Abstract
Hundreds of therapeutic monoclonal antibodies (mAbs) are currently in development, and many companies have multiple antibodies in their pipelines. Current methodology used in recovery processes for these molecules are reviewed here. Basic unit operations such as harvest, Protein A affinity chromatography and additional polishing steps are surveyed. Alternative processes such as flocculation, precipitation and membrane chromatography are discussed. We also cover platform approaches to purification methods development, use of high throughput screening methods, and offer a view on future developments in purification methodology as applied to mAbs.
Key words: monoclonal antibody, recovery, purification, chromatography, membrane, filtration, platform process
Introduction
Hundreds of monoclonal antibodies (mAbs) are either currently on the market or under development.1 With the prominence of mAbs as therapeutic agents, many companies have multiple antibodies in their development pipeline. These molecules are often derived from a common framework, and have a high degree of homology, and therefore they have similar physico-chemical properties. This allows similar purification processes to be employed in the recovery of different products, establishing a platform process that can be used to purify similar molecules with minimal process alterations.
The purification process needs to reliably and predictably produce product suitable for use in humans. Impurities such as host cell protein, DNA, adventitious and endogenous viruses, endotoxin, aggregates and other species must be removed while an acceptable yield is maintained. In addition, impurities introduced during the purification process must be removed as well. These include leached Protein A, extractables from resins and filters, process buffers and agents such as detergents that may have been employed for virus reduction.
The first step in the recovery of an antibody from a mammalian cell culture is harvest—removal of cells and cell debris to yield a clarified, filtered fluid suitable for chromatography, i.e., the harvested cell culture fluid (HCCF). This is generally accomplished through use of centrifugation, depth filtration and sterile filtration, although other approaches may be applicable depending on scale and facility capability.
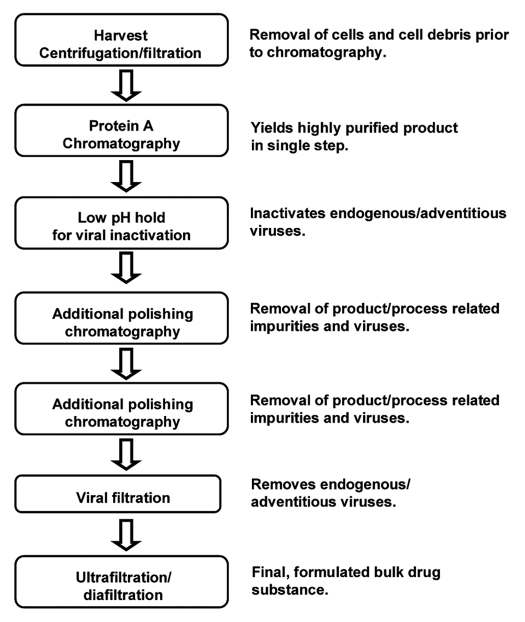
In the biopharmaceutical industry, chromatography is a critical and widely used separation and purification technology due to its high resolution. Chromatography exploits the physical and chemical differences between biomolecules for separation. The majority of purification processes for mAbs involve Protein A-based chromatography, which results in a high degree of purity and recovery in a single step.2 Figure 1 depicts a typical recovery process. Protein A chromatography follows harvest, and yields a relatively pure product that only requires removal of a small proportion of process and product related impurities. One or two additional chromatography steps are employed as polishing steps, generally incorporating cation and anion exchange chromatography, although hydrophobic interaction chromatography, mixed mode chromatography or hydroxyapatite chromatography may be chosen as well. These steps provide additional viral, host cell protein and DNA clearance, as well as removing aggregates, unwanted product variant species and other minor contaminants. Additionally, to achieve sufficient viral clearance, a low pH hold post Protein A chromatography and a viral filtration step are generally included in a process. Lastly, the purified product is concentrated and diafiltered into the final formulation buffer.
Figure 1.
A typical monoclonal antibody recovery process.
In developing an early stage process, speed to the clinic, efficient resource utilization, fit with existing facilities and equipment, minimized introduction of new raw materials, robust viral clearance and simplified viral clearance validation, are all factors to be taken into consideration. Processes developed for early stage clinical trials, including those developed using a platform, may be non-optimal with respect to process economics, yield, pool volumes, throughput and may not be suitable for producing the quantities required for late stage or commercial campaigns. For example, gradient elution may be used instead of step elution because it is more rapid to develop, even though it may result in larger and more dilute pools.
Once the candidate antibody has passed the hurdles of early stage clinical studies, a more optimal process that takes these additional factors into account may be developed. For example, it may be possible to achieve desired purity and viral clearance with fewer chromatography steps, and the use of higher capacity resins and in-line conditioning and buffer dilution may be required to accommodate high titer processes in the plant.
In this review of current methodology used in recovery and purification process development for mAbs, basic unit operations such as harvest, Protein A affinity chromatography and additional polishing steps are surveyed. Alternative processes such as flocculation, precipitation and membrane chromatography are discussed. We also cover platform approaches to purification methods development, use of high throughput screening methods and offer a view on future developments in purification methodology as applied to mAbs.
Primary Recovery Process
The first unit operation in a downstream process is the removal of cells and cell debris from the culture broth and clarification of the cell culture supernatant that contains the antibody product. Given the high cell densities achievable in both mammalian cell culture and microbial processes, primary recovery can be a significant challenge at lab, pilot and commercial manufacturing scales.3
The current trend for cell culture processes is to increase product titer through using enriched culture media, improving cell productivity and increasing cell mass. The high titer is also achieved in many cases through increased culture duration, although this may lead to a significant drop in cell viability. Those factors lead to an increase in the levels of process impurities such as host cell proteins, nucleic acids, lipids, colloids and the generation of a broad particle size distribution in the cell culture fluid (CCF). The current preferred process for accomplishing this initial recovery is to use a disc-stack continuous centrifugation coupled with depth filtration.4–6
Tangential flow micro filtration.
Tangential flow micro filtration has been successfully implemented for mammalian cell harvest since the early days of biotechnology. Here, the CCF flows tangential to the microporous membrane and pressure driven filtrate flow separates the soluble product from the larger, insoluble cells. Membrane fouling is limited by the inertial lift and shear-induced diffusion generated by the laminar flow across the membrane surface.7–10
A high yielding harvest is achieved by a series of concentration and diafiltration steps. In the former, the volume of the CCF is reduced, thereby concentrating the solid mass. The diafiltration step then washes the product from the concentrated CCF mixture.
Ideally, a pore size of 0.22 µm is employed for the TFF membrane as this will produce the target quality HCCF without the need for further clarification. As cell culture technology advanced, debris loading became too challenging and more open pore sizes at the TFF barrier were used to better manage fouling, but the more open pore sizes meant that an additional clarification step, such as normal flow depth filtration, was required downstream of the TFF system. Ultimately, because of the requirement to achieve a high degree of concentration of cell mass to achieve high yields, it became apparent that TFF is best suited for cell cultures with percent solids of <3%. As cell cultures exceeded this level, and as levels of sub-micron debris loads continued to climb, TFF gave way to centrifugation as the harvest method of choice for large scale manufacturing.
Centrifugation.
Centrifugation coupled with depth filtration has been employed for the initial recovery of therapeutic proteins for years and is well suited for pilot and commercial scale manufacturing. Disk-stack continuous centrifuges are capable of removing cells and large cell debris; however, cells can be disrupted during the process, especially when the feedstock is a low cell viability culture fluid. Many particles of submicron size cannot be removed in the centrifuge, increasing the burden on the subsequent depth filtration.
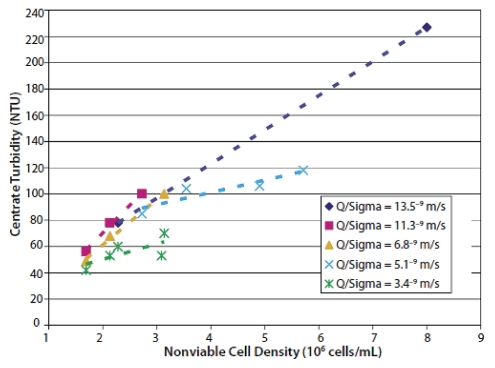
The clarification efficiency of the centrifugation process is affected by harvest parameters such as centrifuge feed rate, G-force, bowl geometry, operating pressures, discharge frequency and ancillary equipment used in the transfer of cell culture fluid to the centrifuge. The cell culture process characteristics such as peak cell density, total cell density and culture viability during the culture process and at harvest will also affect separation performance.11–13 Some optimization of the centrifugation process can be done at laboratory scale and pilot scale to select the feed rate and bowl rotational speed using the scaling factors of feed rate (Q) and equivalent settling area (∑) in the centrifuge. The settling area that can be achieved depends on the centrifuge geometry, number of discs in the stack and rotational speed. As a general rule, the lower the value of Q/∑, the better the clarification that will be achieved.4 It should be kept in mind that Q/∑ only governs the specific separation space in the disc-stack itself and cannot predict effects on the inlet zone or the mechanical effects of equipment upstream of the centrifuge. For a culture broth containing fragile or dead cells, increased rotational speed or increased residence times through the inlet zone may have a negative impact on shear force and cell lysis, generating submicron particles that are difficult to be removed through centrifugation. The affect of nonviable cell density at harvest on centrate quality for different centrifugation conditions is depicted in Figure 2.
Figure 2.
Impact of nonviable cell density at harvest on centrate quality for different disc-stack centrifugation conditions.13
The basic premise for scaling up a centrifugation process is to keep the optimized Q/∑ value constant, although there are practical limitations due to different centrifuge designs that can result in significant differences in performance as a function of process scale. As a consequence, it is often necessary to operate at a lower Q/∑ value at large scale to obtain the desired clarification efficiency.11–13 The centrifugation process developed at laboratory or pilot scale frequently needs to be tested and modified at manufacturing scale before finalizing the operational parameters and downstream filtration requirements. The optimized process will minimize cell lysis and debris generation while maximizing the sedimentation of submicron particles and product yield.
There have recently been further improvements in mammalian cell harvests using fully hermetic centrifuges. In the standard non-hermetic disc-stack centrifuge, an air-liquid interface at both the inlet and outlet zones is managed by applying enough back pressure to achieve an air-free centrate, but avoiding too high of a pressure that would cause overflow of an inner weir, thus sending product to drain. The hermetic design incorporates mechanical seals to fully isolate the product fluid from the outside air. The machine can be completely filled with liquid eliminating any air-liquid interface inside the centrifuge, which is known to be harmful to shear-sensitive materials like mammalian cells. Also, a completely sealed system, along with head pressure applied to the fermenter, can allow the flow to be controlled at the outlet of the machine, thereby removing pumps and flow-control valves from the inlet piping. With these improvements, the hermetically designed centrifuge harvest has been able to reduce the amount of cell lysis that is incurred during this unit operation by more than 50%.14,15
Depth filtration.
Depth filters are traditionally used in the clarification of cell culture broths, to maintain capacity on membrane filters or to protect chromatography columns or virus filters. Depth filters are typically composed of cellulose, a porous filter-aid such as diatomaceous earth and an ionic charged resin binder.16 A binding resin is often added to a small weight percent to covalently bind dissimilar construction materials together, giving the resultant media wet strength and conferring positive charge to the media surfaces.17 Because of this make-up, depth filters rely on both size exclusion and adsorptive binding to effect separation. Considerably thicker than a membrane filter, depth filters are approximately 2–4 mm in thickness. Depth filters are usually given a nominal pore-size rating, but these filters are far from absolute with regard to their particle size retention
For harvesting applications, depth filters can be applied directly with the whole cell broth or in conjunction with a primary separator such as TFF or centrifugation. The whole-cell broth depth filter harvest is common for bench, pilot and smaller commercial-scale applications. For this, a filtration train containing three stages of filters is usually employed. The primary stage uses a coarse or open, depth filter with a pore size range of up to 10 µm and removes whole cells and large particles. The secondary stage uses a tighter depth filter and clears colloidal and submicron particles. The last stage contains a membrane filter that is 0.2 µm pore size in most cases. The filtration process generally scales linearly; however, to ensure adequate filter capacity, a safety factor of 1.5X to >3X can be employed for each stage. Having too much excess capacity leads to large filter housing hold-up volumes, increased preparation time and floor space requirements and, ultimately, increased product loss due to hold-up.
A depth filter is employed most frequently after the centrifugation process because there is a practical lower limit to the particle size that can be removed by centrifugation.13 This type of depth filter has a pore size range of 0.1–4 µm and is usually made of two distinct layers, with the upstream zone being a coarser grade compared with the downstream. The larger particles are trapped in the coarse grade filter media and smaller particles are trapped in the tighter media, reducing premature plugging and increasing filtration capacity.
Cell culture process characteristics, such as cell density through the course of the culture process, cell viability and the centrifugation process itself can significant affect the centrate properties. Typically, a process with higher cell density or lower cell viability at the end of the cell culture process constitutes the greatest challenge with respect to harvest.
