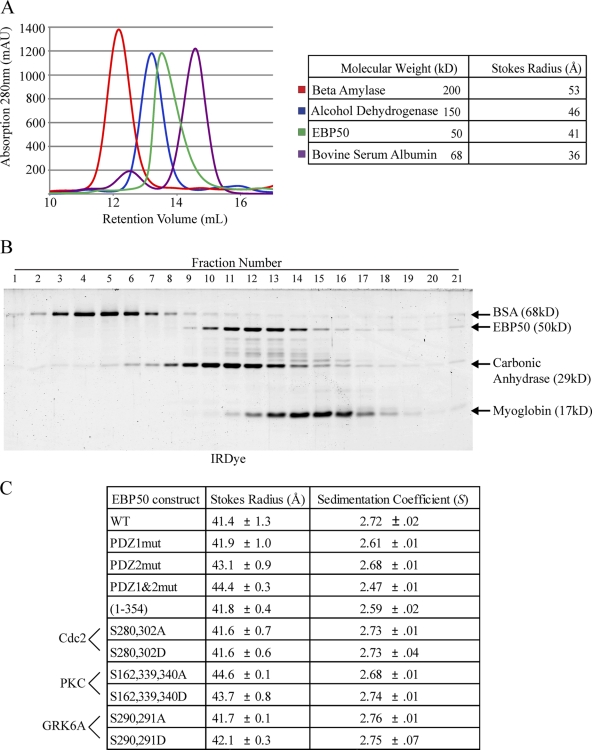
Our results show that EBP50 mutants with phospho-mimetic or -deficient mutations can affect its function in microvillar assembly. Because it has been suggested, based on blot overlay experiments, coprecipitations out of cell lysates, and migration in sizing columns, that EBP50 may be capable of oligomerizing (Fouassier et al., 2000; Lau and Hall, 2001; Shenolikar et al., 2001) and that phosphorylation of EBP50 plays a role in regulating its oligomeric state (Lau and Hall, 2001; Fouassier et al., 2005), we set out to determine the oligomerization state of EBP50 and whether it is affected in our EBP50 mutants. To purify recombinant EBP50, 6xHis-SUMO–tagged versions of wild-type and all the site-directed mutants were expressed in bacteria, purified on Ni2+ resins, and the EBP50 was then cleaved off from the SUMO tag to recover highly pure untagged EBP50 and its mutants (purification of wild-type is shown in Fig. S5). Wild-type EBP50 analyzed on a Superdex 200 10/300 GL gel filtration column ran as a single population with a Stokes radius of 41 Å, which corresponds for a globular protein to a molecular weight of ∼100 kD (Fig. 8 A), a size similar to that reported earlier (Li et al., 2007; Morales et al., 2007). The large Stokes radius suggests that it is either a dimer or a highly elongated monomer. As an independent method for examining its physical properties, the S value of EBP50 was determined by velocity sedimentation in sucrose gradients. Sedimentation together with standards of known S value indicated a sedimentation coefficient of 2.7 S for EBP50 (Fig. 8 C). Using these two physical constants predicts a native molecular weight of ∼47 kD (Siegel and Monty, 1966). Furthermore, using the partial specific volume value for proteins of 0.74 cm3/g, these data reveal a highly asymmetric protein with a frictional coefficient f/fo of 1.8 (a value of 1 represents a spherical protein, whereas a higher number is characteristic of an elongated molecule), thereby accounting for its high Stokes radius and low sedimentation coefficient. Thus, EBP50 is an elongated monomer in solution.
Figure 8.
EBP50 is an elongated monomer. (A) Untagged, full-length EBP50 was run over a Superdex 200 10/300 GL column and compared with size standards β-amylase, alcohol dehydrogenase, and bovine serum albumin (BSA); the Stokes radius of EBP50 was calculated to be 41 Å. (B) EBP50 was run on sucrose gradients with size standards BSA (4.30 S), carbonic anhydrase (3.20 S), and myoglobin (2.04 S); the sedimentation coefficient was calculated to be 2.72 S. (C) Table summarizing gel filtration and sucrose gradient results for all PDZ and phosphorylation mutants of EBP50. The results are from at least two independent determinations each.
Analysis of EBP50 with mutations in either PDZ domain, especially to PDZ2, increased the Stokes radius slightly and decreased the S value, indicating the protein is still monomeric but becomes slightly more elongated (Fig. 8 C), consistent with the finding that the tail normally binds PDZ2 (Morales et al., 2007). Phospho-mimetic and -deficient mutations of either the Cdc2 or GRK6A phosphorylation sites did not affect the overall conformation of EBP50 (Fig. 8 C). Interestingly, mutation of the PKC sites to either phospho-mimetic or -deficient forms resulted in a modest increase in Stokes radius of EBP50, similar to that seen with mutation of the second PDZ domain (Fig. 8 C), and similar to what has been seen before (Li et al., 2007).
Importantly, in none of the 11 EBP50 constructs analyzed by gel filtration and sucrose gradients, each of which was performed at least twice, did we detect any evidence of EBP50 oligomers. Additionally, no evidence of oligomerization was seen when all constructs were sized by blue-native PAGE (data not shown). Collectively, these results indicate that EBP50 is an elongated monomer and phospho-mimetic mutations do not mediate its oligomerization.
