Abstract
The metalloregulatory protein NikR from Helicobacter pylori (HpNikR) is a master regulator of gene expression which both activates and represses specific genes in response to nickel availability. Here, we report the first crystal structure (at 2.37Å resolution) of Ni(II)HpNikR prepared directly from the holo-protein. The protein contains four nickel ions located in two distinct coordination environments. Two nickel ions are bound to sites in a four-coordinate square planar geometry as predicted based on the structures of NikR from E. coli and P. horikoshii. The remaining two nickel ions are bound to sites with unexpected 5 or 6 coordination geometries which were previously thought to be involved in nickel incorporation into the protein. The nickel with 5/6 coordination geometry utilizes three histidines from two separate monomeric HpNikR units along with two or three water molecules as ligands. The spatial location of the nickel in the 5/6 coordinate site is within approximately 5 Å of the expected site if a 4- coordinate square planar geometry occurred. Two of the histidines that participate as ligands in 5/6 coordinate site would also participate as ligands if the 4-coordinate site was occupied making it impossible for both sites to be occupied simultaneously. DFT calculations show that the 5/6 coordinate geometries are energetically favorable when the local protein environment is included in the calculations. The presence of two distinct coordination environments in HpNikR is suggested to be related to the specificity and binding affinity of this transcription factor for DNA.
Introduction
Helicobacter pylori (H. pylori or Hp) is a virulent bacterium that colonizes the highly acidic, mucosal layer (or gastric epithelium) of the human stomach, causing gastritis and in some cases gastric cancer.1–4 H. pylori infection constitutes a significant public health threat with more than half of the world’s population infected. There is an urgent need for the development of novel anti-Helicobacter agents.5 H. pylori are among a select number of bacteria that are able to survive in the highly acidic environment of the gastric epithelium. The pH of the gastric epithelium ranges from 4 to 6.5 with periodic acid shocks of pH < 2.6
One feature that enables H. pylori to survive under acidic conditions is that it releases large quantities of ammonia to neutralize its immediate environment.6,7 Much of this ammonia is produced by urease, which is a nickel dependent enzyme that converts urea to ammonia and bicarbonate 6,8. H. pylori contains high levels of urease (~10% of total protein content) and the metalloregulatory protein NikR regulates its expression.6,9,10 HpNikR activates the transcription of urease in response to nickel by binding to a specific region of the promoter of ureA that encodes for urease. 11–14 In addition to regulating urease production, HpNikR regulates the transcription of a large number of genes either directly or indirectly.15 The genes for which direct regulation have been established contain variable recognition sequences;11–14,16 and the physical basis for DNA recognition and discrimination by HpNikR is currently unresolved.
HpNikR has homologs in both Gram-negative and Gram-positive bacteria.17 The best studied homolog of NikR is from E. coli, EcNikR,18,19 which has low sequence similarity with HpNikR (21% identity and 54% homology). Unlike HpNikR, which regulates multiple genes as either an activator or a repressor,20 EcNikR regulates a single gene as a repressor.21 Several crystal structures of EcNikR have been reported. 22–24 These structures revealed that EcNikR folds as a symmetrical tetramer with a central tetrameric metal binding domain (MBD) flanked by two dimeric DNA binding domains (DBD). Nickel binds to the metal binding domains via three ligands from one monomer (His2Cys) and one ligand from another monomer (His) in a 4-coordinate square planar geometry.22,23,25 The four nickel ions each occupy the analogous site on each monomer. Structures of NikR from Pyrococcus horikoshii have also been reported, and again the protein is a symmetrical tetramer with four nickel ions per tetramer in a square planar geometry.26 The histidine and cysteine ligands that bind nickel in E. coli NikR and P. horikoshii NikR are conserved in H. pylori NikR, thus it was presumed that HpNikR would also bind four nickel ions at four identical square planar sites.
There are no reported structures of holo-Ni(II)HpNikR, although there are two crystal structures of nickel bound to HpNikR that were generated by soaking excess nickel (20–100 mM) into crystals of apo-HpNikR. 27 In one of these structures, named Ni1-HpNikR (PDB code 2CAD), three nickel sites are observed with six nickel atoms per HpNikR tetramer. One of the sites is the predicted square planar site called final (“F”), the second is a novel octahedral nickel site that is thought to be an intermediate nickel binding site (“I”) and the third is another novel octahedral site, referred to as an external binding site (“X”), made up of ligands from the protein and citrate from the crystallization conditions. In the second structure, Ni2-HpNikR (pdb code 2CAJ) nickel is bound at a site between the “I” and the “X’ sites of Ni1-HpNikR with just two histidine ligands. Because these two structures were generated by soaking apo-HpNikR with excess nickel, the unexpected nickel sites were proposed to represent ‘snapshots’ of nickel incorporation serving as sites for nickel to bind enroute to its final holo-Ni(II)HpNikR state. The true holo-Ni(II)HpNikR structure was predicted to house a total of four nickel ions in a square planar coordination geometry at identical sites.