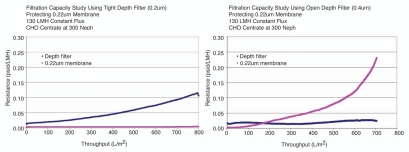
Optimization of filter type, pore size, surface area and flux can be done at lab bench scale and then scaled up to pilot scale. The selection often depends on the centrate turbidity and particle size distribution. As mentioned previously, it is often important to test centrate feedstocks obtained from the production-scale system due to the poor scalability of centrifuge performance in this area. Depth filter sizing experiments are typically done at constant flux using pressure endpoints in any one or combination of filtration stages. Different failure modes of pressure build-up or particle breakthrough can be observed in Figure 3. For this reason it is important to scale-down the entire filtration train to bench scale and test in series. Most often a 0.22 µm grade filter is used to filter the supernatant at the end of harvest process to control bioburden.The 0.22 µm-filtered supernatant can oftentimes be stored at 2–8°C for several days or sometimes longer without changing the antibody product-related variant profile. For antibodies that are susceptible to proteolysis, deamidation, oxidation or other stability-impacting degradation, the supernatant should be processed through the subsequent purification steps as soon as possible. In-process hold times at manufacturing scale are set by supernatant holding time studies performed at lab scale.
Figure 3.
Depth Filter experiments showing different modes of failure. Plot on the left shows a tight depth filter protecting a 0.22 um membrane and resistance builds up on this first layer. Plot on the right shows the case using a more open depth layer and particle breakthrough to membrane layer.
Due to the adsorptive mechanism of depth filters, they have been extensively used as a purification tool to remove a wide range of process contaminants and impurities. Positively charged depth filters were employed for the removal of Escherichia coli-derived and other endogenous endotoxins and viruses many times smaller than the average pore size of the filter.18,19 Zeta Plus® (Cuno) VR series depth filters were found to bind enveloped retrovirus and non-enveloped parvovirus by adsorption.17 The challenge in using these filters in this capacity is a reliable means for integrity testing, both as a filter and as an absorber. This latter aspect is especially important for validating the removal of putative viruses.
Depth filtration was also employed to remove spiked prions from an immunoglobin solution.20 Charged depth filters have been used to remove DNA, and the level of charges on Zeta Plus (Cuno) 90SP was correlated with its ability to remove DNA.21 The ability of depth filters to reduce DNA in actual harvested cell culture fluid was studied more comprehensively for the positively charged Zeta Plus (Cuno) 10SP and 90SP.22 This work demonstrated that not only electrostatic interactions between the positive charges of depth filters and DNA molecules but also hydrophobic interactions between depth filter media and DNA molecules played important roles in the adsorptive reduction of DNA.
This insoluble impurity removal aspect of depth filtration has been exploited to reduce host cell proteins from a recombinant mAb process stream.23 The removal of host cell proteins through depth filtration prior to a Protein A affinity chromatography column was shown to significantly reduce precipitation during the pH adjustment of the Protein A pool.
Depth filtration involves a number of complementary mechanisms that allow contaminant removal, sieving and electrokinetic adsorption, as well as hydrophobic interactions, which makes this technique a powerful tool in antibody purification.
Flocculation/precipitation.
A higher quantity of cell debris and colloids in the cell culture fluid produces an increased burden to the clarification train. This can result in a high depth filter surface area requirement, which can exceed the existing filtration train equipment capability. In recent years, precipitation/flocculation-based pretreatment to circumvent this issue has been studied.
Flocculation of animal cells in suspension culture and selective flocculation of cellular contaminants from soluble proteins using acidic or cationic polyelectrolytes has been a topic of interest for some time.24,25 Polyelectrolytes normally work by adsorbing to a particle to create an oppositely charged patch on the surface. This patch can then adhere to a bare patch on an opposing particle surface due to electrostatic attraction. The bridging mechanism of polymer adsorption to the cell and cell debris is electrostatic attraction in most cases. Strongly cationic polymers are more effective at flocculating cells, whereas neutral and anionic polymers are often ineffective.
New applications of flocculation and acid precipitation during the primary recovery of mammalian cell culture broth have shown promising results to improve clarification efficiency and achieve high recovery yield.26,27 Recently, the addition of “generally recognized as safe” flocculation reagents such as calcium chloride and potassium phosphate has been studied to avoid potential issues related to polymer or residual monomer toxicity and the need for their removal through subsequent purification steps. The amount required can be very low, e.g., 20–60 mM calcium chloride with an equimolar amount of phosphate added to form calcium phosphate.28 The mechanism is thought to be related to the co-precipitation of calcium phosphate with cells, cell debris and impurities.
Chitosan is another promising flocculant for biotechnology application as it is produced from non-mammalian sources (typically arthropod shells), and is inexpensive and available in a highly purified form that is low in heavy metals, volatile organics and microbial materials. It is a cationic linear polymer of beta-(1–4) linked D-glucosamine monomers generated by the chemical deacetylation of chitin. Chitosan has been used for the removal of nucleic acid and endotoxin, and the flocculation of cell and cell debris of yeast and bacteria.29–32 A study conducted by Riske et al. showed that 82–88% deacetylated chitosan, with a molecule weight range of 30–1,000 kDa, improved clarification efficiency by flocculating the cell debris and submicron particles which normally escape sedimentation in the centrifuge and increased the depth filter capacity by several fold.26 They demonstrated that factors such as chitosan concentration, sodium chloride concentration and pH had significant effects on chitosan flocculation of cells, cell debris and particulates. The underlying mechanism of flocculation likely involves electronic interaction between chitosan and charged cellular debris, followed by additional interactive forces such as hydrogen bonding.
If flocculation is being considered for harvest improvement, diligent consideration must be made to verify its impact on downstream chromatography operations as well as ensuring removal of the flocculation agents to acceptable levels in the Drug Product. General guidelines on the removal of contaminants can be found in the FDA's ‘Points to Consider in the Manufacture and Testing of Monoclonal Antibody Products for Human Use’ and ‘Guidance for Industry Q3A Impurities in New Drug Substances’.
Chromatographic Processes
For more than a decade the workhorse of commercial mAb purification has been Protein A chromatography, followed by two or three subsequent chromatographic polishing steps. Column chromatography continues to be favored, and has been the focus of continuous improvements through development of higher capacity resins and use of mixed-mode sorbents. The application of traditional chromatography and newly developed chromatographic resins to mAb purification is reviewed in the following section.
Affinity chromatography.
Affinity separation is the most selective type of chromatography used in biotechnology. It separates proteins on the basis of a reversible interaction between a protein and a specific ligand covalently coupled to a chromatography matrix. The technique makes for an ideal capture step in purification processes.
The high affinity of Protein A for the Fc region of IgG-type antibodies forms the basis for the purification of IgG, IgG fragments and subclasses. The procedure typically employed for Protein A chromatography involves passage of clarified cell culture supernatant over the column at pH 6–8, under which conditions the antibodies bind and unwanted components such as host cell proteins and cell culture media components and putative viruses flow through the column. An optional intermediate wash step may be carried out to remove non-specifically bound impurities from the column, followed by elution of the product at pH 2.5–4.
Because of its high selectivity, high flow rate and cost effective binding capacity and its capacity for extensive removal of process-related impurities such as host cell proteins, DNA, cell culture media components and endogenous and adventitious virus particles, Protein A chromatography is typically used as the first step in an antibody purification process. After this step, the antibody product is highly pure and more stable due to the elimination of proteases and other media components that may cause degradation.
There are currently three major types of Protein A resins, classified based on their resin backbone composition: glass or silica-based, e.g., Prosep vA, Prosep vA Ultra (Millipore); agarose-based, e.g., Protein A Sepharose Fast Flow, MabSelect (GE Healthcare); and organic polymer based, e.g., polystyrene-divinylbenzene Poros A and MabCapture (Applied Biosystems). They are widely used from laboratory bench scale, which is typically a column diameter of less than 1 cm, to industrial manufacturing scale, which includes column diameters of up to 2 meters. All three resin types are resistant to high concentrations of guanidinium hydrochloride, urea, reducing agents and low pH.
The column bed height employed at large scale is between 10 and 30 cm, depending on the resin particle properties such as pore size, particle size and compressibility. Flow rate and column dimensions determine antibody residence time on the column. The typical linear velocity employed for Protein A steps ranges from 300–500 cm/hr. Dynamic binding capacity ranges from 15–50 g of antibody per liter of resin, and depends on the flow rate, the particular antibody to be purified, as well as the Protein A matrix used.33 A method for determining dynamic binding capacities of Protein A resins has been described by Fahrner et al.34 A lower loading flow rate increases antibody residence time and promotes higher binding capacity. It also results in a longer processing time per cycle, requires fewer cycles and consumes less buffer per batch of harvested cell culture fluid.
To develop or design a Protein A chromatography operation, several key factors such as flow rate, dimensions of available columns, buffer solution volume, resin cost and processing time need to be considered. Careful consideration should be given to the large volumes of HCCF to be processed, the resultant impact on processing time and the high cost of the resin before the column size and number of cycles to be run is selected.
The usable lifetime of the resin is commonly beyond 200 cycles.35 The usable lifetime of Protein A resin is affected a number of factors. One factor is the amount of Protein A that is leached with each cycle. A recent study by Carter-Franklin et al.36 demonstrated that intact Protein A leaches when loading either purified antibody or unpurified antibody in harvested cell culture fluid and additional fragments of Protein A leach when loading unpurified antibody in harvested cell culture fluid. The amount of leached Protein A can be reduced by addition of EDTA in harvested cell culture fluid to inhibit proteinases.
The usable lifetime of Protein A resin is also affected by the cleaning procedure, as chemical degradation of the ligand and insufficient cleaning are both important. In general, an acidic solution at low pH (<3) and solutions containing chaotropic reagents such as guanidine hydrochloride and urea can be used to strip off most antibodies and impurities. A well-developed cleaning procedure can ensure removal of impurities and components that degrade the Protein A on the resin, and maintain usable lifetime.
Agarose-based and polymer-based resins can be sanitized after each purification cycle with dilute sodium hydroxide solution (0.05–0.2 N) for a contact time of 30 min. An alkali-stabilized Protein A-derived ligand that can withstand harsh cleaning solutions such as 0.1–0.5 N sodium hydroxide has been developed in the agarose-based MabSelect family (MabSelect SuRe).
A common problem with Protein A resins is non-specific binding of impurities such as host cell protein, DNA and other cell culture-derived impurities. The level of impurities bound depends on the resin, the composition of harvested cell culture fluid and column loading and washing conditions. Many intermediate wash solutions have been developed containing additional components or salts. Several hydrophobic electrolyte solvents such as tetramethylammonium chloride (TMAC) and tetraethylammonium chloride (TEAC), for example, have been developed as efficient wash reagents for glass-based resins to reduce bound host cell protein that can co-elute with the antibody product.37 A buffer composition comprising salt and polymer demonstrated a comparable effect on impurity removal.38 Two chromatography polishing steps are normally used in a platform antibody purification process, but an effective intermediate wash step can eliminate the need for one of these steps.
Antibody elution from Protein A requires low pH (2.5–4), a condition under which enveloped viruses such as murine retrovirus can be effectively inactivated; however, such a harsh condition could potentially alter the biological activity of the antibody or cause antibody aggregation. Several elution buffer components such as acetic acid, citric acid, phosphoric acid, arginine HCl and glycine HCl have been evaluated with respect to antibody quality and elution pool profile.39,40
The selection of elution pH is also dependent on the binding affinity of the antibody to the resin. Some antibodies demonstrate a higher binding affinity than others, requiring a lower elution pH. Higher binding affinity is often contributed through the binding of the Fab region to the Protein A ligand;41 however, the alkali-stabilized Protein A-derived ligand (in MabSelect SuRe) has shown a unique binding affinity for several mAbs studied, and it is believed that the contribution through the Fab region to binding affinity is reduced or eliminated.42
In summary, developing and optimizing a Protein A chromatography process should focus on defining the binding capacity at the operational flow rate range used at manufacturing scale, defining the number of cycles required to process a batch of harvested, clarified cell culture fluid, establishing an intermediate wash step capable of efficiently eliminating host cell protein, DNA and other nonspecific bound impurities from the resin and optimizing elution conditions to ensure product quality and achieve desirable elution pool attributes such as low volume, pH and conductivity of the pool. It is paramount that the process should be repeatable and scalable.
Ion exchange chromatography.
Most mAb purification processes will include at least one ion exchange chromatography step. Separation by this technique is fairly selective, and the resins used are relatively inexpensive, so it can be applied early or late in a purification process. For an antibody having a basic isoelectric point (pI), cation exchange chromatography can even be used as an initial capture step,43 but most frequently ion exchange chromatography is applied as a polishing step(s) after the Protein A step. Ion exchange chromatography is ideal for reducing high molecular weight aggregate, charge-variants, residual DNA and host cell protein, leached Protein A and viral particles.
Several commonly employed ion exchange resins are commercially available. The resin matrix backbone includes agarose and dextran (GE Healthcare), glycidyl methacrylate (Macroprep, Bio-Rad), polystyrenedivinylbenzene (Poros, Applied Biosystems) and polymethacrylate (Fractogel, EMD Chemicals and Toyopearl, Tosoh). These resins are all capable of being cleaned and sanitized with a high concentration of sodium hydroxide. Applicable flow rates range widely from 100 to 500 cm/hr. Ligands available include strong and weak anion and cation exchangers. The selection of the resin backbone and resin ligand will be mainly guided by the required resolution, binding capacity and feasibility of use at manufacturing scale.
Anion exchange chromatography.
