Abstract
Branched disulfide-containing poly(amido ethyleneimines) (SS-PAEIs) are biodegradable polymeric gene carrier analogs of the well-studied, non-degradable and often toxic branched polyethylenimines (bPEIs), but with distinct advantages for cellular transgene delivery. Clinical success of polycationic gene carriers is hampered by obscure design and formulation requirements. This present work reports synthetic and formulation properties for a graft copolymer of polyethylene glycol (PEG) and a branched SS-PAEI, poly(triethylentetramine/cystaminebisacrylamide) (p(TETA/CBA)). Several labs have previously demonstrated the advantages of PEG conjugation to gene carriers, but have also shown that PEG conjugation may perturb plasmid DNA (pDNA) condensation, thereby interfering with nanoparticle formation. With this foundation, our studies sought to mix various amounts of p(TETA/CBA) and p(TETA/CBA)-g-PEG2k to alter the relative amount of PEG in each formulation used for polyplex formation. The influence of different PEG/polycation amounts in the formulations on polymer/nucleic acid nanoparticle (polyplex) size, surface charge, morphology, serum stability and transgene delivery were studied. Polyplex formulations were prepared using p(TETA/CBA)-g-PEG2k, p(TETA/CBA), and mixtures of the two species at 10/90 and 50/50 volumetric mixture ratios (wt/wt %), respectively. As expected, increasing the amount of PEG in the formulation, adversely affects polyplex formation. However, optimal polymer mixtures could be identified using this facile approach to further clarify design and formulation requirements necessary to understand and optimize carrier stability and biological activity. This work demonstrates the feasibility to easily overcome typical problems observed when polycations are modified and thus avoids the need to synthesize multiple copolymers to identify optimal gene carrier candidates. This approach may be applied to other polycation-PEG preparations to alter polyplex characteristics for optimal stability and biological activity.
INTRODUCTION
Gene therapy is a feasible alternative to conventional therapies that simply manage symptoms of diseases and lack an effective treatment. However, gene therapy’s clinical success is impaired by uncertain design and formulation requirements for safe and efficient nucleic acid delivery to cells. Recent research advancements have improved carrier safety and efficacy through carrier chemical modification to alter surface charge and/or tissue specificity using polyethylene glycol (PEG) and/or cell-specific targeting ligands (1). Polymeric non-viral gene carriers have distinct advantages because if designed prudently they are non-immunogenic and are easily modified to exhibit multi-functional properties (2). Non-viral polycations are also relatively cost-effective, easy to produce industrially and can carry relatively large amounts of therapeutic nucleic acid (3,4).
Many structurally different polymers and copolymers consisting of linear, branched or dendritic architectures have been tested for their efficacy and suitability for in vitro and in vivo gene delivery. Polyethylenimine gene carriers (PEIs) have been most rigorously studied and are a standard for polycationic gene carriers because they easily condense DNA into nucleic acid/polycation nanoparticles (polyplexes) that protect nucleic acid from serum nuclease degradation as well as exhibit relatively high transgene delivery and expression in many cell types in vitro and in vivo. Unfortunately, PEIs often exhibit cellular toxicity due to intracellular accumulation of non-degradable polycations (3,5). Increased PEI molecular weight and branching, which influence polycation charge density, correlate with increased transgene expression but also with cellular toxicity. Conversely, low molecular weight PEIs show reduced cellular toxicity that correlate with reduced transgene expression (6,7). As predicted, the design of degradable polycationic gene carriers such as the reducible disulfide-containing poly(amidoamine) (SS-PAA) and poly(amido ethylenimines) (SS-PAEI), as well as hydrolyzable poly(β-amino ester) families have demonstrated comparable or improved cellular gene delivery and less cell toxicity when compared to PEIs (8,9,10). Reducible SS-PAEIs are synthetic analogs of the PEI family but have the aforementioned advantages of improved biological activity and compatibility (11). A recent abstract by Martello et al. showed that hyperbranched, SS-PAAs can condense plasmid DNA (pDNA) into polyplexes with sizes similar to bPEI25kDa, encouraging further functional studies (12).
Often cationic polyplexes interact with net negatively charged proteins found in serum, leading to particle aggregation and reduced efficacy in vitro and in vivo (13,14,15). In order to overcome this hurdle, poly(ethylene glycol) (PEG) conjugation to polycations has been employed and studies have shown that PEGylation often improves carrier function in the presence of serum. However, previous studies have also clearly shown that increasing targeting ligand and/or PEG conjugation to PEIs, especially to low molecular weight (LMW) PEI ~5kDA, adversely affects polyplex formation and carrier function (16, 17).
In order to better design and formulate hyperbranched SS-PAEIs and their corresponding graft PEG copolymers, several SS-PAEI polycationic gene carriers are synthesized and the influence of varying the relative PEG/polycation amounts on polyplex formation, size, surface charge, morphology, serum stability and ultimately biological activity are studied. Polyplex formulations were prepared using a known SS-PAEI grafted PEG, p(TETA/CBA)5k-g-PEG2k, or by mixing the two species at 10/90 and 50/50 volumetric ratios respectively. Altering the formulation mixture was employed to identify a suitable strategy to easily alter the gene carrier’s physiochemical properties and identify a suitable formulation with synthetic ease to enhance carrier efficacy.
EXPERIMENTAL PROCEDURES
Materials
Triethylenetetramine (TETA), tris(2-carboxyethyl)phosphine) (TCEP), N-Ethylemaleimide (NEM), hyperbranched polyethylenimine (bPEI, Mw 25 000 (bPEI25kDa)) and HPLC grade methanol were purchased from Sigma-Aldrich (St. Louis, MO). N’N’-Cystamine bisacrylamide (CBA) was purchased from Polysciences, Inc. (Warrington, PA). Ultrafiltration devices and regenerated cellulose membranes (1kDa and 5kDa) were supplied by Millipore Corporation (Billerico, MA). The reporter gene plasmid, pCMVLuc, was designed previously by insertion of luciferase cDNA into a pCI plasmid (Promega, Madison, WI) driven by the pCMV promoter and was purified using Maxiprep (Invitrogen, Carlsbad, CA) protocols. Dulbecco’s Modified Eagle’s Medium (DMEM), penicillin streptomycin, trypsin-like enzyme (TrypLE Express), and Dubelcco’s phosphate buffered saline were purchased from Gibco BRL (Carlsbad, CA). EBM-2 with EGM-2 singlequots was purchased from Lonza. Fetal bovine serum (FBS) was purchased from Hyclone Laboratories (Logan, UT).