Cdc2 or PKC phosphorylation of EBP50 inhibits ligand accessibility to PDZ1 when PDZ2 is occupied
Our results reveal that EBP50 is monomeric in solution, and that constructs with defects in ezrin binding or PDZ1 binding are not functional for microvillar formation in vivo. Additionally, phospho-mimetics for the Cdc2 and PKC sites, and phospho-deficient for the PKC sites, are also nonfunctional in vivo in terms of the microvilli maintenance assay, but the underlying reasons were not clear. We therefore explored in depth the biochemical properties of wild-type EBP50, the Cdc2 phospho-deficient and -mimetic constructs S280,302A and S280,302D, and the PKC phospho-deficient and -mimetic constructs S162,339,340A and S162,339,340D.
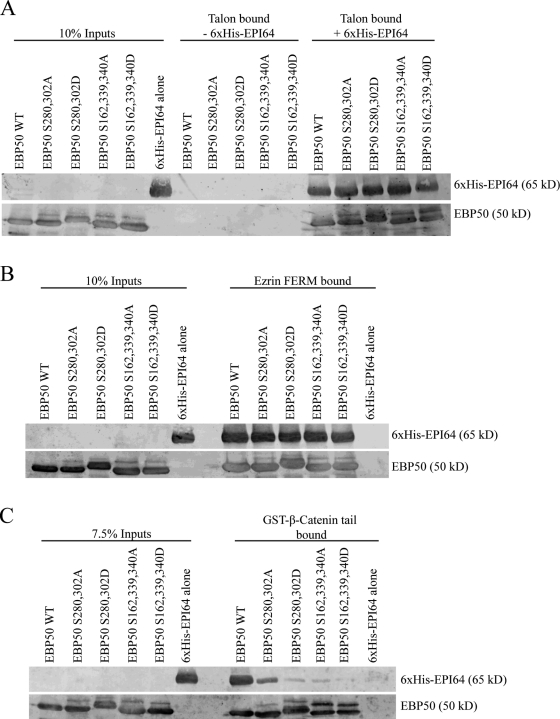
The most likely scenario for the lack of in vivo function of the Cdc2 EBP50 phospho-mimetic mutant and PKC phospho-mimetic and -deficient mutants is that they are compromised in either PDZ1 accessibility or ezrin binding. Therefore, we first examined the mutants’ abilities to bind EPI64, a well-characterized ligand that binds PDZ1 selectively (Reczek and Bretscher, 2001). 6xHis-tagged EPI64 was incubated with untagged EBP50 constructs and precipitated with Talon cobalt beads, which bind the 6xHis tag (Fig. 9 A). All phospho-deficient and -mimetic mutants were able to bind EPI64 as well as wild-type EBP50, indicating that phosphorylation has no pronounced effect on PDZ1 accessibility. Next, we wanted to investigate if phosphorylation could affect the ability of EBP50 to bind ezrin, or PDZ1 ligands when its tail was bound to ezrin. Wild-type and all phospho-deficient and -mimetic EBP50 mutants bound equally to immobilized ezrin FERM domain and simultaneously bound to soluble 6xHis-EPI64 (Fig. 9 B). This eliminated a defect in ezrin binding, and simultaneous PDZ1 and ezrin binding, as causes for their lack of in vivo function. Next, we examined if phosphorylation could regulate simultaneous occupation of both PDZ domains in EBP50. The tail of β-catenin has been shown to bind selectively to PDZ2 of EBP50 (Shibata et al., 2003), so we used immobilized GST–β-catenin tail to precipitate a preincubated mixture of EBP50 and EPI64 (Fig. 9 C). Wild-type EBP50 and the Cdc2 phospho-deficient mutant were retained together with EPI64, although the mutant binding to EPI64 was less efficient. However, although both phospho-mimetic and the PKC phospho-deficient mutants bound β-catenin equivalently to wild type, none were able to retain EPI64. Thus, phosphorylation by either Cdc2 or PKC blocks PDZ1 accessibility specifically when PDZ2 is occupied. Additionally, mutation of the PKC sites to alanine also blocks the accessibility of PDZ1 when PDZ2 is occupied.
Figure 9.
Phosphorylation of EBP50 by PKC or Cdc2 inhibits its ability to bind two PDZ ligands simultaneously. (A) Binding between untagged full-length EBP50 and 6xHis-EPI64 was tested by precipitating the 6xHis tag with Talon beads. (B) EBP50 was preincubated with soluble 6xHis-EPI64 and complexes recovered on FERM beads. (C) EBP50 was preincubated with soluble 6xHis-EPI64 and its ability to simultaneously bind GST–β-catenin tail coupled to glutathione agarose was tested. For A–C, 20% of the bound fraction was run on the gels.
Cdc2 and PKC phosphorylation and PDZ2 occupancy coordinately regulate the microvillar assembly function of EBP50
Our biochemical results show that in the context of Cdc2 or PKC phospho-mimetic mutations, or the PKC phospho-deficient mutation, accessibility of PDZ1 in EBP50 is inhibited by the occupancy of PDZ2. If this in vitro biochemical result is the cause of the inability of the mutants to function in microvillar assembly, introducing an additional mutation to inactivate PDZ2 should render the PDZ1 accessible and the proteins functional. We therefore mutated the PDZ2 domain in these mutants and assayed their ability to restore microvilli in siEBP50-treated cells (Fig. 10). Strikingly, mutation of the PDZ2 domain was able to reverse the defect in microvillar formation for both the Cdc2 site phospho-mimetic and the PKC site phospho-mimetic and -deficient mutants (Fig. 10, A and B). Collectively, these results suggest that EBP50 phosphorylation by either PKC or Cdc2 inhibits simultaneous binding to both PDZ domains and that in cells the presence of a PDZ2-binding partner acts to inhibit PDZ1 binding in this phosphorylated pool of EBP50.