Anion exchange chromatography uses a positively charged group (weakly basic such as diethylamino ethyl, DEAE or dimethylamino ethyl, DMAE; or strongly basic such as quaternary amino ethyl, Q or trimethylammonium ethyl, TMAE or quaternary aminoethyl, QAE) immobilized to the resin. It is a powerful tool to remove process-related impurities such as host cell proteins, DNA, endotoxin and leached Protein A, product-related impurities such as dimer/aggregate, endogenous retrovirus and adventitious viruses such as parvovirus, pseudorabies virus.44–46 It can be used either in flow-through mode or in bind and elute mode, depending on the pI of the antibody and impurities to be removed. For antibodies having a pI above 7.5, which includes most humanized IgG1 and IgG2 antibodies, flow-through mode can be a better choice to remove impurities. In flow-through mode, the impurities bind to the resin and the product of interest flows through. The column loading capacity, i.e., mass of antibody to mass of resin, can be quite high since the binding sites on the resin are occupied only by the impurities. For antibodies having a pI in the acidic to neutral range, which includes most humanized IgG4s, bind and elute mode can be used to remove process-related and product-related impurities from the product of interest.
Flow-through mode. Anion exchange chromatography in flow-through mode has been widely used as a polishing step in mAb purification processes designed with two or three unit operations to remove residual impurities such as host cell protein, DNA, leached Protein A and a variety of viruses. The operating pH is normally 8 to 8.2, with a conductivity of up to 10 mS/cm in the product load and equilibration and wash buffers. Conditions are chosen such that the product does not bind to the column, while acidic impurities such as nucleic acid and host cell proteins do. Depending on the resin, loading conditions and charge variant profile of the antibody product, the amount of product loaded can reach one hundred grams per liter of resin without compromising product quality.2 In general, the amount of product loaded in a flow through mode depends on the impurity species and levels to be removed. A lower level of impurity in the product will result in a higher amount of product loaded.
Bind-and-elute mode. Anion exchange chromatography in bind-and-elute mode has also been implemented in the manufacturing of mAbs. The antibody product pool is first loaded onto an anion exchange column and the product of interest is then eluted with a higher salt concentration in a step or linear gradient, leaving the majority of impurities bound to the column. The impurities are eluted from the column during the cleaning or regeneration step.47,48
In the development and optimization of an anion exchange chromatography in bind-and-elute mode, the operating pH should be above or close to the pI of the product in order to obtain a net negative charge or higher negative charge number on the surface of the antibody molecules, and hence to achieve a higher binding capacity during the chromatography step.
Similarly, to reach a high binding capacity, the ionic strength for the load should be in the low range and the pH should be less than pH 9. Manufacturing issues around the need to achieve lower ionic strength in the load will come into play, including the capability or need for diafiltration, product holding vessel volumes if the load will be diluted manually, and operational lower limits of buffer pumps when running a gradient if the sample is to be diluted in-line.
Optimal elution conditions can be achieved using either a linear gradient or step elution. Step elution can deliver smaller product pool volumes and higher product concentration, while linear gradient elution can provide better process control, process monitoring and reproducibility.49,50 If step elution is chosen for a manufacturing process, a thorough study should be carried out to demonstrate the robustness of process performance with multiple lots of the selected resin.
When developing step elution conditions, results from the initial linear gradient run can be used as a starting point, working down from the salt concentration at which dimer/aggregate begins to elute. Even if a quick or short linear gradient program is performed, the same principal should be applied as used to develop a step elution program in selecting an initial salt concentration and the conductivity of the elution buffer.48
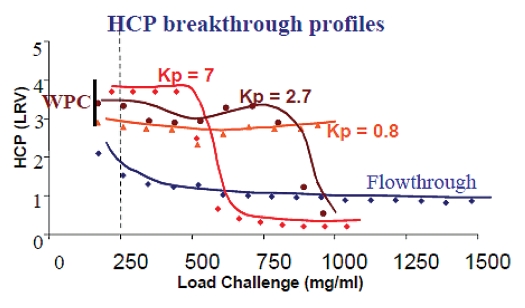
Weak partitioning chromatography. Recently, a mode of anion exchange chromatography termed weak partitioning chromatography (WPC) has been developed. This can enable a two chromatography recovery process comprising Protein A and anion exchange for many antibodies.51 As with flow-through chromatography, the process is run isocratically, but, in contrast to flow-through mode, the conductivity and pH are chosen such that the binding of both the product and impurities are enhanced, attaining an antibody partition coefficient (Kp) between 0.1–20, and preferably between 1 and 3. This takes advantage of the fact that the impurities to be removed are more acidic than the product. Both antibody and impurities bind to the anion exchange resin, but the impurities are much more tightly bound than in flow-through mode, which can lead to an increase in impurity removal (Fig. 4). Thus, weaker binding impurities that are not removed efficiently in flow-through mode can be removed to a greater degree under conditions where their partition coefficient (Kp) has been increased.
Figure 4.
High load capacity runs of monoclonal antibody. Under chromatographic conditions at selected optimal Kp (0.8–7), 3 logs of host cell protein reduction were achieved while the load capacity is higher than 250 mg/mL of resin.51
Product yield in weak partitioning mode is maximized by including a short wash at the end of the load, and averaged 90% for clinical production of a series of products. Load capacity can be quite high, in some cases exceeding 1,500 g/L, which may be beyond the point where such a small column can be used effectively in a recovery process, so this higher capacity may not always be exploited.
Due to the increased clearance of virus, host cell protein and product related species compared to anion exchange chromatography in flow-through mode, weak partitioning chromatography can enable a two column recovery process for many products. One challenge is less efficient removal of high molecular weight species compared to recovery processes employing a third chromatographic step such as cation exchange, so it may not be applicable to all products.
One aspect of weak partitioning chromatography is that the pH and counterion conditions need to be optimized for each product. This is in contrast to some platform chromatography processes that are able to use standardized conditions on an anion exchange matrix (resin or membrane) for most products. In order to rapidly define the optimal operating conditions for WPC, high throughput screening (HTS) methods have been developed.52,53 A robotic system employing 96-well filter plates filled with resin allows rapid optimization in batch mode, allowing the determination of Kp for products and impurities over a range of conditions. This can be refined subsequently through column chromatography or if HTS capability is not available, optimization can be carried out in entirety using columns. Flow-through chromatography conditions can be used as a starting point for determining the optimal mobile phase composition.
Cation exchange chromatography.
Cation exchange chromatography uses a resin modified with negatively charged functional groups. They can be strong acidic ligands such as sulphopropyl, sulfoethyl and sulfoisobutyl groups or weak acidic ligand such as carboxyl group. Cation exchange chromatography has been applied for purification processes for many mAbs with pI values ranging from neutral to basic. Most humanized IgG1 and IgG2 subclasses are perfect candidates for cation exchange chromatography, in which the antibody is bound onto the resin during the loading step and eluted through either increasing conductivity or increasing pH in the elution buffer. The most negatively charged process-related impurities such as DNA, some host cell protein, leached Protein A and endotoxin are removed in the load and wash fraction. Cation exchange chromatography can also provide separation power to reduce antibody variants from the target antibody product such as deamidated products, oxidized species and N-terminal truncated forms, as well as high molecular weight species.
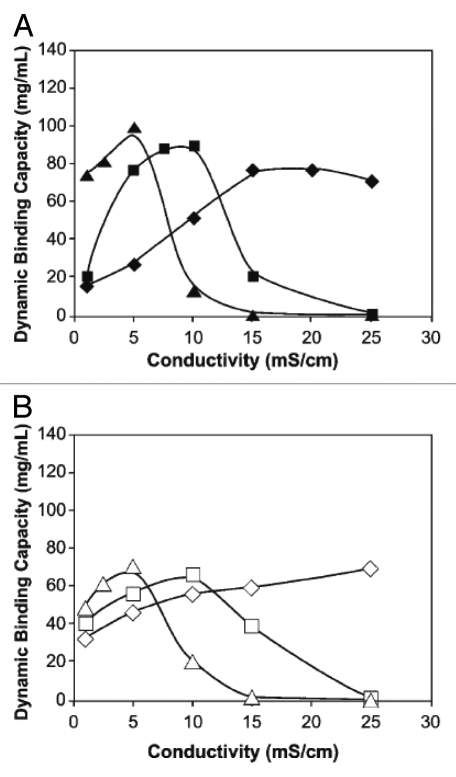
The dynamic binding capacity of mAbs on cation exchange resins depends on pH and conductivity. The impact of these parameters on the adsorption of mAbs to two different commercial cation exchange media (SP Sepharose FF and SP Sepharose XL) was investigated by Harinarayan et al.54 The dynamic binding capacity of the antibodies was shown to be significantly reduced at low conductivities and low pH (high net protein charge; Fig. 5). Confocal scanning laser microscopy indicated that under the conditions studied proteins are unable to penetrate deeply into the porous chromatography beads. It is possible that protein molecules adsorbed at pore channels near the external surface of the media hinder other molecules from entering the pores and such exclusion is reduced at high conductivity, leading to a direct relationship between net protein charge and the solution conductivity yielding optimal dynamic capacity.54
Figure 5.
Dynamic binding capacity of a monoclonal antibody as a function of pH and conductivity displays two regions. In the first domain (positively sloped capacity trend), the capacity increases with increasing conductivity. In the second region (negatively sloped capacity trend), the capacity decreases with increasing conductivity. (A) SP Sepharose XL, pH 4 (◆), 5 (■) and 6 (▲), (B) SP Sepharose FF, pH 4 (◊), 5 (□) and 6 (△).54
The maximum binding capacity attained can be as high as >100 g/L of resin volume depending on the loading conditions, resin ligand and density, but impurity removal depends highly on the loading density. High loading on the resin normally results in higher levels of impurities in the elution pool, although different ligands and resin bead sizes can have significant effects on the resolution of impurities. Therefore, a resin screening study should be performed to select the best resin, i.e., one with a high binding capacity and the best selectivity and resolution. The resin screening study is also linked with the elution condition development. The same principles described for anion exchange chromatography regarding development of the elution program apply to cation exchange chromatography as well.
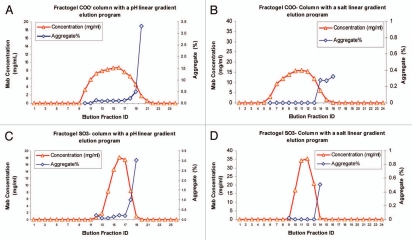
The development of elution conditions is linked to impurity removal and characteristics of the product pool that can be processed easily in the subsequent unit operation. Generally, a linear salt or pH gradient elution program can be conducted to determine the best elution condition.55–58 An elution condition development study conducted for an IgG1 is shown in Figure 6. Four runs were performed in this study with two Fractogel cation exchange resins, Fractogel COO− (weak cation exchanger) and Fractogel SO3− (strong cation exchanger) for the purpose of removing aggregate. Linear gradient elution conditions were from 5 mM to 250 mM NaCl at pH 6 and linear pH gradient elution runs were from pH 6 to pH 8. Aggregate removal was comparable with both resins and both linear gradient programs. Final selection of a weak or strong cation exchanger should take into consideration other factors such as binding capacity, removal of other impurities and the product pool profile. Typically, a smaller pool volume with higher product concentration and lower conductivity is desired if subsequent anion exchange chromatography is to be employed.
Figure 6.
Elution condition development for a monoclonal antibody. (A and B) a pH linear gradient elution program and a salt linear gradient elution program applied on a weak cation exchange column (Fractogel COO− resin), respectively. (C and D) the same linear pH and salt elution programs applied on a strong anion exchange column (Fractogel SO- resin), respectively.
Hydrophobic interaction chromatography.
Hydrophobic interaction chromatography (HIC) is a useful tool for separating proteins based on their hydrophobicity, and is complementary to other techniques that separate proteins based on charge, size or affinity. The sample is typically loaded on the HIC column in a high salt buffer. The salt in the buffer interacts with water molecules to reduce solvation of the protein molecules in solution, thereby exposing hydrophobic regions in the sample protein molecules that consequently bind to the HIC resin. The more hydrophobic the molecule, the less salt is needed to promote binding. A gradient of decreasing salt concentration is usually used to elute samples from the column. As the ionic strength decreases, the exposure of the hydrophilic regions of the molecules increases and molecules elute from the column in order of increasing hydrophobicity.59
HIC is a versatile liquid chromatography technique that is frequently used in a rational purification strategy. HIC resins containing phenyl or butyl ligands are often found in mAb purification processes;60–63 however, due to the high efficiency of Protein A affinity chromatography, HIC is mostly used as an intermediate purification step after Protein A chromatography or as a polishing step after ion exchange chromatography. HIC in flow-through mode is efficient in removing a large percentage of aggregates with a relatively high yield. HIC in bind-and-elute mode normally provides effective separation of process-related and product-related impurities from antibody product. The majority of host cell protein, DNA and aggregates can be removed from the antibody product through selection of a suitable salt concentration in the elution buffer or use of a gradient elution method.
Hydrophobic charge induction chromatography.
Hydrophobic charge induction chromatography (HCIC) is based on the pH-dependent behavior of ligands that ionize at low pH.64 This technique employs heterocyclic ligands at high densities so that adsorption can occur via hydrophobic interactions without the need for high concentrations of lyotropic salts. To overcome the problem of harsh elution conditions, which are typically used with very hydrophobic resins, desorption in HCIC is facilitated by lowering the pH to produce charge repulsion between the ionizable ligand and the bound protein.65 The first commercial HCIC resin was MEP-Hypercel (Pall Corporation). It is a cellulose-based media with 4-mercaptoethyl pyridine as the functional group. The ligand is a hydrophobic moiety with an N-heterocyclic ring that acquires a positive charge at low pH.