In the present manuscript we report the 2.37 Å resolution crystal structure of holo-Ni(II)HpNikR. While initial crystallization efforts targeted the holo-Ni(II)HpNikR-DNA complex structure, we obtained a crystal structure of holo-Ni(II)HpNikR in the absence of DNA. While unexpected, this structure allowed us to test the hypothesis that holo-Ni(II)HpNikR, like its homologs in E. coli and P. horikoshii, contains four square planar nickel sites. Surprisingly, the structure does not contain nickel bound to four identical square planar nickel sites as predicted. Instead, the protein has only two of the nickel ions bound to the predicted square planar coordination sites. The additional two nickel ions, while bound in the vicinity of the two remaining predicted sites, assume a 5 and a 6-coordinate geometry. These nickels are in locations analogous to site “I” in the Ni1HpNikR structure and are coordinated to three histidine ligands and two water molecules forming a square pyramidal geometry in one case and to three histidine ligands and three water molecules forming an octahedral geometry in the other case, respectively. When nickel is bound to these sites in either the 5 or 6-coordinate geometry, it is in a location that is approximately 5 Å removed from where the nickel is predicted to be located if bound in a square planar geometry. To verify the geometry and investigate the energetics of the nickel sites, we have performed DFT calculations. Based on this combination of experimental and computational methods it is shown that two of the four nickel ions found in holo- Ni(II)HpNikR bind in either 5 or 6 coordination geometries, in locations adjacent to but distinct from the predicted square planar site; in the remainder of this manuscript these two sites when discussed collectively will be referred to as the 5/6 site. The implications of the presence of two of the bound nickels in 5 or 6 coordinate geometries in addition to the two nickels in the 4-coordinate geometry on Ni(II)HpNikR’s propensity to recognize multiple, different DNA sequences are discussed.
Materials and Methods
Holo-Ni(II)HpNikR overexpression and purification
A pET15b construct containing HpNikR was transformed into E. coli BL21(DE3) cells and grown in LB medium containing 100 μg/ml ampicillin.13 Purification of Holo-Ni(II)HpNikR was carried out using a nickel-loaded 5 mL Hitrap-HP (Amersham) affinity column followed by further purification using a Hitrap Q-Sepharose (Amersham) column. Holo-Ni(II)HpNikR was exchanged into final buffer (20mM Hepes, 100mM NaCl pH 8.0) and concentrated to 10 mg/mL for initial crystal screening. HpNikR as isolated was tested for metal content with Inductively Coupled Plasma Mass Spectroscopy (ICP-MS) and found to contain 4 nickel ions/tetramer. Oligonucleotides with the sequence, 5′-AATATAACACTAATTCATTTTAAATAATAATTAG-3′ were purchased from Integrated DNA Technologies (IDT) in the desalted form and used in crystallization trials. The oligonucleotides were purified by reverse phase HPLC (Waters) using a Symmetry 300 C18 column and applying a linear gradient. Purified DNA was lyophilized, resuspended in 20mM Hepes, 100mM NaCl pH 7.5 and annealed at 95°C.
Fluorescence Anisotropy to verify DNA Binding
To confirm that holo-Ni(II)HpNikR bound DNA, a fluorescence anisotropy titration of the protein with PureA (which corresponds to the ureA promoter) was performed. A 10 nM solution of fluorescently labeled DNA (PureA-F – 5′-CTTCAAGATATAACACTAATT[F] CATTTTAAATAATAATTAGTTAATGAA-3′) in 20 mM HEPES, 100 mM NaCl, and 20 mM glycine (pH 7.5) was added to a 0.5 cm PL Spectrosil far-UV quartz window fluorescence cuvette (Starna Cells) pretreated with 5mM BSA to prevent adherence of either the protein or DNA to the cuvette walls. The anisotropy, r, of the free DNA oligonucleotide was measured. Holo-Ni(II)HpNikR (4 μM) was titrated and the change in anisotropy measured. Data was fit to a 1:1 binding equilibrium as previously reported.13
Initial Crystal Screening
Crystal screens were performed using 96-well sitting drop crystallization trays (Art Robbins Instruments, Sunnyvale, California, USA) with the OryxNano crystallization robot (Douglas Instruments Ltd, Berkshire, UK) at the X-ray Crystallography Core Facility at the University of Maryland, Baltimore. Crystallization drops were set as 50% protein/DNA (protein:DNA 1:1.2 equivalents) to an initial drop volume of 1.4 μl. A crystal was obtained in condition No. 5 of the NeXtal Nucleix Suite (0.05 M MES, 0.2M potassium chloride, 0.01 M magnesium sulfate and 5% PEG 8000, pH 5.6) after 4 days of incubation at 21.5°C. This crystal was cryoprotected in mother liquor supplemented with 12% glycerol and flash cooled in liquid nitrogen prior to data collection.