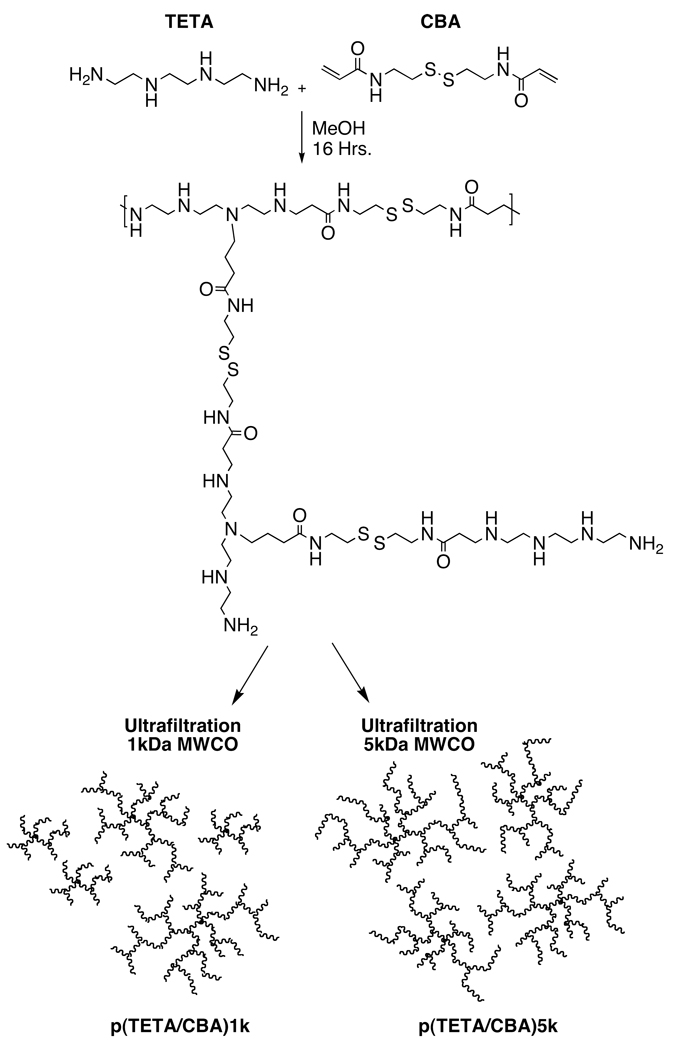
p(TETA/CBA) synthesis
Synthesis of p(TETA/CBA) was performed with minor adaptation of the previously described method at 50°C (11). The polymerization reaction was split in half after the pH was adjusted to 7.0 and purified by ultrafiltration using a 1kDa or 5kDa MWCO regenerated cellulose membrane. The polycations were subsequently lyophilized (Scheme 1). Composition of the polymer was monitored using 1H NMR (400 MHz, D2O). p(TETA/CBA) δ2.61 (COCH2CH2NH, 4H), 2.72 (NHCH2CH2S-S, 4H), 2.90–3.21 (COCH2CH2NHCH2CH2,16H), 3.41 (NHCH2CH2S-S, 4H).
Scheme 1.
Synthesis and Schematics for p(TETA/CBA) fractions.
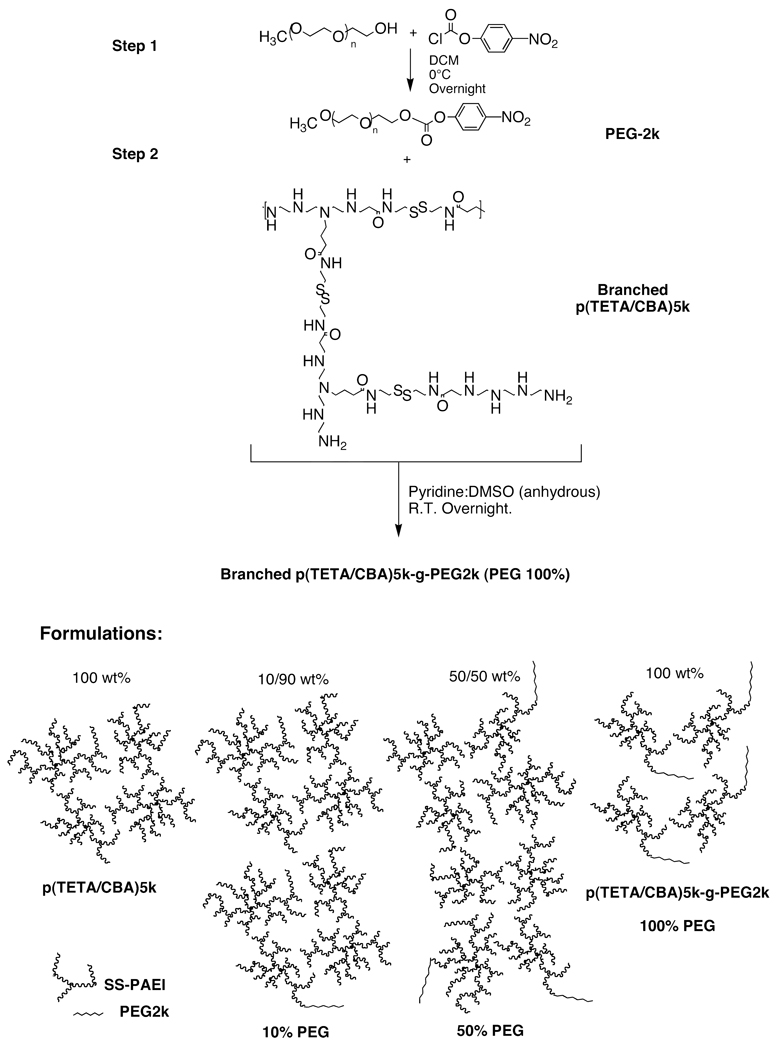
p(TETA/CBA)5k-g-2k synthesis
Methoxy PEG2k was dried using anhydrous toluene and subsequently precipitated in anhydrous ice-cold ether. The white precipitate was collected and dried in vacuo. The mPEG2k was then activated using p-nitrophenyl chloroformate in DCM as solvent and reacted on ice overnight while stirred. The activated PEG product was collected by precipitation in anhydrous ice-cold ether and dried in vacuo. Following NMR analysis to assess the degree of PEG activation, p(TETA/CBA)5k and equal molar active PEG2k were dissolved in anhydrous pyridine/DMSO as solvent and the poly(ethylene glycol)-carbonate solution was added drop wise to the dissolved p(TETA/CBA)5k. The reaction was stirred at room temperature and the release of p-nitrophenolate was monitored optically at 400 nm using UV-Vis. When the reaction was complete at approximately 16 hrs. the sample was purified by ultrafiltration (5kDa MWCO) before being lyophilized. The composition of p(TETA/CBA)-g-PEG2k copolymer conjugates was monitored using 1H NMR (400 MHz, D20). p(TETA/CBA)5k-g-PEG2k δ 2.61 (COCH2CH2NH, 4H), 2.72 (NHCH2CH2S-S, 4H), 2.90–3.21 (COCH2CH2NHCH2CH2,16H), 3.41 (NHCH2CH2S-S, 4H), 3.45–3.7 (CH2CH20, 4H).
Polymer Characteristics
Relative molecular weight analysis for p(TETA/CBA)1k, p(TETA/CBA)5k and p(TETA/CBA)5k-PEG2k was performed using AKTA/FPLC (Amersham Pharmacia Biotech Inc.). A SuperdexPeptide column HR 10/30 was used to analyze p(TETA/CBA)1k (2mg/mL). The calibration curve was prepared using poly(hydroxy propyl methacrylamide) (poly(HMPA) standards ranging from 2 kDa to 10 kDa. p(TETA/CBA)5k and p(TETA/CBA)5k-g-PEG2k were analyzed using a Superose 6 10/300 GL column and poly(HMPA) standards ranging from 40 kDa to 150 kDa. For calibration and analysis, a 0.3 M NaOAc, pH 4.4 with 30% (v/v) acetonitrile eluent buffer was used after it was filtered through a 0.2 µm filter (Nylon, Alltech) and degassed. The flow rate was set to 0.4 mL/min.
Polyplex Formation
In all cases polyplex was formed with a known amount of pDNA and a corresponding and desired amount of polymer. The two solutions were combined, lightly vortexed and allowed to equilibrate for 30 min. When the 10 and 50% polyplex mixtures were formed, a known amount of pDNA was brought up in solution and a corresponding and desired concentration of p(TETA/CBA) and p(TETA/CBA)5k-g-PEG2k were prepared. The polymer solutions were mixed prior to polyplex formation at either 10% p(TETA/CBA)5k-g-PEG2k/ 90% p(TETA/CBA) by volume or 50% p(TETA/CBA)5k-g-PEG2k/ 50% p(TETA/CBA). The mixtures were then added to the pDNA solution and prepared as mentioned above.