Due to the high cost of Protein A resins and their somewhat lower resistance to extreme conditions, HCIC resin has been suggested as a potential alternative to Protein A resins for the initial capture and purification of IgG antibodies. Salt-independent antibody binding and successful elution at a somewhat higher pH range than is possible with Protein A chromatography has been demonstrated;56,66,67 however, one critical drawback of HCIC is that it has stronger non-specific binding and can be less efficient than Protein A chromatography in reducing impurities such as host cell protein. Consequently, use of HCIC in an antibody purification process can be challenging. Continued evaluation of HCIC in combination with other orthogonal purification steps that can provide sufficient removal of residual impurities, such as ion exchange chromatography, precipitation and crystallization, will determine whether it will play a prominent role in antibody purification process development in the future.
Ceramic hydroxyapatite chromatography.
Ceramic hydroxyapatite (Ca5(PO4)3OH)2 is a form of calcium phosphate that can be used for the separation and purification of proteins, enzymes, nucleic acids, viruses and other macromolecules. Hydroxyapatite has unique separation properties and unparalleled selectivity and resolution. It often separates proteins that appear to be homogeneous by other chromatographic and electrophoretic techniques.
Ceramic hydroxyapatite (CHT) chromatography with a sodium phosphate gradient elution has been used as a robust polishing step in mAb purification process to remove dimers, aggregates and leached Protein A.68 Gagnon et al. demonstrated the powerful application of a sodium chloride linear gradient for human antibody IgG4 in CHT chromatography.69 CHT column operation in the bind/elution mode can be used as a powerful polishing step for large-scale mAb production; however, several issues, such as resin lot-to-lot variability, resin life and viral clearance capacity still remain.
Multimodal chromatography.
Resins with multimodal ligands such as Capto MMC and Capto Adhere (GE Healthcare) can be used as polishing steps for antibody purification. These multimodal resins combine different types of interactions such as ionic interaction, hydrogen bonding and hydrophobic interaction. The multimodal functionality of the resin provides selectivity that is different from standard ion exchange ligands, which makes them suitable for solving purification problems at both high and low conductivity or pH. A strong anionic multimodal resin, Capto Adhere, has been evaluated as a second antibody purification step after Protein A chromatography to remove aggregate, host cell protein and leached Protein A.70 Due to their versatile or complex chemistry, a more comprehensive experimental approach is required to achieve optimal separation performance. As for other chromatography resins, one needs to take into account factors such as resin lifetime, lot-to-lot variability, effective cleaning and regeneration, viral clearance, platform suitability and the probability of identifying an alternate resin for second sourcing efforts.
Use of high throughput screening to optimize separations.
High throughput screening (HTS) in robotic format is being used to quickly optimize chromatographic separations.71,72 This methodology can be used to optimize operating conditions for most types of chromatography, including affinity, ion-exchange, hydrophobic interaction and others. Resin selection, binding capacities, binding and elution conditions, impact of intermediate wash steps, yield, as well as resolution of impurities can be determined in automated fashion. Screening can be carried out in batch mode in 96-well plates and 96-well filterplates (Whatman, Innovative Microplate, GE Healthcare), and also in mini-columns in 96-well column formats (Atoll-Bio).73 The difference between the well plate based format and the mini-column based format is that the mini-column is operated more like the preparative scale column. For instance, gradient elution can be more easily implemented for the mini-column format in which multiple step-wise gradient can be used to simulate the linear gradient.
In batch mode, a defined quantity of resin is aliquoted into the well, washed and equilibrated with the appropriate buffer, the protein-containing sample is added and the plate is mixed for an appropriate time and then unbound material is removed either by centrifugation or the use of a vacuum manifold. The resin can subsequently be washed and eluted as appropriate, and all of the various fractions analyzed.
Examples of HTS in batch binding mode applied to HIC,74 hydroxyapatite75 and ion exchange chromatography have been published recently.76 Agreement with results obtained by chromatographic methods is generally good. Kelley et al. demonstrated optimization of a cation exchange capture step for a recombinant human mAb carried out in 8 hrs using 35 mg of protein in total; they evaluated eight different resins and multiple pH and salt conditions for binding and elution.76 Their results clearly demonstrate the potential for rapid screening of initial operating conditions that can be subsequently defined by conventional packed bed chromatography.
Advantages of HTS include the ability to rapidly screen a very large number of conditions, reduced sample requirements and the potential for realizing economic benefits. Since only a single theoretical plate is obtained, this method is not appropriate for evaluation of methods such as SEC that rely on many plates for separation. A consequence of HTS is the generation of a large numbers of samples to be analyzed, generating a substantially increased analytical burden. Without the availability of appropriate analytical methods of sufficient throughput and sensitivity for the quantity and volume of the samples, sample and data analysis will become a bottleneck. Parallel analysis methods are highly preferred. Reduced sample volumes affect the sensitivity and often the time, required for analysis.
Dynamic binding capacity and other parameters such as yield and purity may differ in separations in a packed bed column at full bed height under actual operating conditions. For this reason, HTS process development is frequently followed by confirmation and final optimization using standard packed bed chromatography, with the advantage that the operating range is already well defined. HTS is likely to gain in prominence in the future as the methodology evolves and economic pressures increase.
Membrane and Filtration Technology
Membrane and filtration technologies are used extensively in the isolation and purification of mAb and other recombinant DNA products, from the initial clarification of cell culture broth to the final sterile filtration of purified bulk solutions. This section focuses on several of these technologies used for the purification of mAb products or removal of contaminants and impurities, including depth filtration, membrane chromatography, ultrafiltration, high performance tangential flow filtration using neutral or charged membranes and virus filtration.
Membrane chromatography.
Membrane chromatography or membrane adsorbers, function similarly to packed chromatography columns, but in the format of conventional filtration modules.77 Membrane chromatography uses microporous membranes, usually in multiple layers that contain functional ligands attached to the internal pore surface throughout the membrane structure.
The benefit of membrane chromatography over conventional bead chromatography is the elimination of diffusive pores. For membrane chromatography, binding sites are located along the through pores rather than nestled within long diffusive pores.78 Accordingly, mass transfer of biomolecules to binding sites depends on convection instead of diffusion and binding capacities of membrane chromatography are largely independent of flow rate.
In membrane chromatography, membranes consist of a polymeric substrate to which a functional ligand is chemically coupled. The polymer substrate is composed of multilayers of polyethersulfone, polyvinylidene fluoride and regenerated cellulose membrane. The most widely used functional ligands are the same as those used in chromatography resins, including ion exchange, e.g., quaternary amine (Q), diethylamine (D), polyethyleneimine (E), sulfonic acid (S) and carboxylic acid (C); affinity; reverse-phase, hydrophobic interaction and anionic mixed mode that is made of cross-linked Poly(Allylamine) (PAA).79,80
Among the adsorptive membranes commercially available, the Q membrane adsorber has attracted a lot of industry interest, especially in mAb purification processes.78,81,82 Commercially available Q membranes include Intercept™ (Millipore), Mustang® (Pall) and Sartobind® (Sartorius). Similar to conventional bed chromatography, Q membranes are normally exploited as polishing steps in flow-through mode to remove trace amounts of impurities. Around neutral to slightly basic pH and at low conductivities, viruses, DNA, endotoxin, a large population of host cell proteins and leached Protein A bind to the Q membrane, whereas the typically basic antibody molecules flow through the membrane matrix without being bound.83 Recently available commercial anionic mixed mode membrane such as ChromaSorb (Millipore) improves impurity binding capacity or impurity removal function at a higher loading conductivity range and broadens anionic membrane operation conditions.80
Membrane chromatography is potentially useful for protein purification at laboratory and pilot scale; however, it has limitations that need to be overcome before it can be successfully employed in process-scale production. Major limitations are uneven flow distribution, non-identical membrane size distribution, uneven membrane thickness, the availability of appropriate scale-down devices and low binding capacities.83 Low binding capacity is attributed to low surface area as well as poor flow distribution. Improved binding capacities and flow distribution have been achieved by optimizing pore size, membrane chemistry, membrane thickness or the number of membrane layers and employment of tentacle ligands.77,79 In addition, other important aspects should also be considered for this technology, including membrane lifetime (if membrane is used more than once), integrity testing, lot-to-lot variability and process economics.
Ultrafiltration.
Ultrafiltration is a pressure-driven membrane process that is widely used for protein concentration and buffer exchange. Ultrafiltration is a size-based separation, where species larger than the membrane pores are retained and smaller species pass through freely. Separation in ultrafiltration is achieved through differences in the filtration rates of different components across the membrane under a given pressure driving force.84 Buffer exchange is achieved using a diafiltration mode in which buffer of the final desired composition is added to the retentate system at the same rate in which filtrate is removed, thus maintaining a constant retentate volume.
Ultrafiltration with membrane pores ranging from 1 to 20 nm can provide separation of species ranging in molecular weight from 500 daltons to 1,000 kilodaltons.85 This distinguishes the process from reverse osmosis, which involves membrane pores of less than 1 nm that allows water but not salts to pass, and from microfiltration, which has membrane pores ranging in size from 0.05 to 10 µm that allow proteins to pass through.
Ultrafiltration membranes have a unique “skinned” or anisotropic structure. Membrane selectivity is controlled by a thin skin layer that is approximately 0.5 µm thick, and the mechanical strength is provided by a thicker macroporous substrate.86 This allows retention to occur on the membrane surface rather than within the filter structure.85 Filtration fluxes are also greatly increased since a majority of the flow resistance takes place in the thin skin layer.
Ultrafiltration membranes can be cast from a wide variety of polymers, including polysulfone, polyethersulfone, polyvinylidene fluoride and regenerated cellulose. The synthetic polymers exhibit strong resistance to acids, bases, alcohols and higher temperatures, allowing for effective membrane cleaning. In this manner, ultrafiltration membranes can be reused without deterioration of flow rates and cross contamination.85 Synthetic polymers could provide more attractive chemical and thermal stability but are prone to protein fouling. In contrast, cellulose membranes have low protein binding but can be damaged by harsh cleaning methods. New composite regenerated cellulose membranes have significantly less protein fouling, are more easily cleaned, and have excellent mechanical strength. Due to these favorable properties, cellulose membranes are superior to other membranes in process permeability and retention characteristics for protein ultrafiltration and diafiltration.84
Ultrafiltration is normally carried out in tangential flow filtration (TFF) mode, in which fluid passes across the filter (cross-flow), tangential to the plane of the filter surface. The primary advantage of TFF is that the cross-flow continuously sweeps the filter surface, reducing the extent to which materials accumulate on the filter surface, and increasing filtration throughput.
Ultrafiltration systems can be operated with different control strategies. Ultrafiltration and diafiltration processes are typically developed using constant retentate pressure, constant trans-membrane pressure or constant filtrate flux. As these control methods do not factor in the effects of the protein gel layer at the membrane surface, a method of maintaining constant protein concentration at the membrane surface was introduced.87 The potential benefits of this method include enhanced product yield, minimized membrane area, consistent processing times as the variations in feed and membrane properties are controlled.
Delivery of therapeutic mAbs by subcutaneous administration is convenient for patients, but a high concentration formulation must be used in order to keep the injection volume low.88 A high concentration formulation (183 g/L) was developed for an anti-B. anthracis protective antigen antibody in clinical development.88 The optimal antibody concentration at the diafiltration step was selected to minimize the processing time and required surface area. With high concentration ultrafiltration processes, solutions often become very viscous and this limits the final concentration. Recent work has demonstrated use of elevated temperature as an approach to safely manage the rheological properties of high concentration formulations, as well as to enhance overall mass transfer.88,89 Winter et al. developed a high concentration formulation process for an IgG1.88 The high viscosity of the IgG1 limited the ability to achieve the required high concentration of greater than 200 g/L. The process temperature was increased from 23 to 46°C, which resulted in a 2-fold decrease in viscosity and a 2-fold increase in the filtration flux. Processing at the higher temperature did not impact the product quality.
An important process development aspect of a final UFDF formulation step includes the final sterile filtration or bulk filtration of the product. In general, sterile filtration is an important concern for all intermediate purification pools, but considerably more so at the end of the process where the highest protein concentrations are present and greatest value has been imparted onto the product. Proper scale-up techniques using equipment representative to manufacturing is critical.90 Understanding the need for cost-effective, robust sterile filtration, manufacturers have also developed new classes of composite and asymmetric membranes with higher throughput capacities.91,92
High performance tangential flow filtration.
Ultrafiltration has the inherent nature of high throughput and low resolution. Recent studies have shown that ultrafiltration systems can be used for the separation of proteins of similar and moderately different sizes based on differences in protein charge.93–97 Separation is achieved by exploiting electrostatic interactions between protein molecules and the membrane pores. The emerging technique of high performance tangential flow filtration (HPTFF) is a two-dimensional unit operation in which both size and charge differences are utilized for the purpose of purification and separation.98 In addition, protein concentration and buffer exchange can be accomplished in the same unit operation.