Figure 10.
Mutation of PDZ2 restores the ability of Cdc2 and PKC site EBP50 phospho-mutants to form microvilli. (A) Results from scoring JEG-3 cells for the presence of microvilli; error bars indicate mean ± SD. (B) Representative images from the microvillar rescue assay. Cells were stained for ezrin (red) and actin (blue) and transfected cells identified as those expressing the GFP-tagged construct (green). Bar, 10 µm.
Discussion
Microvilli are striking and dynamic morphological features of the apical aspect of polarized epithelial cells, yet little is known about how the cell assembles and regulates their presence. Because both the ezrin and EBP50 knockout mice have defective intestinal epithelial microvilli (Morales et al., 2004; Saotome et al., 2004), these proteins are potential candidates for regulating microvilli. Ezrin’s activity is well known to be regulated by phosphorylation, and recent studies have indicated that the kinase Mst4 is critical in activating it at the apical aspect of epithelial cells (Bretscher, 2009; ten Klooster et al., 2009). EBP50 has been shown to bind multiple ligands through its PDZ domains to either scaffold functionally related proteins (Takahashi et al., 2006) or regulate the presence of proteins at the cell surface, such as EGFR and CFTR (Short et al., 1998; Lazar et al., 2004). Either of these activities could potentially contribute to microvillar biogenesis. Because EBP50 is the target of several kinases, much effort has been devoted to investigating their regulatory function. Thus, it has been suggested that EBP50 phosphorylation by GRK6A regulates its oligomerization to facilitate cellular signaling complexes (Lau and Hall, 2001; Shenolikar and Weinman, 2001), and PKC phosphorylation modulates accessibility to its second PDZ domain (Li et al., 2007). Here we investigate one major function of EBP50, its contribution to the presence of microvilli on epithelial cells, and further characterize its biophysical properties in detail, as well as biochemical and functional consequences of phosphorylation by PKC and Cdc2.
We first investigated which aspect of the microvillar life cycle is affected by EBP50 depletion. Scanning electron microscopy and live-cell imaging with ezrin-GFP both show that cells depleted of EBP50 show no evidence of even nascent microvilli, thus establishing that EBP50 is involved in microvillar assembly. An interesting aspect of this analysis is that microvilli above cell junctions are less sensitive to EBP50 depletion. A similar phenotype has been seen in cells expressing a dominant-negative ezrin construct (Crepaldi et al., 1997), suggesting that the signaling pathways that drive microvillar assembly might be more potent in this region of the cell.
Using expression of RNAi-resistant GFP-EBP50 in cells depleted of EBP50, we have been able to investigate which biochemical properties of EBP50 are necessary for its function. The requirement for EBP50 to bind ezrin is not surprising, as we have shown that either overexpressing an EBP50 construct lacking the ezrin binding site, or siRNA knockdown of EBP50, results in microvillar loss (Hanono et al., 2006). Given the requirement for EBP50, we also suspected that one or both PDZ domains would be needed, and have shown that the function of PDZ2 is dispensable for the presence of microvilli. At this point, the required PDZ1 ligand that is necessary for microvilli remains unknown. One likely candidate is EPI64, as overexpressing a construct of this protein defective in EBP50 binding also results in loss of microvilli (Hanono et al., 2006). However, mislocalization of EPI64, which is a RabGAP protein and an effector for Arf6, away from EBP50 (Hanono et al., 2006; Itoh and Fukuda, 2006), may perturb the cell in additional ways that could indirectly result in loss of microvilli. Moreover, direct targeting of EPI64 to microvilli by expressing a chimera of EPI64 fused to the EBP50 tail did not rescue the loss of microvilli in EBP50-deficient cells. Although this is an artificial system, a similar experiment targeting the EBP50 ligand PDZK1 by chimeric fusion to the EBP50 tail is able to rescue microvilli in EBP50-deficient cells (LaLonde et al., 2010). Our working hypothesis is that microvilli require an unidentified ligand specific for EBP50’s PDZ1 domain, or in the artificial fusion system through binding one of PDZK1’s four PDZ domains. Our current studies are aimed at identifying the relevant ligand.