Data collection, processing and model building
X-ray diffraction data were collected remotely at the Stanford Synchrotron Radiation Lightsource (SSRL, beamline 12-2). Data processing was done with DENZO/SCALEPACK28. The structure was solved by molecular replacement using the nickel binding and DNA binding domains as separate models 27 (PDB ID 2CA9). PHASER 29 was used for molecular replacement in CCP4.30,31 Surprisingly, no DNA was found in the asymmetric unit, which contains four metal binding domains and only two DNA binding domains. Iterative model building was performed using Coot for manual model building 32 with REFMAC for restrained refinement.33,34 Tight NCS restraints were applied between identical monomers in the nickel binding domain core tetramer. Final rounds of refinement included NCS with TLS/restrained refinement.35–37 Anomalous scattering was performed on a holo-Ni(II)HpNikR crystal in order to confirm the presence of nickel. Distance restraints were applied for nickel coordinating side chains and waters. Anisotropic temperature factors were refined for nickel ions with the rest of the model treated isotropically. Figures were generated using PyMOL (www.pymol.org).
During the refinement of the structure, it was observed that DNA was absent in the crystal. In addition, it appeared that the cleavage of the protein may have occurred. This was confirmed by N-terminal sequencing of a protein/DNA solution that had been used to grow crystals, which revealed that the protein had been cleaved between residues Trp54 and Ala55 (Trp54 is at the interface of the DNA binding domain and the metal binding domain).
pH Dependent Ni(II) Binding Titrations
Apo-HpNikR was prepared as previously described. 13 Metal binding titrations were performed in screw-capped quartz cuvettes using a Perking Elmer Lambda 25 UV-vis spectrometer. Titrations were carried out in 20 mM potassium phosphate (and 20 mM glycine at pH 5.6) using metal free reagents and water that had been purified using a MilliQ purification system and passed over Sigma Chelex resin. Upon their prepration, buffers were purged with helium to degas and transferred into a Coy inert atmospheric chamber. Ni(II)sulfate hexahydrate (99.99% Ni) from Puratrem (Strem Chemicals) was used as the nickel source. The binding interaction between apo-HpNikR and Ni(II) was measured at four pH conditions: pH 5.6, 6.5, 7.5 and 8.5 (typically with 45 μM apo-HpNikR.) Titrations were performed in triplicate under anaerobic conditions.
Density Functional Theory Calculations
Model complexes representative of the coordination modes of the Ni atoms in HpNikR were used to study the electronic and geometric structures of the experimentally determined 5 and 6 coordinate metal sites as well as the 4-coordinate sites. Large clusters obtained from the crystallographic coordinates included all coordinating residues, neighboring residues with a structural role in defining the coordination geometries, for example Gln87, and all residues connecting them as well as the two or three water molecules directly coordinated to the metal when present. Residues that do not play a direct structural role at the Ni centers were modeled as glycines. Anchor atoms were selected and kept frozen during the minimization of the geometries of the large clusters to mimic the structural impact of the protein on the residues adjacent to the nickel ions. These atoms were either the carbonyl C or N of the protein backbone or the terminal –COO− and –NH3+ groups, respectively. Arrows in Figure S5 indicate the anchor atoms. The use of large clusters allows inclusion of the effects of the protein medium in the calculations in a more rigorous way. This comes with the penalty of additional computational cost since the size of largest model is 193 atoms. Smaller complexes of the 5 and 6 coordinated Ni(II) centers consisting of Ni and just the coordinating ligands: two or three waters and three imidazole rings (to represent histidine) were also optimized to study the electronic structures.