Polycation Branching
Relative degree of branching was determined as previously described by the reduction and protection of disulfide bonds using Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and N-ethylmaleimide (NEM), respectively (5). MALDI-TOF analysis was performed on the NEM conjugated polymer repeat units using a Voyager-DE STR Biospectrometry Workstation (PerSeptive Biosystems) in positive-ion mode with delayed extraction. Spectra were externally calibrated using a peptide standard mixture spanning a nominal mass range from 325 to 2465 Da.
Acid-Base Titrations
The buffering capacity of each polycation was determined using a previously established method (11). In brief, 6 mg polymer was dissolved in 30mL NaCl solution (0.1 M) and initially titrated to pH 10 with 0.1M NaOH. The pH was subsequently lowered with the addition of 0.1 M HCl. Because the absolute molecular weight is not known for these polymers, titration values were determined in µmols of HCl required to lower the pH of the polycation solution from 7.4-5.1. Branched PEI25kDa was used as a reference control.
Light Scattering and ζ-Potential Measurements
The surface charge and polymer/pDNA particle (polyplex) diameters were measured at 25°C using a Zetasizer 2000 instrument (DTS5001 cell) and a dynamic light scattering (DLS) unit on a Malvern 4700 system, respectively. Polyplexes were prepared by adding equal volume polymer solution (200µl) at increasing concentrations in HEPES buffer (20 mM, pH 7.4, 5% glucose) to a desired concentration of 8 µg pDNA in HEPES buffer (200µl). Polyplexes were allowed to equilibrate for 30 min. and were subsequently diluted in filtered miliQ water to a final 2 mL volume.
Transmission Electron Microscopy (TEM)
Polyplex was prepared in HEPES buffer (20 mM, pH 7.4, 5% glucose) at 0.05 µg/µl and 5 µl was deposited on TEM copper grid plates to dry. Residual buffer salt was removed by carefully rinsing each grid with filtered deionized water thrice. The samples were then stained with filtered phosphotungstenic acid (PTA) for 1 min. before washing again with filtered deionized water. Images were visualized using a Technai T12 scope (EFM) at 80 kV. Magnification ranging from 20,000 to 200,000x was utilized and the micrograph images were taken at 110,000x. Particle sizes were analyzed using ImageJ software.
Polyplex Stability in 90 % Fresh Rabbit Serum
Polyplex stability and resulting pDNA stability against nuclease activity in serum was evaluated using 500ng free pDNA as a control and 500ng pDNA complexed with polymer mixtures pre-formed in HEPES buffer. Polyplex formation was carried out by combining equal volume solutions of p(DNA) and polymer mixtures using a polymer/pDNA at N/P 50 (24 w/w) and allowed to equilibrate for 30 min. Pre-formed polyplex was then diluted in 90% fresh rabbit serum and incubated at 37°C over time. 25 µl aliquots (125 ng pDNA) were taken at each time point and 10 µl stop buffer (250 mM NaCl, 25 mM EDTA, 2% SDS) was added to each. The samples were frozen at −70°C until further analysis. Once the samples were thawed, they were incubated overnight at 60°C to completely dissociate polycation from the pDNA and 2 µl of 50 mM DTT was added to each sample and incubated at 37°C for an additional 30 min to ensure complete decomplexation. Lastly, the samples were loaded onto a 2% agarose gel stained with ethidium bromide (EtBr) and subjected to electrophoresis at 96 V for 30 min in TAE (40 mM Tris-acetate, 1 mM EDTA) buffer. The gel image was viewed using GelDoc software (n=2).
Cell Culture
Mouse pancreatic islet endothelial cells (SVR) and colon adenocarcinoma cells (CT-26) (ATCC) were cultured in DMEM containing 10% FBS and 1% penicillin-streptomycin at 37°C in a humidified incubator with an atmosphere containing 5% (v/v) CO2. Human Umbilical Vein Endothelial Cells (HUVEC) (Invitrogen) were cultured in EBM-2 media with EGM-2 singlequots at 37°C in a humidified incubator with an atmosphere containing 5% (v/v) CO2.
In Vitro Transgene Expression
Luciferase reporter gene expression in cell culture was performed using each polymer and pCMVLuc plasmid DNA. Cells were plated in 24-well plates containing 0.5mL media. Once the cells were approximately 70% confluent, polyplexes were prepared using 0.5µg pDNA at N/P 50 (24 w/w) in HEPES buffer. Polyplexes were allowed to equilibrate for 30 min. and the cells were transfected in the presence of serum by adding 20 µl polyplex (0.5 µg pDNA) to each well for 4 hrs before replacing with fresh culture media. The cells remained in the incubator for a total of 48 hrs. before they were washed with 1ml PBS and treated with cell culture lysis buffer (Promega). Luciferase quantification was performed using a Luciferase assay system (Promega) on a luminometer from Dynex Technologies, Inc. (Chantilly, VA). The amount of protein in the cell lysate was determined using a standard curve of bovine serum albumin (Sigma) and a BCA protein assay kit (Pierce) (n=6).
Cell Viability Assay
Cells were plated in 24-well plates and gene transfections were carried out when the cells were approximately 70% confluent. Polyplexes were prepared as they were for the luciferase reporter gene assay. Respective cell cultures were transfected in the presence of serum with the addition of 20 µl equilibrated polyplex in HEPES buffer solution (0.5 µg pDNA) to each well. Cells were left to incubate for a total of 18 hrs. before analyzing cell viability using an MTT assay (Sigma). Percent cell viability was determined relative to untreated cells (n=6).
RESULTS
Synthesis and Characterization
p(TETA/CBA). Polymerization of p(TETA/CBA) occurs via Michael-type addition of the amines present in the TETA monomer to the acrylamide group of the CBA (monomer). Because four reactive amine groups exist within the TETA monomer, highly branched products can be obtained during synthesis and differentially purified (Scheme 1). In the present study, polymerization reactions were carried out at different temperatures in 100% MeOH and monitored by 1H NMR. Synthesis temperature was shown to correlate with the degree of branching in each sample (data not shown). Eliminating oligomer polycations from the sample by using a 5kDa MWCO ultrafiltration membrane reduced the sample polydispersity index (PDI), as expected, which further correlates with the relative molecular weight of the sample as determined using AKTA/FPLC. Commercial bPEI25k was also analyzed as an external control for comparison. Because the Mn and Mw values for bPEI25kDa determined using AKTA/FPLC were lower than reported by the commercial provider, extrapolations need to be made to estimate p(TETA/CBA) molecular weight. Moreover, p(TETA/CBA)5k has a similar buffering capacity to the sample obtained using the original purification approach (1kDa MWCO) (Table 1).
Table 1.
Polymer and co-polymer characteristics
| Sample | Mn (kDa)a | Mw (kDa)a | PDI (Mw/Mn) | Titrationb (µmol HCl) |
Degreec Branching |
|---|---|---|---|---|---|
| p(TETA/CBA)1k | 4.2 | 8.2 | 1.95 | 25.2 | 0.68 |
| p(TETA/CBA)5k | 5.8 | 8.85 | 1.53 | 27.6 | 0.91 |
| p(TETA/CBA)5kg-PEG2k | 8.9 | 10.6 | 1.19 | 22.3 | -- |
| bPEI 25 kDa | 16.4 | 21.0 | 1.28 | 32 | -- |
Number average molecular weight (Mn), weight average molecular weight (Mw), and polydispersity (Mw/Mn) determined using FPLC.
Polymer fraction buffer capacity titrations determined by the mol of HCl required to shift pH from 7.4 to 5.1 in 0.1M aqueous NaCl.
Degree branching was determined by reduction of disulfide bonds with TCEP followed by protection of free sulfhydryls with NEM and analyzed by MALDI-TOF.