Because EBP50 is the target of many kinases, we have analyzed phospho-mimetic and -deficient mutants of EBP50 in their ability to restore microvilli. The major in vivo site of phosphorylation, S290, does not appear to affect EBP50’s role in regulating microvilli, although it has been reported to increase oligomerization (Lau and Hall, 2001). PKC phosphorylates S162 in PDZ2 and this reduces its affinity for CFTR (Raghuram et al., 2003). PKC also phosphorylates S339 and S340, and phosphorylation of these sites has been shown to inhibit the tail–PDZ2 interaction and thereby enhance the binding capacity for CFTR (Li et al., 2007). We have found that an EBP50 mutant where we have added a C-terminal alanine to eliminate the EBP50 tail–PDZ2 intramolecular association is functionally capable of microvilli assembly in our assay, implying that the loss of tail–PDZ2 interaction is not the reason our PKC site phospho-mimetic mutant does not support microvilli. Additionally, and quite surprisingly, both the EBP50 S162,339,340A and S162,339,340D mutants were unable to function in this assay. Preliminary experiments with these individual sites mutated gave intermediate results in vivo, and their biochemical properties have not been fully analyzed (unpublished data). Remarkably, simultaneous coexpression of these phospho-mimetic and -deficient mutants restored microvilli. Whether this indicates a requirement for phosphorylation recycling, or of differential binding of distinct ligands to the phospho-mimetic and -deficient forms is not clear. However, the physiological relevance of PKC site phosphorylation is strongly suggested by a similar clustered microvillar phenotype in cells treated with the PKC activator PMA, and expressing either wild-type or the phospho-mimetic construct, but not the phospho-deficient mutant.
The two Cdc2 sites, S280 and S302, become highly phosphorylated in mitosis (He et al., 2001). Expression of the S280,302D phospho-mimetic mutant was not able to support microvilli, whereas the corresponding phospho-deficient forms were. As these sites are located well downstream of the PDZ domains, this was an unexpected finding. Our results show that microvilli on mitotic cells are much shorter than those on interphase cells and do not have the same requirement for EBP50. It appears, therefore, that in mitotic cells the regulation conferred by EBP50 has been inactivated by phosphorylation to allow cells to assemble structurally different microvilli, but that still contain ezrin. Despite not being required, EBP50 is nevertheless present in mitotic cell microvilli, presumably because the phosphorylated EBP50 still retains the ability to bind active ezrin.
Our biochemical and functional analyses of the EBP50 Cdc2 phospho-mimetic and PKC phospho-mimetic and -deficient mutants have uncovered a novel mechanism of regulation explaining why these constructs cannot support the assembly of microvilli in interphase cells. All of these mutants are indistinguishable in vitro from the wild-type protein in their ability to bind ezrin alone, or the PDZ1-specific ligand EPI64 alone, or simultaneously to both ezrin and EPI64. However, just those mutants unable to support microvillar assembly in vivo are also unable to bind a PDZ1 ligand in vitro when PDZ2 is occupied. If this mechanism operates in vivo, we predicted that additionally inactivating PDZ2 should restore the ability of these mutants to support microvillar assembly, and indeed this is precisely what we found. These results reinforce the notion that PDZ1 has to bind a ligand to function in microvillar biogenesis. More significantly, they have uncovered an in vivo regulation of PDZ1 accessibility by occupancy of PDZ2, implying that there is a sufficient level of a PDZ2 ligand in vivo to inhibit binding to PDZ1. Thus, EBP50 is regulated both by phosphorylation and PDZ2 occupancy.
Our biophysical studies on purified wild-type and phospho-mimetic and -deficient mutants show that they are highly elongated monomeric proteins in solution. Our analyses have been performed at concentrations of EBP50 many times higher (2–4 mg/ml) than is encountered in vivo, so the lack of a tendency to form oligomers in vitro suggests that EBP50 is unlikely to form intermolecular associations in vivo. However, this does not exclude the possibility that other proteins could bridge individual EBP50 monomers to generate hetero-oligomeric species in vivo, which may in part explain some of the past results involving precipitation out of cell extracts (He et al., 2001; Shenolikar et al., 2001; Fouassier et al., 2005).
Several studies have suggested that EBP50 is regulated by its oligomeric state. Two properties of EBP50 may have contributed to some of these conclusions. First, it was not initially appreciated that the tail of EBP50 can bind PDZ2 intramolecularly, so some overlay experiments may have been misinterpreted. Second, being unaware of the high asymmetry of EBP50 might have led some authors to misinterpret gel filtration studies, where EBP50 runs at a position typical for a globular protein of ∼100 kD, exactly twice the size it runs on SDS-PAGE.
In summary, EBP50 has previously been shown to be an important scaffolding protein in bringing specific membrane ligands together, or retaining them at the cell surface. Here we show that EBP50 is also a point of regulation for microvilli, and have elucidated its functional requirements and regulatory mechanisms in this process. More than just a “simple scaffolding” protein, EBP50 is emerging as an important nexus for coordinating structure with function at the apical aspect of interphase polarized epithelial cells.
Materials and methods
Antibodies and reagents
Antisera and affinity-purified antibodies against human ezrin and EBP50 were described previously (Bretscher, 1989; Reczek et al., 1997). The mouse antibody against α-tubulin was purchased from Sigma-Aldrich. The mouse antibody against ezrin developed at the National Cancer Institute (Bethesda, MD) was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology at The University of Iowa (Iowa City, IA). All Alexa Fluor–conjugated secondary antibodies, YOPRO-1 DNA stain, and Alexa Fluor 660–conjugated phalloidin were purchased from Invitrogen. IRDye 680– and 800–conjugated secondary antibodies were obtained from LI-COR Biosciences. The siRNAs targeting human EBP50 (5′-CGGCGAAAACGTGGAGAAG-3′) and Luciferase GL2 (5′-CGUACGCGGAAUACUUCGA-3′) were obtained from Thermo Fisher Scientific and Applied Biosystems.
