Abstract
Secondary cell walls, consisting of cellulose, hemicelluloses and lignin, make up the bulk of wood biomass. It is therefore expected that dissection of the molecular mechanisms underlying secondary wall biosynthesis and its regulation will be instrumental to unravel the process of wood formation in tree species. Wood formation requires the coordinated activation of genes in the secondary wall biosynthetic program that is essential for the biosynthesis and assembly of wood components. It has recently been discovered that a group of poplar (Populus trichocarpa) wood-associated NAC domain transcription factors, PtrWNDs, which are functional orthologs of the Arabidopsis SND1, are capable of turning on the entire secondary wall biosynthetic program when expressed in Arabidopsis. In addition, two of the PtrWNDs were found to be able to activate the promoters of poplar wood biosynthetic genes and a number of other poplar wood-associated transcription factors. Further testing reveals that the promoters of these poplar wood-associated transcription factors are also activated by other PtrWNDs. It is therefore proposed that PtrWNDs are master transcriptional switches regulating a cascade of downstream transcription factors and thereby mediate the coordinated activation of wood biosynthetic genes during wood formation.
Key words: NAC domain transcription factor, Populus trichocarpa, secondary wall biosynthesis, transcriptional regulation, wood formation
Wood formation is a complex developmental process encompassing the differentiation of the vascular cambium into secondary xylem mother cells, cell elongation, secondary cell wall deposition, programmed cell death, and finally, heart wood formation (Fig. 1).1 At maturity, wood is mainly composed of the remains of secondary cell walls, which are a composite structure made of polymers of cellulose, hemicelluloses and lignin. Because the quality of wood is largely determined by the amount and the composition of secondary walls and the orientation of cellulose microfibrils in the secondary walls, there has been a strong motivation to unravel the molecular mechanisms underlying secondary wall formation in the hope of genetically modifying wood properties to better suit our needs.2 Because wood formation requires the highly coordinated operation of the biosynthetic pathways for all wood components, it is conceivable that the expression of their biosynthetic genes must be coordinately regulated by specific master transcriptional switches. Discovery of such switches will provide novel molecular tools to genetically improve wood quantity and quality. The recent report by Zhong et al.3 indicates that a group of wood-associated NAC domain transcription factors, PtrWNDs, are likely such master transcriptional switches involved in the coordinated activation of secondary wall biosynthetic genes during wood formation in poplar (Populus trichocarpa).
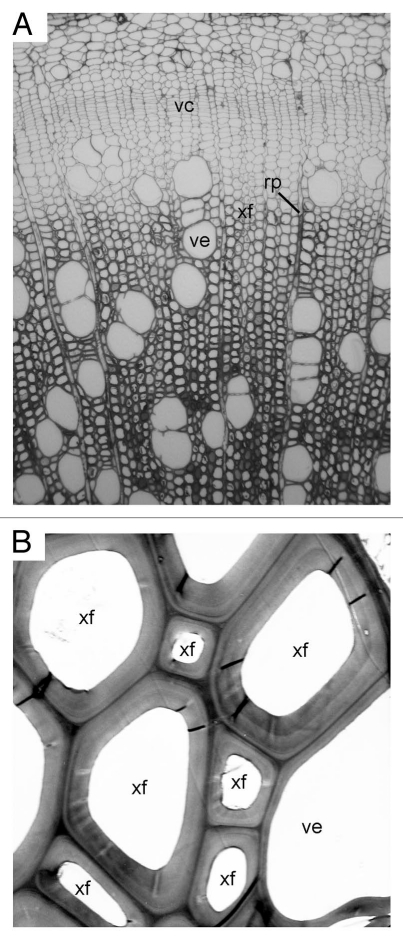
Figure 1.
Complexity of wood structure. (A) Cross section of a stem of poplar (Populus trichocarpa) showing the differentiation of vascular cambium (vc) into secondary xylem (wood) cells, including vessels (ve), xylary fibers (xf) and ray parenchyma cells (rp). Note the gradient in the degree of secondary wall thickness from developing to mature secondary xylem. (B) Transmission electron micrograph of secondary walls of the mature secondary xylem of poplar.
PtrWNDs are homologs of a group of closely related Arabidopsis (Arabidopsis thaliana) NAC domain transcription factors, including SND1, NSTs and VNDs, which have been shown to be master switches activating the biosynthetic pathway of cellulose, xylan and lignin during secondary wall formation.4–8 PtrWNDs are highly expressed in developing secondary xylem cells, including vessels, xylary fibers and ray parenchyma cells, which is consistent with their possible involvement in wood formation. When expressed in the Arabidopsis snd1 nst1 double mutant, PtrWNDs can effectively rescue the secondary wall defects caused by the loss of SND1 and NST1, indicating that PtrWNDs are functional orthologs of SND1 and NST1.