Charged proteins in electrolyte solutions are surrounded by a diffuse ion cloud or electrical double layer due to electrostatic interactions with the counter-ions and co-ions.86 In HPTFF, by optimizing buffer pH and ionic strength, the difference in hydrodynamic size between product and impurities can be increased. The hydrodynamic size increases with charges and decreases with ionic strength since higher conductivities shield charges on proteins.99 High selectivity is achieved by increasing the electrostatic exclusion of the more highly charged protein relative to the neutral protein. In addition, membrane charge could further enhance the resolution between charged and neutral molecules.95 The use of charged membranes provides an additional degree of robustness to the process compared to uncharged membranes. A positively charged membrane could provide much greater retention of a positively charged protein than a negatively charged or neutral membrane.
In HPTFF, membrane pore size distribution affects selectivity by altering solute sieving coefficients and filtrate flow distribution. Eliminating large defects and controlling the pore size distribution can significantly improve the performance of the HPTFF membrane.86
As in conventional ultrafiltration processes, the successful implementation of HPTFF processes relies on optimizing operational fluxes, transmembrane pressure, module and flow path design to enhance selectivity and reduce protein fouling on membrane surfaces to reach an optimal performance.96
HPTFF has been reported to achieve removal of host cell proteins and host cell DNA impurities in mAb purification processes utilizing non-affinity chromatography steps combined with HPTFF.100 HPTFF was used in diafiltration mode in which product was retained while impurities were removed from the retentate. One example resulted in the reduction of Chinese hamster ovary proteins (CHOP) from 530 ppm (parts per million, i.e., nanogram of CHOP per milligram of mAb product) after cation exchange purification, to 15 ppm after anion exchange purification and then to less than 0.6 ppm after HPTFF.100
Virus filtration.
Mammalian cells used in the manufacture of mAbs and other therapeutic recombinant proteins produce endogenous retroviruses and are occasionally infected with adventitious viruses during processing.101 Due to safety requirements, mammalian cell-derived products may contain less than one virus particle per million doses. This typically translates to approximately 12–18 log10 clearance of endogenous retroviruses and 6 log10 clearance for adventitious viruses.102 The inclusion of virus removal and inactivation steps in designing purification processes is an integral component of product safety assurance strategies.101 It is generally required that orthogonal virus clearance steps are used that employ complementary mechanisms, so that viruses not cleared by one mechanism may be cleared by the other.103
Virus filtration can provide a size-based viral clearance mechanism that complements other virus clearance steps. Since the presence of only a small number of abnormally large pores will permit excessive virus leakage, virus filters must be manufactured so as to eliminate these large porous defects.86 This is typically accomplished through use of composite membranes that provide the required combination of virus retention and mechanical stability.86
Current virus-retentive filters are ultrafilters or microfilters with very small pores.104 Virus filtration membranes are made from hydrophilic polyethersulfone (PES), hydrophilic polyvinylidene (PVDF) and regenerated cellulose. According to the size distribution of viruses that are removed, virus filters can be categorized into retrovirus filters and parvovirus filters. Commercially available virus filters, their construction materials and virus retention are summarized in Table 1.102
Table 1.
Commercially available virus filtration products
| Company | Product | Material | Virus or bacteriophage retention claimed by filter manufacturer | Virus size (nm) |
| Asahi-Kasei | Planova 15N | cuprammonium regenerated cellulose | >6.2 log parvovirus | 18–26 |
| >6.7 log poliovirus | 28–30 | |||
| Planova 20N | cuprammonium regenerated cellulose | >4.3 log parvovirus | 18–26 | |
| >5.4 log encephalomyocarditis | 28–30 | |||
| Planova 35N | cuprammonium regenerated cellulose | >5.9 log bovine viral diarrhea virus | 40–70 | |
| >7.3 log HIV | 80–130 | |||
| Planova BioEX | PVDF | >4.8 log minute virus of mice | 18–24 | |
| >5.2 log amphotropic murine leukemia virus | 80–130 | |||
| Millipore | Viresolve NFP | PVDF | >4 log φX-174 bacteriophage | 28 |
| Viresolve NFR | PES | >6 log retrovirus | 80–130 | |
| Viresolve Pro | PES | ≥4 log minute virus of mice | 18–24 | |
| ≥5 log xenotropic murine leukemia virus | 80–110 | |||
| Pall | Ultipor DV 20 | PVDF | >3 log PP7 bacteriophage | 26 |
| >6 log PR772 bacteriophage | 76–88 | |||
| Ultipor DV 50 | PVDF | >6 log PR772 bacteriophage | 76–88 | |
| Sartorius | Virosart CPV | PES | >4 log PP7 bacteriophage | 26 |
| >6 log retrovirus | 80–130 |
Parvoviruses have a diameter of 18–26 nm, and a typical mAb has a hydrodynamic diameter of 8–12 nm. To achieve efficient retention of the viruses and passage of the mAb, parvovirus filters are required to have a very narrow pore size distribution.102 Therefore, they are generally sensitive to the presence of impurities in the feed solution.
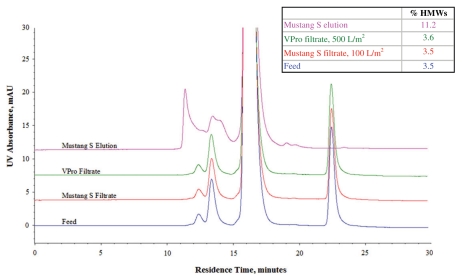
During virus filtration, fouling is typically caused by the presence of protein aggregate, DNA, partially denatured product and other debris. Fouling can be significantly reduced using appropriate prefilters, and prefiltration of the feed solution can have a dramatic impact on virus filtration performance.102 Larger impurities can be removed using 0.1–0.2 µm microfilters, but impurities that are only marginally larger than the protein product are not easily removed using size-based methods. Prefiltration through adsorptive depth filters and charged membranes have been observed to provide significant protection to the virus filters.105–108 Figure 7 illustrates how a negatively charged membrane (Pall Mustang S), used as a prefilter before a Viresolve Pro (VPro) virus filter, removed high molecular weight species (HMWS) in the feedstock.106 Consequently, the capacity of the Viresolve Pro filter was significantly increased (Fig. 8).
Figure 7.
High performance size exclusion chromatography analysis does not show a reduction of HMWS across Mustang S/VPro filtrates. The lack of any significant change suggests the levels of HMWS (as parvovirus filter foulants) in the feedstream are low. However, the enrichment of HMWS is presented in the Mustang S elution pool.106
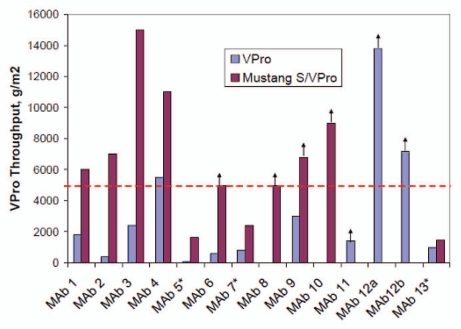
Figure 8.
Viresolve Pro filter throughput improvement through coupling Mustang S membrane as a pre-filter.106
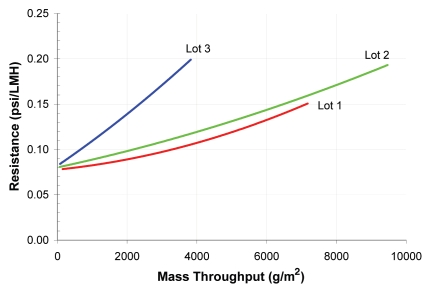
Different lots of viral filters and feed solutions used in viral filtration processes can give different filtration throughputs, whereas manufacturing variability of filter membrane permeability can be controlled with an acceptable range. The impurity content in feed streams from different manufacturing lots is often variable and freshness and storage conditions for feed solutions can also significantly affect throughput. This can be more pronounced if an in-process product pool from an earlier purification step is used as feed solution for viral filtration. The difference of filtration throughput of an in-process pool on a VPro viral filter is shown in Figure 9. The lower throughput of the third lot was due to slightly higher percentage of HMW impurities present in the pool.108 When developing a large scale viral filtration process, the throughput obtained with the representative feed solutions, including the worse case purity pool, should be considered when selecting viral filter membrane surface area.
Figure 9.
Filtration throughput variation of a mAb in-process purification pool on Viresolve Pro filter.108 Viresolve Pro filter resistance (psi/LMH) increases faster with the Lot 3 in-process pool. It results a lower filtration throughput with the Lot 3 in-process pool.
As a routine practice, virus filters are integrity tested pre- and post-use to ensure that the filter achieves the required level of performance.109 To facilitate this, filter manufacturers have developed a variety of destructive and non-destructive physical integrity tests.102 The purposes of these physical integrity tests are to confirm that: (1) the virus filter is properly installed; (2) the filter is free from defects and damages; and (3) the performance of the filters is consistent with both manufacturers' specifications and end-user virus retention studies.102 The most commonly used nondestructive tests include the bubble point test, the forward flow test, the water intrusion test and the binary gas test. Both the bubble point test and the forward flow test evaluate a wet membrane as a barrier to the free flow of a gas.110 The water intrusion test, also called HydroCorr test, uses a dry hydrophobic membrane as a barrier to the free flow of water, a non-wetting fluid.110 The binary gas test uses a mixture of two gases with high differences in permeability, and the test is based on measurement of the composition of the gas mixture upstream and downstream of a water-wetted membrane. The gold particle test, the post-use integrity test used with Planova (Asahi-Kasei) filters, is a destructive integrity test. Generally, nondestructive tests are preferred for end-users due to the option of retesting filter integrity if the initial test fails. In practice, if the post-use integrity tests and retests fail, refiltration is a common practice for virus filtration steps.
Platform Purification Processes
The use of a platform for cell culture and purification processes allows rapid development of a suitable process for generation of early phase clinical supplies. A platform process incorporates experience gained from working with a number of antibodies and thus includes defined purification steps and resins or membrane with operating conditions chosen to work with the majority of new products. Table 2 provides a reference list for selecting primary recovery and purification unit operations that can be used to build a platform process.
Table 2.
Unit operations that can be used in mAb purification process
| Unit operation | Matrix | Function | Operation mode | Limitation | Place in purification process | Recommendation for current process use |
| Centrifugation | Mechanical equipment | Centrifugal separation of host cells, leaving product in clarified supernatant | Solid/Liquid separation | Proper system design required to minimize shear damage | Primary harvest step | high |
| Depth filtration | Filter | Used to remove cells and cell debris as well as host cell proteins and DNA | Size exclusion, adsorption | Handling, disposal, flow distribution | Used as primary and secondary harvest step | high |
| Flocculation | Chemical | Aggregation of solids to aid in solid and liquid separation (centrifuge, TFF, filtration) | N/A | Shear sensitivity, residual flocculent removal and impact downstream | Harvest | medium |
| Protein A affinity chromatography | Resin | Used as a primary purification step to capture Mab product from HCCF Very effective step to remove most process-related impurities generated during cell culture such as HCP, DNA and other unwanted components | BE | Mab binding capacity. High cost. Multiple cycles on smaller column may reduce cost and risk. | 1st chromatography step | High |
| Cation exchange chromatography | Resin | Used as a polishing step to remove unwanted product variants and aggregates, residual amount of HCP, DNA and other unwanted impurities | BE | Mab binding capacity, cost or plant constraints may require multiple cycles to processing a single batch | 2nd or 3rd chromatography step | high |
| Resin | Use as a capture step to capture Mab from HCCF. Reduce process-related impurities and product-related impurities Enables building a process without protein A affinity chromatography | BE | Mab binding capacity, cost or plant constraints may require multi-cycles to processing a single batch. Dirty feed streams may limit regeneration and reuse. | 1st chromatography step | low | |
| Membrane | Used as a polishing step to bind process-relate impurities and aggregates | FT | Impurity binding capacity | As a prefilter for parvo virus filtration | high | |
| Anion exchange chromatography | Resin | Used as a polishing step to remove residual HCP, DNA, viruses and other unwanted impurities derived from cell culture process, leached protein A if protein A affinity chromatography is used as the capture step and aggregates | FT, BE | The loading conductivity can not be high. May require pre-dilution of the load pool. Mab binding capacity constraint in BE mode may require multiple cycles to process a single batch. | 2nd or 3rd chromatography step | high |
| Membrane | Used as a polishing step to remove residual HCP, DNA, viruses and other unwanted impurities derived from cell culture process, leached protein A if protein A affinity chromatography is used as the capture step | FT | Requires low load conductivity. May require pre-dilution of the load pool. Higher impurity level in load can limit throughput and increase process cost | 2nd chromatography step | low | |
| 3rd chromatography step | medium | |||||
| Hydrophobic interaction chromatography | Resin | Used as a polishing step to remove unwanted product variants and aggregates, viruses, residual HCP, DNA and other unwanted impurities | BE, FT | Mab binding capacity, cost or manufacturing plant constraints may require multiple cycles to process a single batch. | 2nd or 3rd chromatography step | medium |
| Anionic hydrophobic mixed mode | Resin | Used as a polishing step to remove residual HCP, DNA and other unwanted impurities derived from cell culture process and aggregates | BE | Mab binding capacity or manufacturing plant constraints may require multiple cycles to process a single batch. | 2nd or 3rd chromatography step | medium |
| Anionic mixed mode | Membrane | Used as a polishing step to remove residual amounts of HCP, DNA, viruses and other unwanted impurities derived from cell culture process | FT | Higher impurity level in load can limit throughput and increase process cost | 3rd chromatography step | medium |
| Ceramic Hydroxyapatite Chromatography | Resin | Used as a polishing step to remove residual amounts of HCP, DNA and other unwanted impurities derived from cell culture process, leached protein A if protein A affinity chromatography is used as the capture step and aggregates | BE | Mab binding capacity or manufacturing plant constraints may require multiple cycles to process a single batch. Not compatible with citrate salt and EDTA (chelating agent). Resin lifetime needs to be evaluated. | 2nd or 3rd chromatography step | low |
| Hydrophobic Charge Induction Chromatography | Resin | Used as a capture step to capture Mab from HCCF Used as a polishing step to remove residual impurities | BE | Less effective for impurity removal compared with protein A affinity chromatography as a capture step. Limited Mab binding capacity | 1st or 2nd or 3rd chromatography step | low |
| Viral filtration | Membrane | Removes a broad size of viruses depending on pore size of filter | FT | High cost. Often prone to fouling, limiting throughput | After 2nd or 3rd chromatography step | high |
| TFF—Microfiltration | Membrane | Used as a primary cell culture harvest step | Size exclusion | Prone to fouling with cell culture fluid containing high solid contents Prone to higher shear and cell lysis | Harvest step | medium |
| TFF—Ultrafiltration | Membrane | Concentrate and buffer exchange of intermediate pools and purified bulk | Size exclusion | Diligent system design required for complete buffer exchange, high product recovery and high concentration formulation | Various intermediate pools and final formulation | high |
| TFF—High performance TFF | Membrane | Two-dimensional unit operation in which both size and charge differences are utilized for the purpose of purification and separation | Size exclusion | Highly complex. Not commercially available | yet Used for both purification and concentration and buffer exchange | low |
| Sterile filtration | Membrane | Bioburden reduction, particulate removal. Effective in providing a sterile barrier for intermediate hold steps or final storage | Size exclusion | Typically not suitable for handling large particulate loads. Prone to pore plugging. | All points in the purification process | High |
Platforms are designed to allow rapid development of a process generating a final product with the purity required for use in humans with use of minimal resources.111,112 Additionally, cost, time and labor savings may be realized in raw materials introduction and testing, documentation, process transfer activities and viral validation.113 If similar cell lines and cell culture processes are used and cell viability at the end of the culture process falls within a consistent range, it may be possible to develop a generic harvest process. Centrifugation and filtration parameters are chosen that can cover the expected range conservatively to maximize the probability of success for this step.