Cell culture and transfection
JEG-3 cells (American Type Culture Collection) were maintained in a 5% CO2 humidified atmosphere at 37°C in MEM with 10% FBS (Invitrogen). Cells were transfected with polyethylenimine (PEI; Polysciences, Inc.) and DNA as described previously (Hanono et al., 2006). For microvillar rescue assays, cells were first transfected with 1–2 µg of DNA at low confluence, allowed to recover for 24 h, then transfected again with 10 nM of siRNA using Lipofectamine RNAiMax, allowed to grow for another 48 h, then processed for immunofluorescence and counting, or Western blot analysis. Transfected cells expressed RNAi-resistant GFP-tagged EBP50 at levels 70–300% of endogenous in nonsiRNA–treated cells as determined by quantitative Western blot. For mitotic arrest studies cells were incubated with 50 ng/ml nocodazole (Sigma-Aldrich) ± 2 µM roscovitine (Sigma-Aldrich) for 18–20 h before preparation for analysis. For PKC activation experiments, cells were serum starved overnight, then treated with 1 µM PMA for 30 min before preparation for analysis.
DNA constructs
All EBP50 constructs were created in pEGFP-C2 (Takara Bio Inc.), pMal-C2X (New England Biolabs, Inc.), and pE-SUMO (LifeSensors Inc.). TagRFPT-EBP50 constructs were created by PCR from a vector expressing TagRFPT that was generously provided by Roger Tsien (University of California, San Diego, La Jolla, CA; Shaner et al., 2008). Ezrin-GFP was created in pEGFP-N2 (Takara Bio Inc.). The β-catenin tail cDNA was obtained from Thermo Fisher Scientific and tail residues 695–781 were inserted into pGEX-6P1 (GE Healthcare) using PCR. Untagged ezrin FERM in pQE16 (QIAGEN) was described previously (Reczek et al., 1997). Full-length 6xHis-tagged EPI64 was created in pFastBac-HT (Invitrogen).
Western blotting and immunofluorescence
Protein samples were separated by SDS-PAGE and transferred to Immobilon-FL (Millipore). All Western blots were imaged using an Odyssey infrared imaging system (LI-COR Biosciences). For immunofluorescence, cells grown on glass coverslips were fixed in 3.7% formaldehyde/PBS for 10 min at room temperature. Cells were permeabilized in 0.2% Triton X-100/PBS for 5 min at room temperature, rinsed in PBS, and incubated in primary antibodies in 3% FBS in PBS. After washing in PBS, secondary antibodies and additional markers (phalloidin, DNA counterstain) were added in 3% FBS in PBS. After further washing in PBS, coverslips were mounted onto glass slides using Vectashield (H-1000; Vector Laboratories), and images were then acquired on a CSU-X spinning disk microscope (Intelligent Imaging Innovations) with spherical aberration correction device, 63x 1.4 NA objective on an inverted microscope (model DMI6000B; Leica), and an HQ2 CCD camera (Photometrics) using Slidebook 5.0 (Intelligent Imaging Innovations). Images were also processed using Slidebook 5.0 (Intelligent Imaging Innovations).
Scoring the apical surface phenotype of transfected cells
Transfected JEG-3 cells were scored as described previously (Hanono et al., 2006). In brief, cells were stained for ezrin and F-actin to indicate the apical membrane phenotype, whereas the GFP-tagged constructs marked transfected cells. At least 250 cells were counted for each condition in each of three replicates. Cells were scored as having normal microvilli, no microvilli, lacking/few microvilli, or ruffles (with PMA treatment cells were also scored as having microvilli clusters).
Scanning electron microscopy
JEG-3 cells were fixed by removing cell culture media and rapid addition of 2.5% glutaraldehyde in PO4 buffer for 45 min. After fixation, cells were rinsed three times in PO4 buffer and dehydrated in a graded ethanol series. To minimize drying artifacts samples were first dehydrated in 20% ethanol and then increasing amounts for 10 min each followed by two changes in 100% for 10 min each. After dehydration, samples were critical point dried and sputter coated with gold and palladium. Samples were imaged on a scanning electron microscope (Stereoscan 440; Leica) operated at 25 kV; images were obtained at 4,000 and 20,000x and processed with ImageJ.
Live-cell imaging
Transfected JEG-3 cells grown in 35-mm glass-bottom dishes (MatTek Corp) were washed twice in PBS and then maintained in low sodium bicarbonate phenol red–free MEM (Sigma-Aldrich) supplemented with 25 mM Hepes, pH 7.4, with 10% FBS and glutamax (Invitrogen). Live cells were imaged by time-lapse microscopy on a CSU-X spinning disk (Intelligent Imaging Innovations) with spherical aberration correction device, 63x 1.4 NA objective on an inverted microscope (DMI6000B; Leica) and QuantEM EMCCD camera (Photometrics) at 37°C in an environmental chamber (Okolab) controlled by Slidebook 5.0 (Intelligent Imaging Innovations). Movies were processed using Slidebook 5.0 (Intelligent Imaging Innovations) and ImageJ.