Mixture Formulations to Alter the Relative Amount of PEG for Polyplex Formation
PEGylation may often improve polycationic carrier function in the presence of serum both in vitro and in vivo, which is largely due to its ability to reduce polyplex surface charge and maintain a steric barrier against protein adsorption (23). Particles with a surface charge close to neutral tend to aggregate in solution due to their reduced ionic repulsive forces influenced by the isoelectric point, molecular weight and pH of the solution (24). Therefore, we sought to synthesize a p(TETA/CBA)5k-PEGylated product that could be mixed in conjunction with p(TETA/CBA)5k, if necessary, to easily control the amount of PEG used in the formulations in order to examine the effects on particle characteristics and functionality with a non-toxic branched polycation as a model system (Scheme 2). PEG conjugation to the polycation was monitored by following the release of p-nitrophenolate at 400 nm with a UV-Vis spectrophotometer. Reactions were complete by 16 hrs. NMR analysis and comparison of peak AUC suggested approximately 0.96/1 mol PEG:(TETA/CBA)5k and is in good agreement with the AKTA/FPLC analysis (Table1).
Scheme 2.
p(TETA/CBA)5k-g-PEG2k Synthesis and illustrations of evaluated formulations.
Influence of p(TETA/CBA) PDI and Molecular Weight on Biological Activity
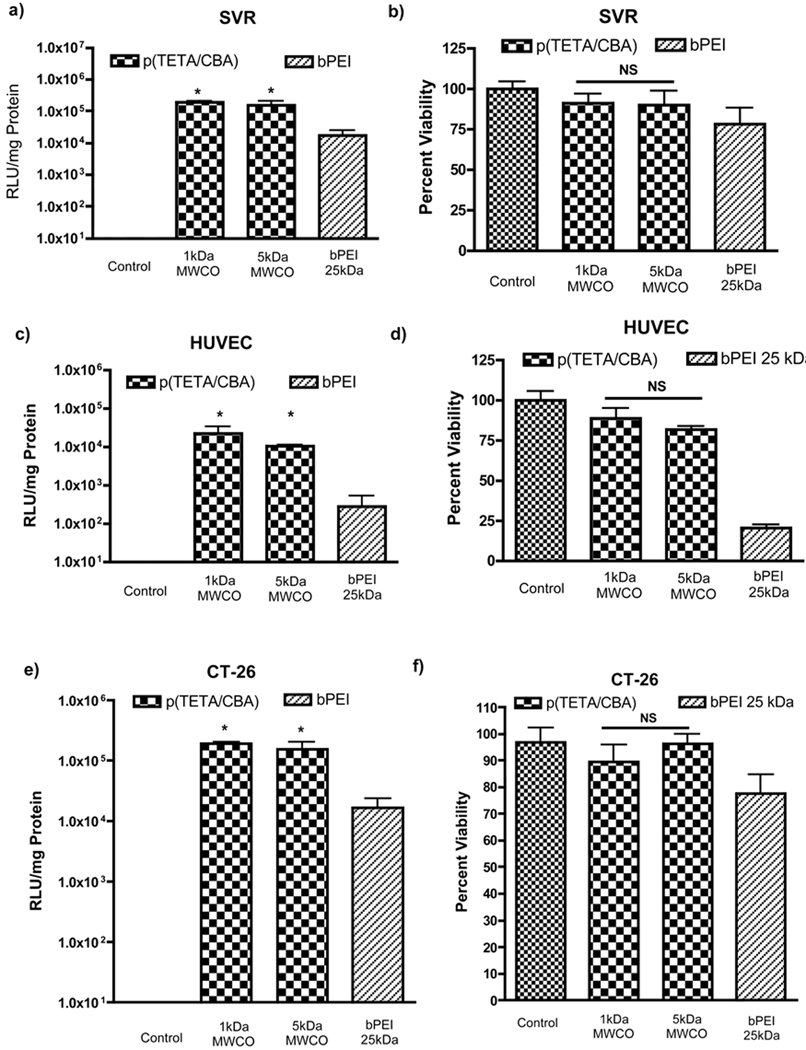
As mentioned previously, LMW PEI exhibits limited pDNA condensation at low N/P ratios and is often perturbed by PEG conjugation. Thus, reducing the PDI of p(TETA/CBA) by eliminating destabilizing oligomers and increasing the average molecular weight without perturbing carrier performance is preferred (17). As seen in Figure 1, a reduced p(TETA/CBA) PDI and correlating molecular weight increase has no adverse effects on carrier performance. It performs similarly to the original polycation, p(TETA/CBA)1k. More importantly, p(TETA/CBA)5k is significantly less toxic in primary HUVEC endothelial cells than bPEI 25kDA, as well as providing greater luciferase transgene expression in HUVEC and SVR endothelial cells, as well as the colon adenocarcinoma cell line, CT-26. The toxicity of bPEI25kDa is likely due to the intracellular accumulation of high molecular weight polycationic species (3). Three cell lines were used because trends seen in multiple cell lines confirms that any trends seen in gene transfection are justifiable and secondly, the endothelial cells (SVR and HUVEC) as well as adenocarcinoma cells (CT-26) comprise the tumor micro-environment in many malignant tumors that are best treated with systemic administration of therapeutic gene. The PEGylation of polycations is most often employed for systemic delivery of gene and drug carriers. Therefore, we tested the biological activity of our gene carriers using both endothelial cells often exposed during systemic administration to examine potential activity, and the adenocarcinoma cell line (CT-26) that is used in our lab for a mouse tumor model when testing therapeutic gene activity in vivo. Often, polycationic species can interact with and disrupt cell membrane function, and/or they can interact with intracellular proteins and nucleic acids, thereby perturbing intracellular and nuclear processes such as cellular trafficking and gene transcription and translation (18, 19). The bioreducible polycation, p(TETA/CBA), likely mitigates these intracellular interactions as it degrades to small molecular weight species intracellularly thereby limiting toxicity in the primary endothelial cell line, irrespective of its relative molecular weight (20). The enhanced transgene expression observed using p(TETA/CBA) is also likely explained by this phenomenon and its ability to more easily release pDNA (6, 9).
Figure 1.
Transfection efficiency (a,c and e) and cell viability (b, d and f) in SVR, HUVEC and CT-26 cells, in the presence of 10% serum for different p(TETA/CBA) molecular weight analogs compared to the positive control bPEI 25kDa. pCMVLuc was used as the reporter gene. Commercial bPEI 25 kDA and p(TETA/CBA) polyplexes were prepared at N/P 10 (1.3 w/w) and N/P 50 (24 w/w), respectively (*p < 0.05, n=6).