The choice of chromatography resins may be limited or fixed in a platform process. Likewise, buffer compositions for wash and elution buffers and even the final formulation buffer, may be predefined or only require optimization within a defined range. In an ideal case, downstream process development for a new molecule entails verifying process fit, establishing dynamic binding capacities and loading parameters for the chromatography steps, defining the pH and conductivity of the buffers involved and verifying impurity clearance and product quality. In practice, this is often successful, but pipelines may include molecules that aren't fully compatible with the platform for one or more steps, and in those instances additional development will be required.
Perspectives on the Future
Downstream processing practices and platform technologies will need to advance to keep pace with increases in cell culture titers, increasing regulatory requirements, changes in the market and the need to control costs. In the near term, downstream processes must be adapted to cope with higher titer cell culture processes in existing plant infrastructure. Future plants will need to take into account current and anticipated advances in expression systems, cell culture and downstream processes. An excellent perspective on future directions in mAb purification has recently been published.114
As titers have increased, the need to recover an entire batch using existing infrastructure has presented new challenges. Constraints include working with existing chromatography column dimensions, buffer and hold tank volumes and skid capabilities in the manufacturing plant. The use of high capacity resins can allow the use of existing chromatography columns to process a greater mass of material. Opportunities for minimizing pool volumes to use existing hold tanks include optimizing elution conditions and taking a close look at step order and load conditioning requirements. The use of inline dilution or conditioning to achieve the target pH, conductivity and buffer composition may be employed. Likewise, using inline dilution and buffer concentrates can allow existing buffer tanks to be employed.
Advances in purification technology that may be considered for incorporation in future recovery processes include use of high-flow high capacity chromatography resins, non-chromatographic processes such as membrane adsorbers, precipitation, flocculation and crystallization, as well as streamlined processes and methods development.
Another possible development in mAb production is the possibility of using expression systems other than CHO, which is the predominant cell line for expressing mAbs. One reason is their capacity for glycosylation, which is critical for antibodies that require effector function to be efficacious. NS0 cells are used to a lesser extent. Engineered yeast cells are capable of producing antibodies with desired glycosylation and may have advantages in both cell line development and engineering the expression of particular glycoforms.115 E. coli can be used to produce full length aglycosylated antibodies for use in indications where effector function is undesirable or for making antibody-drug conjugates. Lastly, transgenic animals and plants have been extensively explored as alternatives to mammalian cell culture.116,117
Recovery of mAbs from different expression systems can be quite variable, e.g., recovery from NS0 cells is nearly identical to recovery from CHO, but drastically different when compared with recovery from transgenic plants or animals. Initial harvest from transgenic plants may involve homogenization and the removal of a large quantity of solids, oils and other substances; in the case of transgenic expression in milk, removal of milk fat and casein is necessary. Even when the product may be secreted, e.g., in expression in yeast, fungi, E. coli, a downstream process is likely to differ substantially from one developed for CHO-expressed antibodies due to the large difference in host and media derived impurities.
Due to the long timelines associated with transgenic expression, the current high titers being achieved in CHO and the fact that the capacity crunch in biologics manufacturing has eased, at present there is less of a drive towards transgenic expression. Expression systems such as yeast (Pichia pastoris) and microbes (E. coli) have advantages compared to CHO in the speed of development of a banked cell line starting from DNA; these systems thus may become more prevalent in the future.
Future purification processes and manufacturing plants are likely to make use of the new high capacity/high flow chromatography resins coming to market; however, even though many of the current limitations in chromatographic processes are being addressed, many people are reconsidering traditional non-chromatographic processes such as crystallization, precipitation and flocculation,118,119 as well as newer alternatives such as membrane adsorbers.78,83
Flocculation is a method that has been widely used in other fields and is gathering interest in the recovery of proteins from mammalian cell culture. One of its most tangible advantages is the reduction of membrane surface area in harvest depth filtration.24,26,120 Salts or synthetic and natural polymers are added to cell culture fluid and cells and particles flocculate, enhancing removal by both centrifugation and filtration. It is also possible that flocculation could provide substantial impurity clearance, opening the door to alternative or streamlined processes downstream.
Since Protein A chromatography is the most expensive chromatographic step in the downstream process, lower cost alternatives are being sought. It is possible to achieve the desired clearance of host cell proteins using only ion exchange and hydrophobic interaction chromatography,56 or even cation and anion exchange chromatography followed by HPTFF.100 It is possible that precipitation with charged polymers may substitute for ion exchange chromatography steps.121 Alternatively, use of solvent fractionation analogous to the Cohn process as an initial purification step can replace Protein A chromatography in a typical three-step purification while providing purity and yield that is at least equivalent.122
Crystallization is a method that has been used for protein purification since the 1920s.123 It is currently employed as an isolation step for industrial enzymes at ton scale and in the purification of several non-antibody recombinant products such as insulin and Apo2L,124 and aprotinin.125 Crystallization of antibodies is less straightforward due to heterogeneity, conformational flexibility and other factors, but nonetheless is an attractive avenue for research.
It is possible that an entirely chromatography-free process that can provide adequate impurity clearance might be available at some point. One factor that needs to be considered is viral clearance—replacing chromatographic steps that provide a high degree of viral clearance with steps that may or may not provide a similar degree of viral clearance may entail the use of new modes of viral filtration and inactivation. Current processes provide viral clearance primarily through the Protein A/low pH hold step, anion exchange chromatography and a dedicated virus filter. Other available methods in common use in the blood products industry such as solvent/detergent inactivation may come to be employed. Newer alternatives being developed include inactivating viruses with UV or broad spectrum light,126–128 gamma irradiation,129,130 heat131,132 and caprylic acid.133,134
Streamlined processes with fewer conventional steps are also gaining prominence. As outlined previously, it is possible to eliminate the cation exchanger in the typical Protein A/cation exchanger/anion exchanger process by the use of careful optimization of the anion exchange step using weak partitioning chromatography.51,135 This method may prove to be applicable to purification of many antibodies, but may not be applicable for processes that require a cation exchanger for substantial aggregate removal or for use with antibodies with a relatively high isoelectric point that would require operation at a basic pH, where degradation due to deamidation is a risk. As a counterpoint to the perceived need for alternatives to chromatography at high titers, Kelley also concludes that it is entirely feasible for a single plant to produce 10 tons of a mAb per year using conventional methods and highlights potential issues that may arise from the adoption of non-conventional methods.131
Over the short term, the primary changes compared to today are likely to be in the adoption of chromatography resins with improved properties and the use of HTS for optimizing chromatographic operations. Time will tell if Q membrane adsorbers gain a significant foothold in replacing a conventional anion exchange chromatography step. The adoption of non-chromatographic recovery schemes will require additional data on viral clearance, ability to be applied to a wide range of antibodies, impact on development time and economic benefit and likely some plant redesign as well.
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/12645
References
- 1.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Discov. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 2.Fahrner RL, Knudsen HL, Basey CD, Galan W, Feuerhelm D, Vanderlaan M, et al. Industrial purification of pharmaceutical antibodies: development, operation and validation of chromatography processes. Biotechnol Genet Eng Rev. 2001;18:301–327. doi: 10.1080/02648725.2001.10648017. [DOI] [PubMed] [Google Scholar]
- 3.Birch JR, Racher JR. Antibody production. Adv Drug Delivery Rev. 2006;58:671–685. doi: 10.1016/j.addr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Kempken R, Pressman A, Berthold W. Assessment of a disc stack centrifuge for use in mammalian cell separation. Biotechnol Bioeng. 1995;46:132–138. doi: 10.1002/bit.260460206. [DOI] [PubMed] [Google Scholar]
- 5.Pham C, Lee M, Westoby M, Thommes J. Solid-liquid separation of mammalian cell culture suspensions using a continuous disk stack centrifuge; 3rd International IBC Recovery and Purification Conference; 2002; San Diego, CA. [Google Scholar]
- 6.Shpritzer R. Evaluation of a continuous disk stack centrifuge for the clarification of mammalian cell cultures; 225th ACS National Meeting; 2003; New Orleans, LA. [Google Scholar]
- 7.Altena FW, Belfort G. Lateral migration of spherical particles in porous flow channels: application to membrane filtration. Chem Eng Sci. 1984;39:343–355. [Google Scholar]
- 8.Davis RH, Leighton DT. Shear-induced transport of a particle layer along a porous wall. Chem Eng Sci. 1987;42:275–281. [Google Scholar]
- 9.Drew DA, Schonberg JA, Belfort G. Lateral inertial migration of a small sphere in fast laminar flow through a membrane duct. Chem Eng Sci. 1991;46:3219–3224. [Google Scholar]
- 10.Romero CA, Davis RH. Global model of crossflow microfiltration based on hydrodynamic particle diffusion. J Membr Sci. 1988;39:157–185. [Google Scholar]
- 11.Bender J, Brown A, Winter C. Scale-up of a disc stack centrifuge for CHO harvest, Downstream Gab '02 abstracts. Amersham Biosciences. 2002:10–11. [Google Scholar]
- 12.Hutchison N, Bingham N, Murrell N, Farid S, Hoare M. Shear stress analysis of mammalian cell suspensions for prediction of industrial centrifugations and its verification. Biotechnol Bioeng. 2006;95:483–491. doi: 10.1002/bit.21029. [DOI] [PubMed] [Google Scholar]
- 13.Iammarino M, Nti-Gyabaah J, Chandler M, Roush D, Goklen K. Impact of cell density and viability on primary clarification of mammalian cell broth. Bioprocess Int. 2007;5:38–50. [Google Scholar]
- 14.Laird M. Antibody disulfide reduction in CHO production process; Williamsburg Bioprocessing Foundation's 15th International Waterside Conference; 2009; South San Francisco CA. [Google Scholar]
- 15.Schmidt M. Antibody degradation (disulfide reduction) in a CHO production process; IBC Antibody Development & Production Conference; 2009; Carlsbad CA. [Google Scholar]
- 16.Knight RA, Ostreicher EA. In: Charge-modified filter media. Filtration in the biopharmaceutical industry. Meltez TH, Jornitz MW, editors. New York, NY: Marcel Dekker, Inc.; 1998. pp. 95–125. [Google Scholar]
- 17.Tipton B, Boose JA, Larsen W, Beck J, O'Brien T. Retrovirus and parvovirus clearance from an affinity column product using adsorptive depth filtration. BioPharm Int. 2002;15:43–50. [Google Scholar]
- 18.Gerba CP, Hou K. Endotoxin removal by charge-modified filters. Appl Environ Microbiol. 1985;50:1375–1377. doi: 10.1128/aem.50.6.1375-1377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou K, Gerba CP, Goyal SM, Zerda KS. Capture of latex beads, bacteria, endotoxin and viruses by charge-modified filters. Appl Environ Microbiol. 1980;40:892–896. doi: 10.1128/aem.40.5.892-896.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.vanHolten RW, Autenrieth SM. Evaluation of depth filtration to remove prion challenge from an immunoglobulin preparation. Vox Sang. 2003;85:20–24. doi: 10.1046/j.1423-0410.2003.00312.x. [DOI] [PubMed] [Google Scholar]
- 21.Dorsey N, Eschrich J, Cyr G. The role of charge in the retention of DNA by charged cellulose-based depth filters. BioPharm. 1997;10:46–49. [Google Scholar]
- 22.Charlton HR, Relton JM, Slater NKH. Characterisation of a generic monoclonal antibody harvesting system for adsorption of DNA by depth filters and various membranes. Bioseparation. 1999;8:281–291. doi: 10.1023/a:1008142709419. [DOI] [PubMed] [Google Scholar]
- 23.Yigzaw Y, Piper R, Tran M, Shukla AA. Exploitation of the adsorptive properties of depth filters for host cell protein removal during monoclonal antibody purification. Biotechnology Progress. 2006;22:288–296. doi: 10.1021/bp050274w. [DOI] [PubMed] [Google Scholar]
- 24.Aunins JG, Wang DI. Induced flocculation of animal cells in suspension culture. Biotechnol Bioeng. 1989;34:629–638. doi: 10.1002/bit.260340507. [DOI] [PubMed] [Google Scholar]
- 25.Salt DE, Hay O, Thomas ORT, Hoare M, Dunnill P. Selective flocculation of cellular contaminants from soluble proteins using polyethyleneimine: a study of several organisms and polymer molecular weights. Enzyme Microb Technol. 1995;17:107–113. [Google Scholar]
- 26.Riske F, Schroeder J, Belliveau J, Kang X, Kutzko J, Menon MK. The use of chitosan as a flocculant in mammalian cell culture dramatically improves clarification throughput without adversely impacting monoclonal antibody recovery. J Biotechnol. 2007;128:813–823. doi: 10.1016/j.jbiotec.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Romero J. Disc stack centrifugation 29,000 L clarification scale-up evaluation: harvest optimization for combined cell removal and purification; BioProcess International Conference; 2007; Boston MA. [Google Scholar]
- 28.Coffman J, Shpritzer R, Vicik S. Recovery of Biological product XII. Litchfield AZ: 2006. Flocculation of antibody producing mammalian cells with precipitating solutions of soluble cations and anions. [Google Scholar]
- 29.Hashimoto M, Yamamoto T, Kawachi T, Kuwashima J, Kitaoka H. EP Patent 240348. 19931027 Low molecular weight chitosans for the removal of nucleic acids and/or endotoxins from liquid samples on a small or large scale.