Purification of recombinant proteins
Ezrin FERM was purified as described previously (Reczek et al., 1997). In brief, induced cells were lysed in 180 mM KH2PO4, pH 7.0, with complete protease inhibitor (Roche), lysed by sonication (Branson Ultrasonics), centrifuged and run over a hydroxyapatite column (Pall), and eluted by linear gradient of 180–800 mM KH2PO4. Ezrin FERM–containing fractions were pooled and dialyzed against 20 mM MES, 150 mM NaCl, pH 6.7, and applied to an S-sepharose column (GE Healthcare) and eluted in a linear gradient of 0.15–1.0 M NaCl. To conjugate purified FERM to cyanogen bromide (CNBr)–activated Sepharose (Sigma-Aldrich) it was dialyzed into C Buffer (0.1 M NaHCO3 and 0.5 M NaCl, pH 8.3) and then added to hydrated CNBr beads for a final concentration of 2 mg/ml on the resin overnight. Beads were then washed, and blocked in 0.25 M glycine, pH 8.0, overnight. Beads were washed again, and then stored as a 50% slurry in H buffer (50 mM Tris, and 150 mM NaCl, pH 7.4).
Maltose-binding protein (MBP) fused to the last 39 amino acids of the EBP50 tails with various mutations were purified as described previously (Finnerty et al., 2004). In brief, induced cells were lysed in PBS with 1 mM DTT and complete protease inhibitor by sonication, centrifuged and run over amylose resin column (New England Biolabs, Inc.), washed with PBS, and then eluted in PBS with 10 mM maltose.
Bacterical pellets induced to express GST–β-catenin tail were lysed in 150 mM NaCl, 20 mM Tris, pH 7.4, 0.1% BME, and 0.1% Triton X-100 with complete protease inhibitor by sonication, centrifuged and added to hydrated glutathione agarose (Sigma-Aldrich), washed, and left as a 50% slurry for binding assays.
SUMO-EBP50 constructs were purified as described previously (LaLonde et al., 2010). In brief, induced bacterial pellets were lysed in binding buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4, and 1% Triton X-100) by sonication, centrifuged and run over a His gravitrap column (GE Healthcare), washed, and eluted in elution buffer (20 mM sodium phosphate, 500 mM NaCl, and 500 mM imidazole, pH 7.4). Eluate was dialyzed into binding buffer, cleaved with 6xHis-Ulp1 for 40 min at 30°C to remove the SUMO tag, and then run over another His gravitrap column and flowthrough containing untagged EBP50 was collected and dialyzed into 150 mM NaCl, 10 mM Tris, pH 7.4, with 1 mM DTT.
6xHis-tagged EPI64 expressed in SF9 insect cells was lysed in 150 mM NaCl, 1% NP-40, and 50 mM Tris, pH 8.0 with complete protease inhibitor by sonication, centrifuged, and incubated with Ni2+-NTA agarose (QIAGEN) for 1 h. Beads were washed in 500 mM NaCl, 5 mM imidazole, and 50 mM Tris, pH 8.0, and then 6xHis-EPI64 was eluted in wash buffer with 1 M imidazole.
Gel filtration and sucrose gradients
1–2 mg of untagged full-length EBP50 was run over a superdex 200 10/300 GL column on an AKTA FPLC (GE Healthcare) in 150 mM NaCl and 10 mM Tris, pH 7.4, with 1 mM DTT. Stokes radii were calculated as described previously (Begg et al., 2001). Full-length EBP50 was run on 5–20% sucrose gradients with size standards BSA (4.3 S), carbonic anhydrase (3.2 S), and myoglobin (2.04 S) in a rotor (model SW60Ti; Beckman Coulter) at 50,000 rpm for 26 h at 4°C. Fractions were collected manually, run out by SDS-PAGE, and stained with IRDye blue protein stain (LI-COR Biosciences) for band densitometry and sedimentation coefficient calculation. Frictional ratios were calculated from ƒ/ƒo = Rs(4πN/3ŪMr)1/3, where Rs is the experimentally determined Stokes radius, N is Avogadro’s number, Ū is the partial specific volume (taken to be 0.74 cm3/g for protein), and Mr is the calculated molecular mass.
In vitro–binding assays
For EBP50-tail to ezrin-FERM binding assays, 7 µg of MBP-EBP50 tail constructs were incubated with 7 µl of ezrin FERM-CNBr bead slurry (coupled to be 2 µg/µl on the beads) in 300 mM NaCl, 0.1% Triton X-100, 5% glycerol, and 20 mM Tris, pH 7.4, for 1 h at 4°C, washed, and then boiled in SDS sample buffer. Samples were run on SDS-PAGE, stained with IRDye blue protein stain and scanned using an Odyssey infrared imaging system.
All other binding assays were incubated in binding buffer (150 mM NaCl, 0.1% Triton X-100, 5% glycerol, and 25 mM Tris, pH 7.4) at 4°C, washed, and boiled in sample buffer. For experiments testing the ability of EBP50 to bind soluble EPI64 simultaneously to either ezrin FERM-CNBr beads or GST–β-catenin tail beads, EBP50 and EPI64 were preincubated for 20 min on ice before addition of the beads. Binding conditions were set so the molarity of all proteins (both soluble and coupled to beads) were kept equal at 500 nM. Beads were boiled in SDS sample buffer and 20% of the sample was run on SDS-PAGE and transferred to Immobilon-FL membrane for Western blot analysis.