NWChem Version 5.1.1, as developed and distributed by Pacific Northwest National Laboratory, P. O. Box 999, Richland, Washington 99352 USA, and funded by the U. S. Department of Energy, was used to obtain the computational results.38,39 All calculations were based on Density Functional theory (DFT) 40–42 and employed the B3LYP (Becke 3-parameter, Lee, Yang and Parr, exchange-correlation) hybrid functional 43–45 with the LANL2TZ basis set which includes effective core potentials for Ni 46 and the 6-31G* and 3-21G basis sets, respectively, for the lighter atoms (H, C, N, O, S). 47–50 In the smaller model complexes, all lighter atoms were described by the 6-31G* basis set. In the large clusters only atoms coordinating Ni or participating in hydrogen bonds with coordinating ligands were described by the 6-31G* basis, the remaining being described by the 3-21G basis. All structures were fully geometry optimized at the B3LYP level of theory until convergence to a root-mean-square value of the energy gradient and nuclear step of 4.5×10−4 hartree/bohr and 1.8×10−3 bohr, respectively. Contour plots were made with gOpenMol 3.00 (gOpenMol can be downloaded from http://www.csc.fi/english/pages/g0penMol) and optimized geometries were plotted using a version of Ortep-3 for Microsoft Windows.51 VMD was used to plot structures of the large models superimposed on the protein.52
Results and Discussion
Overall Structure
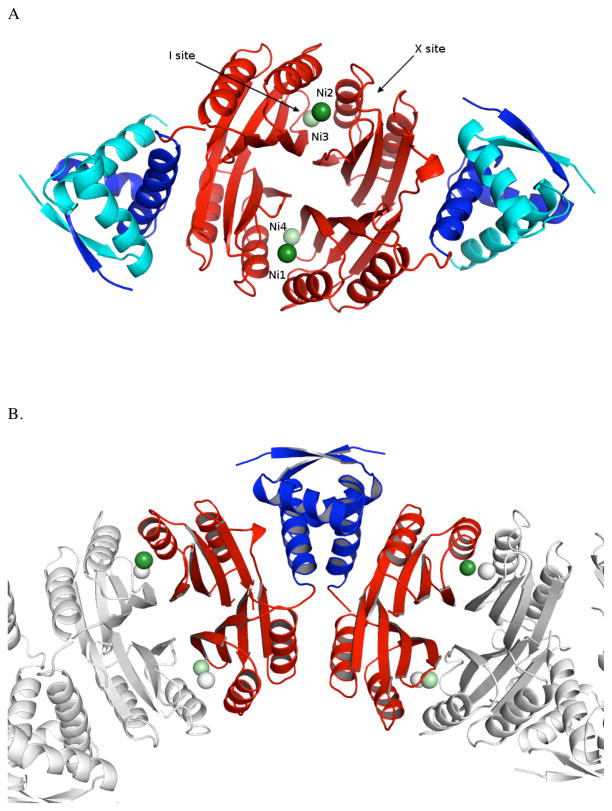
Crystal trays were set up with holo-Ni(II)HpNikR and PureA DNA in a 1:1.2 ratio. The crystals that formed were found to contain just the protein (Figure 1A). A similar result was reported by O’Halloran and Mondragon when they sought to obtain crystals of the zinc metalloregulators CueR and ZntR in complex with their DNA partners.53 The structure of holo-Ni(II)HpNikR was determined at 2.37Å resolution, and data processing and refinement statistics are shown in Table 1. The asymmetric unit (Figure 1B) contained four NikR monomers, two of which were missing the N-terminal fragment that forms the DNA binding domains (DBD). The metal binding domain (MBD) is of ferrodoxin like (βαββαβ) fold and the DBD may be described as a ribbon helix helix fold. 22,26,27,54
Figure 1.
(A) Ribbon diagram of holo-Ni(II)HpNikR tetramer with the metal binding domains (MBD) colored red and the DNA binding domains (DBD) colored blue (dark blue DBD from one asymmetric unit and teal blue froma second asymmetric unti). The 4-coordinate, square planar nickel atoms are colored dark green and labeled Ni1 and Ni2. The 5/6-coordinate nickel atoms colored light green and labeled Ni3 for the 6-coordinate site and Ni4 for the 5-coordinate site. The locations of the sites named “I’ and “X” are indicated by arrows. (B) Asymmetric unit of holo-Ni(II)HpNikR. Only the symmetry mates completing the MBDs are shown. (C) Structure of holo-HpNikR showing domain swapping phenomenon at the DBD. All figures were prepared using PyMol.