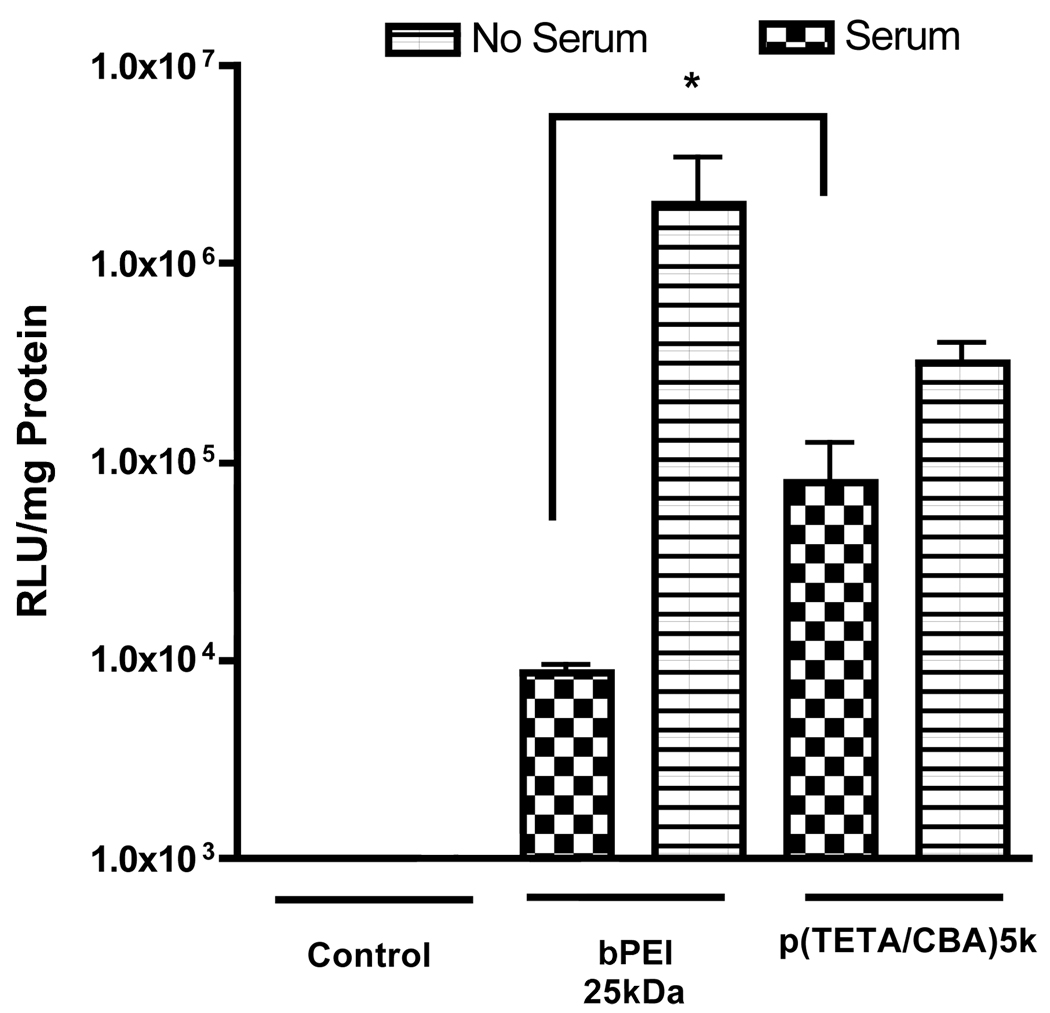
Serum effects on p(TETA/CBA)
Serum-containing media and serum encountered when polyplex is administered in vivo often reduces polycationic performance through particle destabilization and nuclease degradation of therapeutic gene or uptake by the reticular endothelial system in vivo. The data presented in figure 2 is consistent with previously published findings where transgene expression is reduced in the presence of serum compared to non-serum conditions. In addition, the biological activity of p(TETA/CBA) on colon adenocarcinoma cells (CT-26) in serum-containing media is significantly better than that with bPEI25kDa (Figure 2). Evaluation of serum effects on gene transfection was performed solely on CT-26 cells because the data presented in figure 1 showed similar trends as in the other two cell lines. Combined, these results demonstrate the advantages of p(TETA/CBA)5k over bPEI25kDa and suggest a need to PEGylate p(TETA/CBA)5k for improved gene delivery in the presence of serum.
Figure 2.
Comparison of p(TETA/CBA)5k/pCMVLuc transfection efficiency in the presence and absence of 10% serum in culture media. Transfection efficiency was evaluated by luciferase trangene expression in CT-26 cells. p(TETA/CBA) at N/P 50 (w/w 24) exhibits greater reporter transgene expression than bPEI 25kDa N/P 10 (w/w 1.3) in serum-containing media but is still perturbed compared to its performance in the absence of serum (*p < 0.05, n=6).
Polyplex Characterization
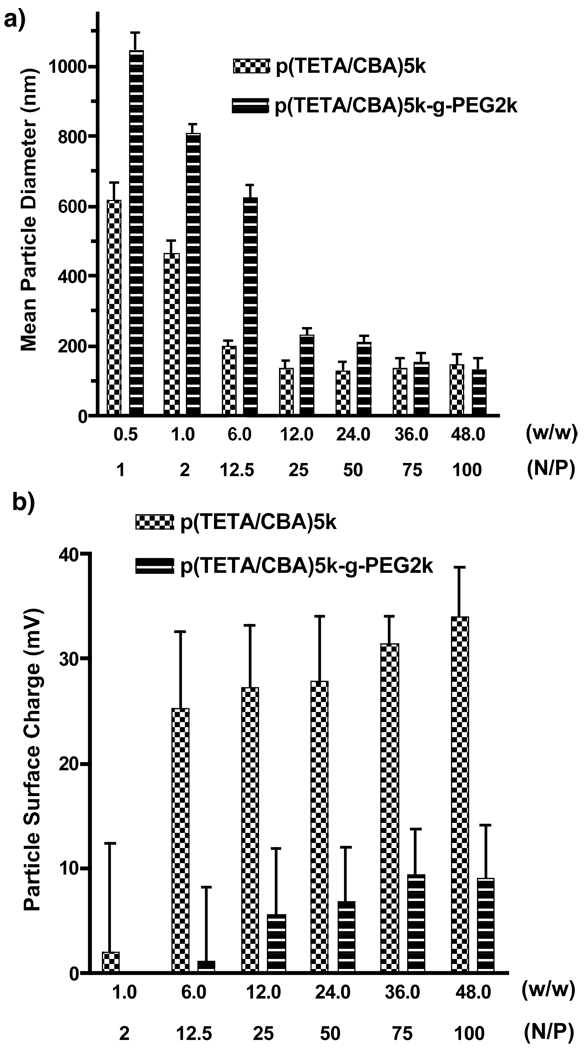
The ability of p(TETA/CBA)5k and p(TETA/CBA)5k-g-PEG2k to form condensed polyplexes was investigated by particle size analysis and ζ-potential measurements. Indeed, nanosized particles below or near 100 nm in diameter were formed for both potential gene carriers. As expected, PEG conjugation interfered with polyplex formation at preferred, low polymer concentrations (Figure 3a). PEG conjugation sufficiently decreased polyplex surface charge. However, higher polymer concentrations are required in this case in order to condense p(DNA) into 200nm polyplexes (Figure 3b).
Figure 3.
Physiochemical characteristics of p(TETA/CBA)5k and p(TETA/CBA)5k-g-PEG2k/pCMVLuc polyplexes. a) Particle size and b) ζ-potential measurements of each respective gene carrier at increasing polymer concentrations using a known amount of pDNA. PEG2k conjugation to p(TETA/CBA) reduces polyplex surface charge, but interferes with pDNA condensation.
PEG Effects on Polyplex Characteristics
Our findings coincide with previous studies that used PEGylated polyethyleneimine gene carriers (17) and show that PEGylation of the p(TETA/CBA) polycation perturbs nucleic acid condensation and polyplex size. To overcome this problem and validate the possibility of premixing polymer/PEG-copolymer solutions in order to control the relative PEG/polycation amounts, as well as identify an ideal formulation that maintains homogenous and stable polyplex with reduced surface charge, polyplexes were prepared using p(TETA/CBA)5k-g-PEG2k, p(TETA/CBA)5k and mixtures of the two molecular entities at 10/90 and 50/50 volumetric ratios, respectively, at a summed polycation/pDNA N/P = 50 (24 w/w). Mixtures are schematically represented in Scheme 2.