Online supplemental material
Fig. S1 shows Western blots of lysates from microvillar rescue experiments. Fig. S2 shows that an EPI64-EBP50 tail chimera is unable to restore microvilli in siEBP50-treated cells. Fig. S3 shows that phosphorylation of EBP50 by GRK6A does not affect its ability to form microvilli. Fig. S4 shows that EBP50 is phosphorylated by Cdc2 during mitosis. Fig. S5 shows purification of SUMO-EBP50. Video 1 shows normal microvilli in control cells. Video 2 shows defects in microvilli formation in siEBP50 cells. Video 3 shows RNAi-resistant EBP50 restores microvilli in siEBP50-treated cells. Video 4 shows that the RNAi-resistant EBP50-PDZ1/2 mutant is unable to form microvilli in siEBP50-treated cells. Video 5 shows that RNAi-resistant EBP50-S280,302D is unable to form microvilli in siEBP50-treated cells. Video 6 shows that RNAi-resistant EBP50-S162,339,340D is unable to form microvilli in siEBP50-treated cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004115/DC1.
Acknowledgments
We are indebted to Bret Judson for assistance with scanning electron microscopy. We are very grateful to Janet Ingraffea for providing the 6-His-tagged EPI64 purified from insect cells and for helpful discussions. We also thank Jessica L. Wayt for creating the GFP-tagged GRK6A and Cdc2 phospho-mutants of EBP50, and Raghuvir Viswanatha for creating GFP-tagged ezrin and for helpful discussions. We are also grateful to Dr. Roger Tsien for generously providing the plasmid encoding TagRFP-T.
This work was supported by National Institutes of Health grant GM-036652 to A. Bretscher, and National Research Service Award Postdoctoral Fellowship GM-080847 to D.P. LaLonde. D. Garbett was partially supported by National Institutes of Health Training Grant 5T32GM007273.
Footnotes
Abbreviations used in this paper:
- CNBr
- cyanogen bromide
- EBP50
- ERM-binding phosphoprotein of 50 kD
- EPI64
- EBP50 PDZ interactor of 64 kD
- ERM
- ezrin/radixin/moesin
- FERM
- 4.1 ERM
- MBP
- maltose-binding protein
- NHERF
- Na+-H+ exchanger regulatory factor
- PDZ
- postsynaptic density 95/discs large/zona occludens-1
References
- Begg G.E., Harper S.L., Speicher D.W. 2001. Characterizing recombinant proteins using HPLC gel filtration and mass spectrometry. Curr. Protoc. Protein Sci. Chapter 7:Unit 7.10 [DOI] [PubMed]
- Bretscher A. 1989. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J. Cell Biol. 108:921–930 10.1083/jcb.108.3.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. 2009. Epithelial polarity: dual Lkb1 pathways regulate apical microvilli. Dev. Cell. 16:491–492 10.1016/j.devcel.2009.03.012 [DOI] [PubMed] [Google Scholar]
- Cao T.T., Deacon H.W., Reczek D., Bretscher A., von Zastrow M. 1999. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 401:286–290 10.1038/45816 [DOI] [PubMed] [Google Scholar]
- Cheng H., Li J., Fazlieva R., Dai Z., Bu Z., Roder H. 2009. Autoinhibitory interactions between the PDZ2 and C-terminal domains in the scaffolding protein NHERF1. Structure. 17:660–669 10.1016/j.str.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepaldi T., Gautreau A., Comoglio P.M., Louvard D., Arpin M. 1997. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J. Cell Biol. 138:423–434 10.1083/jcb.138.2.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 85:1067–1076 10.1016/S0092-8674(00)81307-0 [DOI] [PubMed] [Google Scholar]
- Fehon R.G., McClatchey A.I., Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11:276–287 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty C.M., Chambers D., Ingraffea J., Faber H.R., Karplus P.A., Bretscher A. 2004. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 117:1547–1552 10.1242/jcs.01038 [DOI] [PubMed] [Google Scholar]
- Fouassier L., Yun C.C., Fitz J.G., Doctor R.B. 2000. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J. Biol. Chem. 275:25039–25045 10.1074/jbc.C000092200 [DOI] [PubMed] [Google Scholar]
- Fouassier L., Nichols M.T., Gidey E., McWilliams R.R., Robin H., Finnigan C., Howell K.E., Housset C., Doctor R.B. 2005. Protein kinase C regulates the phosphorylation and oligomerization of ERM binding phosphoprotein 50. Exp. Cell Res. 306:264–273 10.1016/j.yexcr.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Georgescu M.M., Morales F.C., Molina J.R., Hayashi Y. 2008. Roles of NHERF1/EBP50 in cancer. Curr. Mol. Med. 8:459–468 10.2174/156652408785748031 [DOI] [PubMed] [Google Scholar]
- Gorelik J., Shevchuk A.I., Frolenkov G.I., Diakonov I.A., Lab M.J., Kros C.J., Richardson G.P., Vodyanoy I., Edwards C.R., Klenerman D., Korchev Y.E. 2003. Dynamic assembly of surface structures in living cells. Proc. Natl. Acad. Sci. USA. 100:5819–5822 10.1073/pnas.1030502100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Spurney R.F., Premont R.T., Rahman N., Blitzer J.T., Pitcher J.A., Lefkowitz R.J. 1999. G protein-coupled receptor kinase 6A phosphorylates the Na(+)/H(+) exchanger regulatory factor via a PDZ domain-mediated interaction. J. Biol. Chem. 274:24328–24334 10.1074/jbc.274.34.24328 [DOI] [PubMed] [Google Scholar]