- 30.Hughes J, Ramsden DK, Symes KC. The flocculation of bacteria using cationic synthetic flocculants and chitosan. Biotechnol Tech. 1990;4:55–60. [Google Scholar]
- 31.Persson I, Lindman B. Flocculation of cell debris for improved separation by centrifugation. Amsterdam: Elsevier Science Publishers B.V.; 1987. Flocculation in biotechnology and separation systems; pp. 457–466. [Google Scholar]
- 32.Weir S, Ramsden DK, Hughes J, LeThomas F. The flocculation of yeast with chitosan in complex fermentation media: the effect of biomass concentration and mode of flocculant addition. Biotechnol Tech. 1993;7:199–204. [Google Scholar]
- 33.Ghose S, Hubbard B, Cramer SM. Binding capacity differences for antibodies and Fc-fusion proteins on protein A chromatographic materials. Biotechnol Bioeng. 2007;96:768–779. doi: 10.1002/bit.21044. [DOI] [PubMed] [Google Scholar]
- 34.Fahrner RL, Whitney DH, Vanderlaan M, Blank GS. Performance comparison of protein A affinity-chromatography sorbents for purifying recombinant monoclonal antibodies. Biotechnol Appl BioChem. 1999;30:121–128. [PubMed] [Google Scholar]
- 35.O'Leary RM, Feuerhelm D, Peers D, Xu Y, Blank GS. Determining the useful lifetime of chromatography resins. BioPharm Int. 2001:10–18. [Google Scholar]
- 36.Carter-Franklin JN, Victa C, McDonald P, Fahrner R. Fragments of protein A eluted during protein A affinity chromatography. J Chromatogr A. 2007;1163:105–111. doi: 10.1016/j.chroma.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Blank G. Protein purification. United States Patent No: US 6797814 2004
- 38.Breece T, Fahrner R, Gorrell JR, Lazzareschi KP, Lester P, Peng D. United States Patent No: US 6870034 B2 2005 Protein purification.
- 39.Arakawa T, Philo JS, Tsumoto K, Yumioka R, Ejimac D. Elution of antibodies from a Protein A column by aqueous arginine solutions. Protein Expr Purif. 2004;36:244–248. doi: 10.1016/j.pep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Shukla AA, Hinckley PJ, Gupta P, Yigzaw Y, Hubbard B. Strategies to address aggregation during protein A chromatography. Bioprocess Int. 2005;3:36–44. [Google Scholar]
- 41.Ghose S, Allen M, Hubbard B, Brooks C, Cramer SM. Antibody variable region interactions for the development of generic purification process. Biotechenol Bioeng. 2005;92:665–673. doi: 10.1002/bit.20729. [DOI] [PubMed] [Google Scholar]
- 42.Gronberg A, Monié E, Murby M, Rodrigo G, Wallby E, Johansson H. A strategy for developing a monoclonal antibody purification platform. Bioprocess Int. 2007;5:48–55. [Google Scholar]
- 43.Wan M. Process mapping—A necessary prelude for process changing; IBC's BioProcess International Conference; 2004; Boston MA. [Google Scholar]
- 44.Curtis S, Lee K, Blank GS, Brorson K, Xu Y. Generic/matrix evaluation of SV40 clearance by anion exchange chromatography in flow-through mode. Biotechnol Bioeng. 2003;84:179–186. doi: 10.1002/bit.10746. [DOI] [PubMed] [Google Scholar]
- 45.Norling LA, Lute S, Emery R, Khuu W, Voisard M, Xu Y, et al. Impact of multi re-use of anion exchange chromatography media on virus removal. J Chromatogr A. 2005;1069:79–89. doi: 10.1016/j.chroma.2004.09.072. [DOI] [PubMed] [Google Scholar]
- 46.Zhou JX. Development of future downstream process for commercial monoclonal antibody production; Presentation at Asia bioLogic; 2007; Beijing, China. [Google Scholar]
- 47.Liu H, Kellogg T. Development of an anion exchange chromatographic process in bind-elute and bind-washout modes; BPI International Conference & Exhibition; 2008; Anaheim, CA. [Google Scholar]
- 48.Liu H, Tang K. A method for the removal of aggregate proteins from recombinant samples using ion exchange chromatography. 2005 Patent application WO 2005/077130 A2.
- 49.Blank GS. The future of process development for Mab manufacturing; IBC's 14th International Antibody Production & Downstream Processing; 2005; San Diego CA. [Google Scholar]
- 50.Gagnon P. The question for a generic IgG purification procedure; WilBio-Europe, Antibodies & Recombinant Protein 2nd Annual Meeting; 2005; Amsterdam, The Netherlands. [Google Scholar]
- 51.Kelly BD, Tobler SA, Brown P, Coffman JM, Godavarti R, Iskra T, et al. Weak partitioning chromatography for anion exchange purification of monoclonal antibodies. Biotechnol Bioeng. 2008;101:553–566. doi: 10.1002/bit.21923. [DOI] [PubMed] [Google Scholar]
- 52.Coffman J, Kramarczyk J, Kelley B. High-throughput screening of chromatographic separations: 1. Method development & column modeling. Biotechnol Bioeng. 2008;100:605–618. doi: 10.1002/bit.21904. [DOI] [PubMed] [Google Scholar]
- 53.Kelley B, Switzer M, Booth J, Coffman J. Application of high throughput screening to a Mab purification platform; IBC conference; 2005; Boston MA. [Google Scholar]
- 54.Harinarayan C, Mueller J, Ljunglof A, Fahrner R, Van Alstine J, van Reis R. An exclusion mechanism in ion exchange chromatography. Biotechnol Bioeng. 2006;95:775–787. doi: 10.1002/bit.21080. [DOI] [PubMed] [Google Scholar]
- 55.Ansaldi DA, Lester P. United States Patent No: US 6620918. Separation of polypeptide monomers. 2003
- 56.Follman D, Fahrner R. Factorial screening of antibody purification process using three chromatography steps without protein A. J Chromatogr A. 2004;1024:79–85. doi: 10.1016/j.chroma.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 57.Mhatre R, Nashabeh W, Schmalzing D, Yao X, Fuchs M, Whitney D, et al. Purification of antibody Fab fragments by cation-exchange chromatography and pH gradient elution. J Chromatogr A. 1995;707:225–231. [Google Scholar]
- 58.Zhou J, Dermawan S, Soloamo F, Flynn G, Stenson R, Tressel T, et al. pH-conductivity bybrid gradient cation-exchange chromatography for process-scale monoclonal antibody purification. J Chromatogr A. 2007;1175:69–80. doi: 10.1016/j.chroma.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 59.Porath J. Salt-promoted adsorption: recent developments. J Chromatog. 1986;376:331–341. doi: 10.1016/s0378-4347(00)80850-6. [DOI] [PubMed] [Google Scholar]
- 60.Guse A, Milton A, Schulze-Koops H, Muller B, Roth E, Simmer B. Purification and analytical characterization of an anti-CD4 monoclonal antibody for human therapy. J Chromatogr A. 1994;661:13–23. doi: 10.1016/0021-9673(94)85173-5. [DOI] [PubMed] [Google Scholar]
- 61.Kato Y, Nakamura K, Kitamura T, Hasegawa M, Sasaki H. Hydrophobic interaction chromatography at low salt concentration for the capture of monoclonal antibodies. J Chromatogr A. 2004;1036:45–50. doi: 10.1016/j.chroma.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Manzke O, Tesch H, Diehl V, Bohlen H. Single-step purification of bispecific monoclonal antibodies for immunotherapeutic use by hydrophobic interaction chromatography. J Immunol Methods. 1997;208:65–73. doi: 10.1016/s0022-1759(97)00129-4. [DOI] [PubMed] [Google Scholar]
- 63.Rinderknecht E, Zapata G. United States Patent Application 2002/0002271. Antibody purification. 2002
- 64.Burton SC, Harding DRK. Hydrophobic charge induction chromatography: salt independent protein adsorption and facile elution with aqueous buffers. J Chromatogr A. 1998;814:71–81. doi: 10.1016/s0021-9673(98)00436-1. [DOI] [PubMed] [Google Scholar]
- 65.Yon RJ. Chromatography of lipophilic protein on adsorbents containing mixed hydrophobic and ionic groups. Biochem J. 1972;126:765–767. doi: 10.1042/bj1260765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guerrier GP, Schwartz W, Boschetti E. New method for the selective capture of antibodies under physiological conditions. Bioseparation. 2000;9:211–221. doi: 10.1023/a:1008170226665. [DOI] [PubMed] [Google Scholar]
- 67.Schwartz W, Judd D, Wysocki M, Guerrier L, Birck-Wilson E, Boschetti E. Comparison of HCIC with affinity chromatography on protein A for harvest and purification of antibody. J Chromatogr A. 2001;908:251–263. doi: 10.1016/s0021-9673(00)01013-x. [DOI] [PubMed] [Google Scholar]
- 68.Franklin SG. Removal of aggregate from an IgG 4 product using CHT ceramic hydroxyapatite. BioProcess Int. 2003;1:50–51. [Google Scholar]
- 69.Gagnon P, Ng P, Zhen J, Aberin C, He J, Mekosh H, et al. A ceramic hydroxyapatite based purification platform. BioProcess Int. 2006;4:50–60. [Google Scholar]
- 70.Brekkan E. Capto™ Adhere—optimization of loading conditions by use of design of experiments; IBC's 18th International Antibody Development & Production; 2007; Carlsbad, CA. [Google Scholar]
- 71.Bensch M, Wierling PS, von Lieres E, Hubbuch J. High throughput screening of chromatographic phases for rapid process development. Chem Eng Technol. 2005;28:1274–1284. [Google Scholar]
- 72.Coffman J, Kramarczyk JF, Kelley BD. High-throughput screening of chromatographic separations: I. Method development and column modeling. Biotechnol Bioeng. 2008;100:605–618. doi: 10.1002/bit.21904. [DOI] [PubMed] [Google Scholar]
- 73.Wierling PS, Bogumil R, Knieps-Grunhagen E, Hubbuch J. High-throughput screening of packed-bed chromatography coupled with SELDI-TOF MS analysis: monoclonal antibodies versus host cell protein. Biotechnol Bioeng. 2007;98:440–450. doi: 10.1002/bit.21399. [DOI] [PubMed] [Google Scholar]
- 74.Kramarczyk JF, Kelley BD, Coffman JL. High-throughput screening of chromatographic separations: II. Hydrophobic interaction. Biotechnol Bioeng. 2008;100:707–720. doi: 10.1002/bit.21907. [DOI] [PubMed] [Google Scholar]
- 75.Wensel DL, Kramarczyk JF, Kelley BD, Coffman JL. High-throughput screening of chromatographic separations: III. Monoclonal antibodies on ceramic hydroxyapatite. Biotechnol Bioeng. 2008;100:839–854. doi: 10.1002/bit.21906. [DOI] [PubMed] [Google Scholar]
- 76.Kelley BD, Switzer M, Bastek P, Kramarczyk JF, Molnar K, Yu T, et al. High-throughput screening of chromatographic separations: IV. Ion-exchange. Biotechnol Bioeng. 2008;100:950–963. doi: 10.1002/bit.21905. [DOI] [PubMed] [Google Scholar]
- 77.Roper DK, Lightfoot EN. Separation of biomolecules using adsorptive membranes. J Chromatogr A. 1995;702:3–26. [Google Scholar]
- 78.Knudsen HL, Fahrner R, Xu Y, Norling L, Blank G. Membrane ion-exchange chromatography for process-scale antibody purification. J Chromatogr A. 2001;907:145–154. doi: 10.1016/s0021-9673(00)01041-4. [DOI] [PubMed] [Google Scholar]
- 79.Ghosh R. Protein separation using membrane chromatography: opportunities and challenges. J Chromatogr A. 2002;952:13–27. doi: 10.1016/s0021-9673(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 80.Phillips M, Moya W, Kozlov M, Yang T. A fundamental understanding of a novel membrane adsorber technology; Recovery of Biological Products XIII; 2008; Québec City, Canada. [Google Scholar]
- 81.Phillips M, Cormier J, Ferrence J, Dowd C, Kiss R, Lutz H, et al. Performance of a membrane adsorber for trace impurity removal in biotechnology manufacturing. J Chromatogr A. 2005;1078:74–82. doi: 10.1016/j.chroma.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 82.Zhou JX, Tressel T, Gottschalk U, Solamo F, Pastor A, Dermawan S, et al. New Q membrane scale-down model for process-scale antibody purification. J Chromatogr A. 2006;1134:66–73. doi: 10.1016/j.chroma.2006.08.064. [DOI] [PubMed] [Google Scholar]
- 83.Zhou JX, Tressel T. Basic concepts in Q membrane chromatography for large-scale antibody production. Biotechnol Progr. 2006;22:341–349. doi: 10.1021/bp050425v. [DOI] [PubMed] [Google Scholar]
- 84.vanReis R, Zydney AL. Protein ultrafiltration. In: Flickinger MC, Drew SW, editors. Encyclopedia of Bioprocess Technology-Fermentation, Biocatalysis and Bioseparation. John Wiley & Sons; 1999. pp. 2197–2214. [Google Scholar]
- 85.Tutunjian RS. Ultrafiltration processes in biotechnology. In: Cooney CL, Humphrey AE, editors. Principles of Biotechnology: Engineering Considerations. Elmsford, New York: Pergamon Press; 1985. pp. 411–437. [Google Scholar]
- 86.vanReis R, Zydney A. Bioprocess membrane technology. J Membr Sci. 2007;297:16–50. [Google Scholar]
- 87.vanReis R, Goodrich EM, Yson CL, Frautschy LN, Whiteley R, Zydney AL. Constant Cwall ultrafiltration process control. J Membr Sci. 1997;130:123–140. [Google Scholar]
- 88.Luo R, Waghmare R, Krishnan M, Adams C, Poon E, Kahn D. High-concentration UF/DF of a monoclonal antibody. Bioprocess Int. 2006;4:44–46. [Google Scholar]
- 89.Winter C, Mulherkar P, Thai B, Liu J, Kanai S, Shire SJ. Formulation strategy and high temperature UF to achieve high concentration rhuMAbs; Recovery of Biological Products XIII; 2008; Quebec City, Quebec, anada. [Google Scholar]
- 90.Rajniak P, Tsinontides S, Pham D, Hunke W, Reynolds S, Chern R. Sterilizing filtration—principles and practice for successful scale-up to manufacturing. J Membr Sci. 2008;325:223–237. [Google Scholar]
- 91.Dixit M. High-performance sterile filtration. BioProcess Int (Industry Yearbook) 2009;7:92–93. [Google Scholar]
- 92.Millipore Corporation, author. Data sheet: Millipore express® SHF hydrophilic filters. Lit. No. DS1426EN00.