Serum Stability
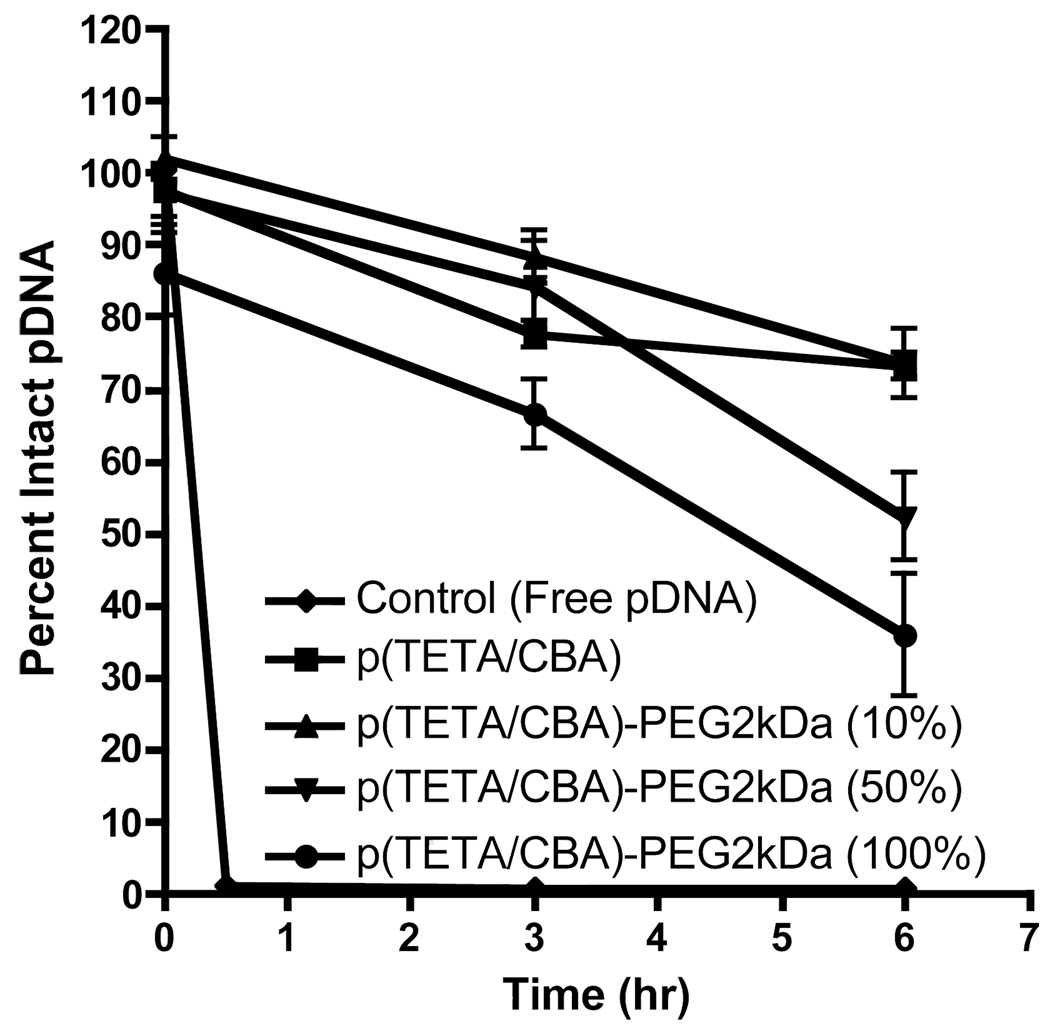
In order to test the influence of increasing PEG content on polyplex stability and pDNA protection against serum nuclease activity, polyplexes were formed and following a 30 min. equilibration time, were added to fresh rabbit serum to a final serum concentration equal to 90 % at 37°C. Aliquots were electrophoresed on an agarose gel to visualize intact pCMVLuc at each time point and further compared to untreated control at zero hours. Figure 4 shows that p(TETA/CBA)5k and 10 % p(TETA/CBA)5k-g-PEG2k protect pDNA from nuclease degradation nearly 80 % or more at 6 hrs. Increasing p(TETA/CBA)5k-g-PEG2k to 50 or 100% reduces particle stability and offers less pDNA protection where approximately 60 % and 40 % pDNA is preserved, respectively, at 6 hrs. incubation time.
Figure 4.
Polyplex stability in 90% rabbit serum at 37°C for p(TETA/CBA)5k, poly(TETA/CBA)5kg-PEG2k, 10/90 (10% PEG) and 50/50 (50% PEG) wt/wt % formulations for p(TETA/CBA)5k-g-PEG2k and p(TETA/CBA)5k, respectively. 500ng pCMVLuc was complexed with each formulation N/P 50 (w/w 24). The relative percent of intact pDNA over time was determined by band pixel intensity using GelDoc software. Increasing the PEG wt% in the polyplex formulations inversely correlates with pDNA protection (n=2).
Polyplex Analysis
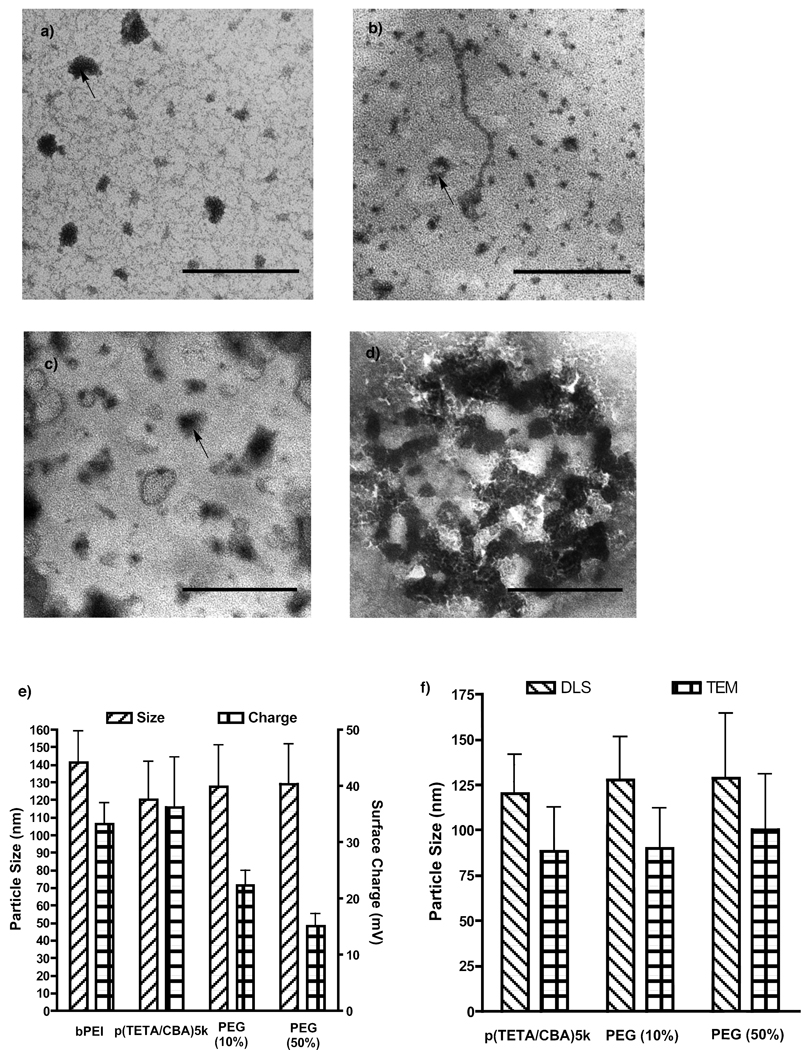
For formulation ease and improved carrier function, stable polyplex was formed using different amounts of PEG. Formulations should display unimodal polyplex size and surface charge with uniform morphology. Polyplex size for each formulation was visualized using TEM (Figure 5a–d) and their size and distribution were compared to measurements provided by dynamic light scattering (DLS). The arrows contained in Figure 5a–d show a relevant particle and the particle size was analyzed using ImageJ software. The values between TEM and DLS are in close agreement with each other. The TEM particle sizes are smaller than those determined using DLS because the polyplexes examined using TEM are in a dehydrated state, whereas polyplex analyzed by DLS are in aqueous buffer and are thus hydrated lending to a larger particle size. Figures 5a–d reveal morphological changes and less compact polyplex with translucent outer shells as the amount of PEG in the formulation increases. These translucent outer shells are thought to be from increasing the amount of PEG. p(TETA/CBA)5k-g-PEG2k exhibited aggregation as seen in (Figure 5d). This aggregation was also noted when analyzed using DLS and adversely influenced the data. Therefore this formulation is excluded from the analysis and not shown in Figure 5e. p(TETA/CBA)5k,10 and 50% PEG formulations generate sub-150nm polyplexes in solution and the amount of PEG inversely correlates with polyplex surface charge as expected (Figure 5e).