- Hanono A., Garbett D., Reczek D., Chambers D.N., Bretscher A. 2006. EPI64 regulates microvillar subdomains and structure. J. Cell Biol. 175:803–813 10.1083/jcb.200604046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Lau A.G., Yaffe M.B., Hall R.A. 2001. Phosphorylation and cell cycle-dependent regulation of Na+/H+ exchanger regulatory factor-1 by Cdc2 kinase. J. Biol. Chem. 276:41559–41565 10.1074/jbc.M106859200 [DOI] [PubMed] [Google Scholar]
- Hernando N., Déliot N., Gisler S.M., Lederer E., Weinman E.J., Biber J., Murer H. 2002. PDZ-domain interactions and apical expression of type IIa Na/P(i) cotransporters. Proc. Natl. Acad. Sci. USA. 99:11957–11962 10.1073/pnas.182412699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Fukuda M. 2006. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J. Biol. Chem. 281:31823–31831 10.1074/jbc.M603808200 [DOI] [PubMed] [Google Scholar]
- LaLonde D.P., Garbett D., Bretscher A. 2010. A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol. Biol. Cell. 21:1519–1529 10.1091/mbc.E10-01-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.G., Hall R.A. 2001. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry. 40:8572–8580 10.1021/bi0103516 [DOI] [PubMed] [Google Scholar]
- Lazar C.S., Cresson C.M., Lauffenburger D.A., Gill G.N. 2004. The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol. Biol. Cell. 15:5470–5480 10.1091/mbc.E04-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Poulikakos P.I., Dai Z., Testa J.R., Callaway D.J., Bu Z. 2007. Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J. Biol. Chem. 282:27086–27099 10.1074/jbc.M702019200 [DOI] [PubMed] [Google Scholar]
- Li J., Callaway D.J., Bu Z. 2009. Ezrin induces long-range interdomain allostery in the scaffolding protein NHERF1. J. Mol. Biol. 392:166–180 10.1016/j.jmb.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F.C., Takahashi Y., Kreimann E.L., Georgescu M.M. 2004. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. USA. 101:17705–17710 10.1073/pnas.0407974101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F.C., Takahashi Y., Momin S., Adams H., Chen X., Georgescu M.M. 2007. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol. 27:2527–2537 10.1128/MCB.01372-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram V., Hormuth H., Foskett J.K. 2003. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc. Natl. Acad. Sci. USA. 100:9620–9625 10.1073/pnas.1633250100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D., Bretscher A. 2001. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J. Cell Biol. 153:191–206 10.1083/jcb.153.1.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D., Berryman M., Bretscher A. 1997. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 139:169–179 10.1083/jcb.139.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J.W., Sanger J.M. 1980. Surface and shape changes during cell division. Cell Tissue Res. 209:177–186 10.1007/BF00237624 [DOI] [PubMed] [Google Scholar]
- Saotome I., Curto M., McClatchey A.I. 2004. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell. 6:855–864 10.1016/j.devcel.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Lin M.Z., McKeown M.R., Steinbach P.A., Hazelwood K.L., Davidson M.W., Tsien R.Y. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 5:545–551 10.1038/nmeth.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S., Weinman E.J. 2001. NHERF: targeting and trafficking membrane proteins. Am. J. Physiol. Renal Physiol. 280:F389–F395 [DOI] [PubMed] [Google Scholar]
- Shenolikar S., Minkoff C.M., Steplock D.A., Evangelista C., Liu M., Weinman E.J. 2001. N-terminal PDZ domain is required for NHERF dimerization. FEBS Lett. 489:233–236 10.1016/S0014-5793(01)02109-3 [DOI] [PubMed] [Google Scholar]
- Shenolikar S., Voltz J.W., Cunningham R., Weinman E.J. 2004. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology (Bethesda). 19:362–369 [DOI] [PubMed] [Google Scholar]
- Shibata T., Chuma M., Kokubu A., Sakamoto M., Hirohashi S. 2003. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 38:178–186 10.1053/jhep.2003.50270 [DOI] [PubMed] [Google Scholar]
- Short D.B., Trotter K.W., Reczek D., Kreda S.M., Bretscher A., Boucher R.C., Stutts M.J., Milgram S.L. 1998. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem. 273:19797–19801 10.1074/jbc.273.31.19797 [DOI] [PubMed] [Google Scholar]
- Siegel L.M., Monty K.J. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362 10.1016/0926-6585(66)90333-5 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Morales F.C., Kreimann E.L., Georgescu M.M. 2006. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 25:910–920 10.1038/sj.emboj.7600979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster J.P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M.M., Hornbeck P., Clevers H. 2009. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev. Cell. 16:551–562 10.1016/j.devcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Terawaki S., Maesaki R., Hakoshima T. 2006. Structural basis for NHERF recognition by ERM proteins. Structure. 14:777–789 10.1016/j.str.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Weinman E.J., Steplock D., Wang Y., Shenolikar S. 1995. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na(+)-H+ exchanger. J. Clin. Invest. 95:2143–2149 10.1172/JCI117903 [DOI] [PMC free article] [PubMed] [Google Scholar]