- 93.Ebersold MF, Zydney AL. Separation of protein charge variants by ultrafiltration. Biotechnol Progr. 2004;20:543–549. doi: 10.1021/bp034264b. [DOI] [PubMed] [Google Scholar]
- 94.Saksena S, Zydney AL. Effect of solution pH and ionic strength on the separation of albumin from immunoglobulins (IgG) by selective filtration. Biotechnol Bioeng. 1994;43:960–968. doi: 10.1002/bit.260431009. [DOI] [PubMed] [Google Scholar]
- 95.vanReis R, Brake JM, Charkoudian J, Burns DB, Zydney AL. High-performance tangential flow filtration using charged membranes. J Membr Sci. 1999;159:133–142. [Google Scholar]
- 96.vanReis R, Gadam S, Frautschy LN, Orlando S, Goodrich EM, Saksena S, et al. High performance tangential flow filtration. Biotechnol Bioeng. 1997;56:71–82. doi: 10.1002/(SICI)1097-0290(19971005)56:1<71::AID-BIT8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 97.Wan Y, Vasan S, Ghosh R, Hale G, Cui Z. Separation of monoclonal antibody alemtuzumab monomer and dimers using ultrafiltration. Biotechnol Bioeng. 2005;90:422–432. doi: 10.1002/bit.20416. [DOI] [PubMed] [Google Scholar]
- 98.vanReis R, Zydney AL. Membrane separations in biotechnology. Curr Opin Biotechnol. 2001;12:208–211. doi: 10.1016/s0958-1669(00)00201-9. [DOI] [PubMed] [Google Scholar]
- 99.Burns DB, Zydney AL. Effect of solution pH on protein transport through ultrafiltration membranes. Biotechnol Bioeng. 1999;64:27–37. doi: 10.1002/(sici)1097-0290(19990705)64:1<27::aid-bit3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 100.Fahrner R, Follman D, Lebreton B, Van Reis R. United States Patent 2008; 7323553 Non-affinity purification of proteins.
- 101.Brorson K, Krejci S, Lee K, Hamilton E, Stein K, Xu Y. Bracketed generic inactivation of rodent retroviruses by low pH treatment for monoclonal antibodies and recombinant proteins. Biotechnol Bioeng. 2003;82:321–329. doi: 10.1002/bit.10574. [DOI] [PubMed] [Google Scholar]
- 102.Kern G, Krishnan M. Virus removal by filtration: points to consider. BioPharm Int. 2006:19. [Google Scholar]
- 103.FDA, author. Points to Consider in the Manufacture and Testing of Monoclonal Antibody products for Human Use. 1997. Points to Consider in the Manufacture and Testing of Monoclonal Antibody products for Human Use. [DOI] [PubMed] [Google Scholar]
- 104.Carter J, Lutz H. An overview of viral filtration in biopharmaceutical manufacturing. Eur J Parenteral Sci. 2002;7:72–78. [Google Scholar]
- 105.Bolton GR, Spector S, LaCasse D. Increasing the capacity of parvovirus-retentive membranes: performance of the viresolve prefilter. Biotechnol Appl Biochem. 2006;43:55–63. doi: 10.1042/BA20050108. [DOI] [PubMed] [Google Scholar]
- 106.Brown A. Use of charged membrane to identify soluble protein foulants in order to facilitate parvovirus filtration. IBC's 20th antibody development and production conference; 2008; San Diego, CA. [Google Scholar]
- 107.Ireland T, Bolton G, Noguchi M. Optimizing virus filter performance with prefiltration. BioProcess Int. 2005;3:44–47. [Google Scholar]
- 108.Liu H, Hove S, Brown A, Amanullah A, Chang D. Development of a high throughput viral filtration process; 236th ACS National Meeting & Exposition; 2008; Philadelphia, Pennsylvania. [Google Scholar]
- 109.Kuriyel R, Zydney AL. Sterile Filtration and Virus Flitration. In: Desai MA, editor. Downstream Processing of Proteins. Humana Press; 2000. pp. 185–194. [Google Scholar]
- 110.Morris GM, Rozembersky J, Schwartz L. Validation of Filtration. In: Sofer G, Zabriskie DW, editors. Biopharmaceutical Process Validation. Marcel Dekker; 2000. pp. 213–233. [Google Scholar]
- 111.Kelley B, Blank G, Lee A. Downstream Processing of Monoclonal Antibodies: Current Practices and Future Opportunities. In: Gottschalk U, editor. Process Sacle Purification of Antibodies. John Wiley & Sons, Inc; 2009. pp. 1–23. [Google Scholar]
- 112.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies—Application of platform approaches. J Chromatogr B. 2007;848:28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 113.Chen Q. Viral Clearance Design Space for mAb Purification Process. CMC Strategy Forum. 2007. www.casss.org/cmc/PDFs/ChenPPSlidesJuly2007.pdf.
- 114.Low D, O'Leary R, Pujar NS. Future of antibody purification. J Chromatogr B. 2007;848:48–63. doi: 10.1016/j.jchromb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 115.Gerngross TU. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat Biotechnol. 2004;11:1409–1412. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 116.Hellwig S, Drossard J, Twyman RM, Fischer R. Plant cell cultures for the production of recombinant proteins. Nat Biotechnol. 2004;11:1415–1422. doi: 10.1038/nbt1027. [DOI] [PubMed] [Google Scholar]
- 117.Nikolov ZL, Woodard SL. Downstream processing of recombinant proteins from transgenic feedstock. Curr Opin Biotechnol. 2004;15:479–486. doi: 10.1016/j.copbio.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 118.Przybcien TM, Pujar NS, Steele LM. Alternative bioseparation operations: life beyond packed-bed chromatography. Curr Opin Biotechnol. 2004;15:469–478. doi: 10.1016/j.copbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 119.Thommes J, Etzel M. Alternatives to chromatographic separations. Biotechnol Progr. 2007;23:42–45. doi: 10.1021/bp0603661. [DOI] [PubMed] [Google Scholar]
- 120.Shan JG, Xia J, Guo YX, Zhang XQ. Flocculation of cell, cell debris and soluble protein with methacryloyloxyethyl trimethylammonium chloride-acrylonitrile copolymer. J Biotechnol. 1996;49:173–178. [Google Scholar]
- 121.Fahrner RL, Franklin J, McDonald P, Peram T, Sisodiya V, Victa C. US Patent Application 2008; 20080193981 Polyelectrolyte precipitation and purification proteins.
- 122.Ramanan S. Presentation to IBC Antibody Development and Production conference. 2007. [Google Scholar]
- 123.Sumner JB. The isolation and crystallization of the enzyme urease. J Biol Chem. 1926;69:435–441. [Google Scholar]
- 124.Flores H, Lin TP, Matthews TC, Pai R, Shahrokh Z. US patent application 2006; 20060009387 Apo-2 ligand/trail formulations.
- 125.Peters J, Minuth T, Schroder W. Implementation of a crystallization step into the purification process of a recombinant protein. Protein Expression Purif. 2005;39:43–53. doi: 10.1016/j.pep.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 126.Drohan WN, Miekka SI, Griko YV, Forng RY, Stafford RE, Hill CR, et al. Gamma irradiation of intravenous immunoglobulin. Dev Biol. 2004;118:133–138. [PubMed] [Google Scholar]
- 127.Hart H, Reid K, Hart W. Inactivation of viruses during ultraviolet light treatment of human intravenous immunoglobulin and albumin. Vox Sang. 1993;64:82–88. doi: 10.1111/j.1423-0410.1993.tb02523.x. [DOI] [PubMed] [Google Scholar]
- 128.Roberts P, Hope A. Virus inactivation by high intensity broad spectrum pulsed light. J Virol Methods. 2003;110:61–65. doi: 10.1016/s0166-0934(03)00098-3. [DOI] [PubMed] [Google Scholar]
- 129.Li Q, MacDonald S, Bienek C, Foster PR, MacLeod AJ. Design of a UV-C irradiation process for the inactivation of viruses in protein solutions. Biologicals. 2005;33:101–110. doi: 10.1016/j.biologicals.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 130.Tran H, Marlowe K, McKenney K, Petrosian G, Griko Y, Burgess W, et al. Functional integrity of intravenous immunoglobulin following irradiation with a virucidal dose of gamma radiation. Biologicals. 2004;32:94–104. doi: 10.1016/j.biologicals.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 131.Charm SE, Landau S, Williams B, Horowitz B, Prince AM, Pascual D. High temperature short-time heat inactivation of HIV and other viruses in human blood plasma. Vox Sang. 1992;62:12–20. doi: 10.1111/j.1423-0410.1992.tb01160.x. [DOI] [PubMed] [Google Scholar]
- 132.Charm SE, Landau S, Zarrineghbal H, Golden RF. US patent 1995; 5,389,335 High temperature, short time microwave heating system and method of heating heat-sensitive material.
- 133.Korneyeva M, Hotta J, Lebing W, Rosenthal RS, Franks L, Petteway SR. Enveloped virus inactivation by caprylate: a robust alternative to solvent-detergent treatment in plasma derived intermediates. Biologicals. 2002;30:153–156. doi: 10.1006/biol.2002.0334. [DOI] [PubMed] [Google Scholar]
- 134.Lebing W, Remington KM, Schreiner C, Paul HI. Properties of a new intravenous immunoglobulin (IGIV-C, 10%) produced by virus inactivation with caprylate and column chromatography. Vox Sang. 2003;84:193–201. doi: 10.1046/j.1423-0410.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 135.Kelley B. Very large scale monoclonal antibody purification: the case for conventional unit operations. Biotechnol Progr. 2007;23:995–1008. doi: 10.1021/bp070117s. [DOI] [PubMed] [Google Scholar]