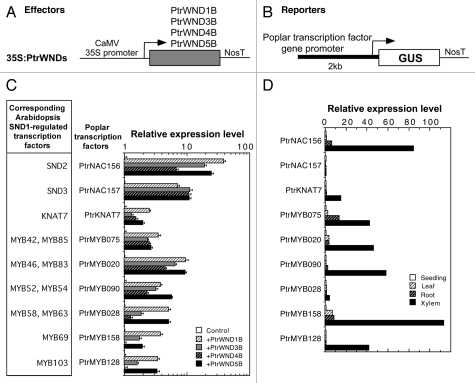
Overexpression of PtrWND2 and PtrWND6 in Arabidopsis leads to the activation of secondary wall biosynthetic genes and a concomitant ectopic deposition of secondary walls. Their overexpression in Arabidopsis also results in an upregulation in the expression of SND1-regulated downstream transcription factors.3,9 Furthermore, it was demonstrated that PtrWND2 and PtrWND6 were able to activate the promoters of poplar wood biosynthetic genes and a number of poplar wood-associated transcription factors. These results indicate that similar to the Arabidopsis SND1, the poplar PtrWNDs might mediate a transcriptional network regulating wood formation. This hypothesis is further supported by the fact that in addition to PtrWND2 and PtrWND6, other PtrWNDs are also capable of activating the promoters of the poplar wood-associated transcription factors that are close homologs of Arabidopsis SND1-regulated transcription factors (Fig. 2). Taken together, these findings suggest that PtrWNDs are master transcriptional switches regulating a cascade of other transcription factors, which leads to activation of wood biosynthetic genes and thereby wood formation.
Figure 2.
PtrWNDs are capable of activating the promoters of a number of poplar wood-associated transcription factors. (A) Diagram of the effector constructs consisting of the full-length cDNA of PtrWNDs driven by the CaMV 35S promoter. NosT, nopaline synthase terminator. (B) Diagram of the GUS reporter constructs driven by the 2-kb promoters of poplar transcription factor genes. (C) Transactivation analysis showing the PtrWNDs-activated expression of the GUS reporter gene driven by the promoters of poplar transcription factor genes. Poplar transcription factors are listed together with their corresponding Arabidopsis secondary wall-associated transcription factors regulated by SND1. The effector and reporter constructs were cotransfected into Arabidopsis protoplasts for the activation analysis.3 The GUS activity in protoplasts transfected with the reporter construct alone was used as a control and was set to 1. Error bars represent the SE of three biological replicates. (D) Wood-associated expression of the poplar transcription factors used in this study based on the PopGenExpress transcript abundance data.20 The expression level of each gene in the seedlings was taken as 1.
The finding that PtrWNDs regulate the expression of other wood-associated transcription factors opens a new avenue in dissecting the molecular mechanisms underlying the transcriptional regulation of wood formation. In will be imperative to identify all the players in the PtrWND-mediated transcriptional network, investigate their inter-relationships, and finally unravel how the combinatorial activities of these transcriptional factors activate wood biosynthetic genes. A number of wood-associated transcription factors have previously been identified in several tree species and some of them are implicated in the regulation of lignin biosynthesis.10–20 The identification of PtrWNDs as potential master transcriptional switches of wood formation will aid in the functional characterization of these wood-associated transcription factors by testing whether they are downstream targets of PtrWND orthologs in different tree species.
The study of PtrWNDs provides genetic evidence indicating that the molecular mechanisms underlying the transcriptional regulation of secondary wall biosynthesis is evolutionarily conserved between the herbaceous Arabidopsis and woody tree species. PtrWND homologs exist in a number of other tree species, such as eucalyptus, pine and spruce, and it is likely that they are also master transcriptional regulators of wood formation. Because wood is such an important raw material for a myriad of industrial uses and a potential source for biofuel production, it is imperative to unravel the transcriptional network regulating wood formation. The knowledge gained from such studies may help us design novel strategies to genetically modify wood properties in tree species.
Acknowledgements
This work was supported by the National Science Foundation (ISO-0744170) and the U.S. Department of Agriculture National Institute of Food and Agriculture AFRI Plant Biology Program (#2010-65116-20468).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11400
References
- 1.Plomion C, Leprovost G, Stokes A. Wood formation in trees. Plant Physiol. 2001;127:1513–1523. [PMC free article] [PubMed] [Google Scholar]
- 2.Mellerowicz EJ, Sundberg B. Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr Opin Plant Biol. 2008;11:293–300. doi: 10.1016/j.pbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhong R, Lee C, Ye Z-H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010;152:1044–1055. doi: 10.1104/pp.109.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong R, Demura T, Ye Z-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong R, Richardson EA, Ye Z-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225:1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- 9.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, et al. Characterization of a pine MYB that regulates lignification. Plant J. 2003;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 11.Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, et al. Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol Biol. 2003;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- 12.Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, et al. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol. 2004;56:255–270. doi: 10.1007/s11103-004-3354-5. [DOI] [PubMed] [Google Scholar]
- 13.Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16:2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 15.Prassinos C, Ko JH, Yang J, Han KH. Transcriptome profiling of vertical stem segments provides insights into the genetic regulation of secondary growth in hybrid aspen trees. Plant Cell Physiol. 2005;46:1213–1225. doi: 10.1093/pcp/pci130. [DOI] [PubMed] [Google Scholar]
- 16.Andersson-Gunneras S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, et al. Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 2006;45:144–165. doi: 10.1111/j.1365-313X.2005.02584.x. [DOI] [PubMed] [Google Scholar]
- 17.Bedon F, Grima-Pettenati J, Mackay J. Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca) BMC Plant Biol. 2007;7:17. doi: 10.1186/1471-2229-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legay S, Lacombe E, Goicoechea M, Briere C, Seguin A, Mackay J, Grima-Pettenati J. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 2007;173:542–549. [Google Scholar]
- 19.Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JE, et al. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot. 2008;59:3925–3939. doi: 10.1093/jxb/ern234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]