Figure 5.
Polyplex formulations visualized with TEM (a–d) and studied using DLS (e) at 24 w/w. a) p(TETA/CBA)5k b) 10% PEG c) 50% PEG d) p(TETA/CBA)5k-g-PEG2k. Scale bars equal 200nm. e) Particle size and ζ-potential of polyplex formulations. F) Comparison of polyplex size using TEM and DLS.
PEG formulations on carrier function and in vitro biological activity
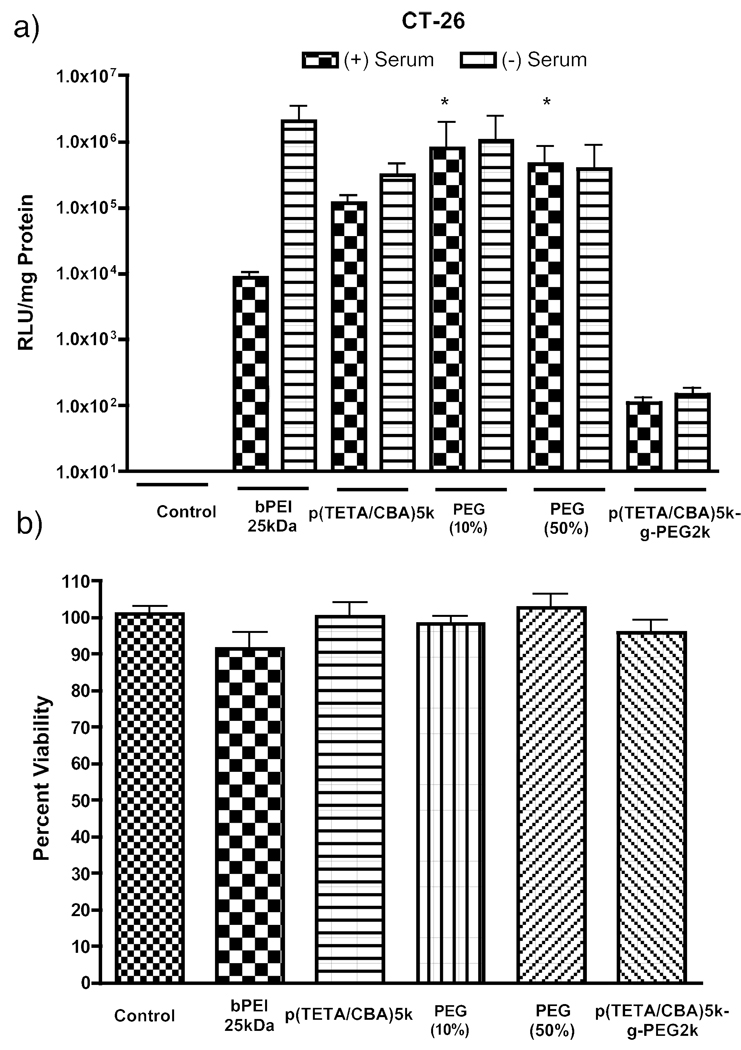
To investigate the potential advantages of PEG-copolymer formulations for gene delivery in the presence of serum, a luciferase transgene assay was performed using colon adenocarcinoma cells (CT-26). The 10 and 50% PEG formulations have significantly better (p < 0.05; n = 6) transgene expression than bPEI 25kDa and p(TETA/CBA) in the presence of serum (Figure 6a). However, these PEG formulations are not significantly different from each other. Moreover, these formulated polyplexes are non-toxic to these cells (Figure 6b).
Figure 6.
a) Transfection efficiency of p(TETA/CBA)5k,10/90, 50/50 and 0/100% p(TETA/CBA)5k/p(TETA/CBA)5k-g-PEG2kpolyplex formulations with CMVLuc in the presence and absence of serum. The 10 and 50% PEG formulations are significantly better gene carriers than bPEI 25kDa and p(TETA/CBA) in the presence of serum. However these two formulations are not significantly different from each other. b) Cell viability with analogous polyplex formulations in serum-containing media (p < 0.05, n=6).
DISSCUSION
Clinical advancements of polycationic gene carriers is hampered by uncertain design and formulation requirements. In the present work, we show that a graft copolymer of polyethylene glycol (PEG) and a branched SS-PAEI can be synthesized and used in formulation mixtures to alter the relative amount of PEG, thereby altering the physiochemical characteristics of the gene carrier in order to easily study the design and formulation requirements to improve gene carrier function. Knowing that PEG conjugation can interfere with polyplex formation and carrier function, this work demonstrates the feasibility to easily overcome these issues by preparation of homogenous polyplexes that alter the relative amount of PEG using a mixture of un-PEGylated p(TETA/CBA) and the PEGylated counterpart, p(TETA/CBA)5k-g-PEG2k, to evaluate and identify products that are functionally viable. More importantly, this approach may be applied to other polycation-PEG preparations to easily alter polyplex characteristics to optimize polyplex stability and biological activity in vitro. Further studies need to be performed to understand the feasibility of using this approach for in vivo screening and evaluation of gene carriers.
When synthesizing p(TETA/CBA)5k-g-PEG2k, reducing the PDI of p(TETA/CBA) by removing polycationic oligomers with a limited ability to properly condense pDNA is preferred given that PEG conjugation may interfere with polyplex formation as it is. Therefore, in order to reduce the PDI of p(TETA/CBA) following the Michael-addition of the bisacrylamide group with TETA, ultrafiltration was performed using a higher molecular weight cut-off membrane (5 kDa) than was used previously (11). As expected, this approach was effective in reducing the polycation’s PDI and correlates with a relative increase in molecular weight. Prior research has shown that increasing the molecular weight and branching profile of polyethyleneimines correlates with increased transgene expression, but also cellular toxicity. Therefore, the present study investigated this putative effect with regards to p(TETA/CBA) and found no significant influence on its biological activity in primary and immortalized cell lines (6,7). These results are explained by the gene carrier’s ability to exploit the intracellular redox potential and avoid disruption of intracellular function through relatively high molecular weight polycationic species (21).
While p(TETA/CBA) demonstrated significantly better transgene expression than bPEI 25kDa in serum-containing media, p(TETA/CBA) delivery capacity was noticeably lower when compared to its activity in the absence of serum. Therefore, to reduce p(TETA/CBA)/pDNA polyplex interactions with serum proteins and thus improve carrier function in the presence of serum, polyethylene glycol was conjugated to p(TETA/CBA)5k at an equimolar ratio and confirmed by 1H NMR following purification. The corresponding relative molecular weight was also in agreement with what is expected for equimolar conjugation when analyzed using AKTA/FPLC. Conjugating PEG2k to p(TETA/CBA)5k reduced polyplex surface charge, however, adversely affected nucleic acid condensation. This result coincides with prior findings by our lab and others (16, 22). Because polyethylene glycol and/or ligand conjugation for cell-specific gene delivery commonly mitigates nucleic acid condensation, synthesis of multiple co-polymeric gene carriers is required to ascertain optimal ratios for maximal carrier performance. In an attempt to overcome this problem and avoid the need to synthesize multiple carriers for screening, this study investigated the feasibility of altering and optimizing PEG/polycation amounts by formulating mixtures of a polycation and its corresponding PEGylated counterpart. Polyplex stability in serum was evaluated in this study for p(TETA/CBA)5k-g-PEG2k alone, p(TETA/CBA)5k alone, and 10/90 or 50/50 volumetric mixtures of p(TETA/CBA)5k-g-PEG2k/p(TETA/CBA)5k, respectively. Polyplex formed using p(TETA/CBA) and 10/90%, sufficiently protects up to 70% of the pDNA from serum nuclease degradation over 6 hr. Increasing the p(TETA/CBA)5k-g-PEG2k amount to 50 and 100% reduced the relative pDNA protection in serum, which correlates with the capability of each formulation to condense pDNA into nano-sized polyplex determined using DLS and TEM. One potential concern is that if polyplex is formed at a unified 50 N/P (24 w/w) the N/P ratios could be significantly different between p(TETA/CBA) alone and the mixtures. However, NMR results demonstrate that there is approximately 1 PEG chain per TETA/CBA molecule. If a 50 N/P (24 w/w) is used for formulations, there are approximately 1.2% of the amines PEGylated in the 100% p(TETA/CBA)(5k)-g-PEG(k) formulation, ~0.6% for the 50 % formulation, ~0.12% for the 10% formulation and 0% for p(TETA/CBA)(5k). This being the case, the difference in N/P ratios between the formulations is insignificant. Moreover, the 10% formulation that demonstrated significantly better transgene expression than p(TETA/CBA)(5k) in the presence of serum has only 0.12% less amines for use in formulating polyplex.
Luciferase transgene expression and cell viability was investigated in cell culture using the aforementioned formulations to evaluate their bioactivity. As expected, PEG was able to improve gene delivery in serum-containing media compared to p(TETA/CBA) alone. However, this improvement was observed only at specific PEG ratios. These results provide evidence that PEG/polycation ratios can be easily altered in order to easily evaluate and optimize PEG ratios for improved gene carrier function. In doing so, one can avoid synthesis of multiple bio-reducible co-polymers with different physiochemical characteristics currently employed for gene carrier optimization.
CONLUSIONS
In the present study, we developed a novel gene carrier comprised of an efficient and non-toxic bioreducible polycation in conjunction with its PEGylated counterpart to improve carrier performance in the presence of serum. In addition we provide a feasible and facile approach to tailor polycationic-PEG copolymer formulations to alter the relative amount of PEG in several formulations in order to obtain optimal physiochemical properties for ideal gene carrier function. By doing so, synthesis of multiple candidate copolymers for gene delivery is avoided when designing a gene carrier with preferred physiochemical properties, which may also be employed for facile in vivo evaluation.
ACKNOWLEDGEMENTS
This work was supported by NIH grant CA107070. We thank Jindrich Kopecek’s group for their assistance with AKTA/FPLC analysis (Department of Pharmaceutics, University of Utah), as well as the University of Utah Core Facilities for assistance with Maldi-TOF analysis and TEM imaging.
REFERENCES
- 1.Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv. Drug Delivery Rev. 2003;55:199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 2.Christiano RJ, Roth JA. Molecular conjugates: a targeted gene delivery vector for molecular medicine. J. Mol. Med. 1995;73:479. doi: 10.1007/BF00198899. [DOI] [PubMed] [Google Scholar]
- 3.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Delivery Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Lee M, Kim SW. Polyethylene glycol-conjugated copolymers for plasmid DNA delivery. Pharm. News. 2004;9:597. doi: 10.1007/s11095-004-9003-5. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee P, Reichardt W, Weissleder R, Bogdanov A. Novel Hyperbranched Dendron for Gene Transfer in Vitro and in Vivo. Bioconjugate Chem. 2004;15:960–968. doi: 10.1021/bc0342128. [DOI] [PubMed] [Google Scholar]
- 7.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 8.Luten J, Van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. Journal of Controlled Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 9.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjugate Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 10.Anderson DG, Lynn DM, Langer R. Semiautomated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew. Chem., Int. Ed. Engl. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 11.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, Engbersen JF, Feijen J. Reducible poly(amido ethylenimine) designed for triggered intracellular gene delivery. Bioconjugate Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 12.Martello F, Engbersen JFJ, Ferruti P. Abstracts/Journal of Controlled Release. 2008;132:e1–e18. [Google Scholar]
- 13.Plank C, Mechtler K, Szoka FC, Jr, Wagner E. Activation of the complement system by synthetic DNA complexes: potential barrier for intravenous gene delivery. Hum. Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 14.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel ML. Influenza virus hemagglutinin HA-2 N-terminal fusogenic peptides augment gene transfer by transferrin-polylysine-DNA complexes: toward a synthetic virus-like gene-transfer vehicle. Proc. Natl. Acad. Sci. USA. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbaan FJ, Oussoren C, Van Dam IM, Takakura Y, Hashida MD, Crommelin JA, Hennink WE, Storm G. The fate of poly(2-dimethyl amino ethyl)methacrylate-based polypolylysine as a synthetic vector for gene transfer into plexes after intravenous administration. Int. J. Pharm. 2001;214:99–101. doi: 10.1016/s0378-5173(00)00642-6. [DOI] [PubMed] [Google Scholar]
- 16.Suh W, Han S, Yu L, Kim SW. An Angiogenic, Endothelial-Cell-Targeted Polymeric Gene Carrier. Molecular Therapy. 2002;6:664–672. [PubMed] [Google Scholar]
- 17.Merkel OM, Germershaus O, Wada CK, Tarcha PJ, Merdan T, Kissel T. Integrin αvβ3 Targeted Gene Delivery Using RGD Peptidomimetic Conjugates with Copolymers of PEGylated Poly(ethylene imine) Bioconjugate Chemistry. 2009;20:1270–1280. doi: 10.1021/bc9001695. [DOI] [PubMed] [Google Scholar]
- 18.Burckhardt BC, Thelen P. Effect of primary, secondary and tertiary amines on membrane potential and intracellular pH in Xenopus laevis oocytes. Pflugers Arch. 1995;429:306–312. doi: 10.1007/BF00374144. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Wang Y, Yan H. Binding of Sodium Dodecyl Sulfate with Linear and Branched Polyethyleneimines in Aqueous Solution at Different pH Values. Langmuir. 2006;22:1526–1533. doi: 10.1021/la051988j. [DOI] [PubMed] [Google Scholar]
- 20.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 21.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: Synthesis and in vitro gene transfer properties. Journal of Controlled Release. 2006;116:130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Kunath K, Merdan T, Hegener O, Häberlein H, Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. The Journal of Gene Medicine. 2003;5:588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 23.Pasche S, Voros J, Griesser HJ, Spencer ND, Textor M. Effects of Ionic Strength and Surface Charge on Protein Adsorption at PEGylated Surfaces. J. Phys. Chem. B. 2005;109:17545–17552. doi: 10.1021/jp050431+. [DOI] [PubMed] [Google Scholar]
- 24.Ga Q, Wang T, Cochrane C, McCarron P. Modulation of Surface Charge, Particle Size and Morphological Properties. Colloids and Surfaces B: Biointerfaces. 2005;44:65–73. doi: 10.1016/j.colsurfb.2005.06.001. [DOI] [PubMed] [Google Scholar]