Table 1.
Data Collection and Model Refinement Statistics
| Parameter | HpNikR PDB code 3LGH |
|---|---|
| Data collection | |
| Wavelength | 0.97946 |
| Space group | P212121 |
| Unit-cell a(Ǻ), b(Ǻ), c(Ǻ) | 89.5, 105.1, 48.2 |
| Resolution range (Å) | 50.0–2.37 |
| Redundancy | 4.3 (1.8) |
| No. of unique reflections | 16660 (1,316) |
| Rmerge/Rpim (%)61–62 | 18.6/9.4 |
| Completeness (%) | 90.5 (73.6) |
| Mean [I/σI] | 7.0 (1.0) |
| Matthews coefficient | 2.22 |
| Refinement statistics | |
| Resolution range | 39.3–2.37 |
| No. of reflections | 16595 |
| Rcryst/Rwork (%)61,62 | 27.1/26.8 |
| Rfree (No. of reflections) | 31.8 (857) |
| No. of protein atoms | 3472 |
| No. of water molecules | 62 |
| Average Bf (Ǻ) | 42.8 |
| Ramachandran statistics | |
| Favored (%) | 95.7 |
| Outliers (%) | 0.9 |
| RMSD from Ideality | |
| Bond lengths (Ǻ) | 0.011 |
| Bond angles (deg.) | 1.326 |
Domain Swapping
The unit cell contained density for a tetrameric metal binding subunit but only two of the four DNA binding domains, which was unexpected. The two missing DNA binding domains had been cleaved during crystallization. A complete tetrameric structure is observed due to domain swapping of the DNA binding domains between adjacent unit cells. The tetrameric structure is seen by taking two MBDs from one asymmetric unit and two MBDs from the adjacent unit cell with two symmetrical faces of the DBDs that make identical contacts with respective MBDs. This strings together unit cells throughout the crystal yielding the pseudo tetrameric structures (Figure 1C). The domain swapping does not appear to affect the structure of the metal binding domains as we observe low RMSDs (< 1.3 Å for Cα atoms) with respect to the MBDs of previously reported apoHpNikR structures. In addition, we have recently solved the X-ray structure of just the metal binding domain of holo-Ni(II)HpNikR (data not shown) to 3.1 Å resolution. From this structure, we are able to determine that the metal binding domains and the nickel sites are identical to those in the crystal structure of holo-Ni(II)HpNikR that we report here.
Ni(II) Binding Sites
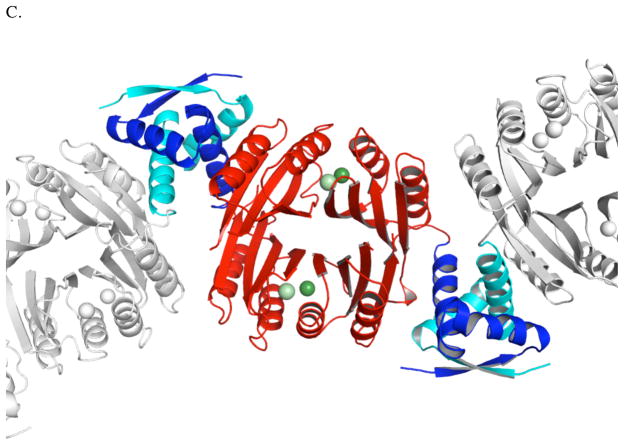
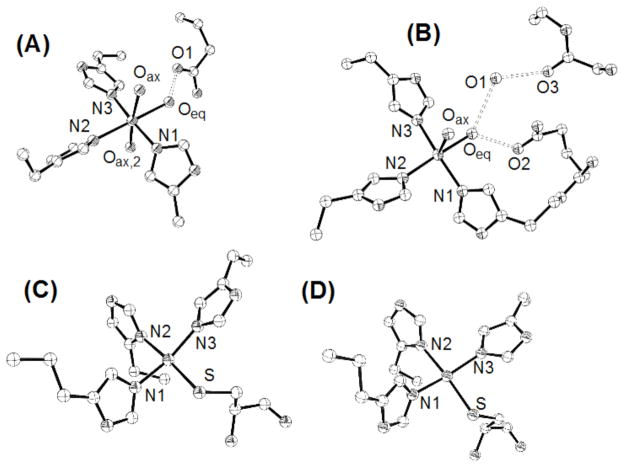
In the presence of four ligands, Ni(II) preferentially adopts a square planar coordination geometry while most other divalent metal ions adopt a tetrahedral geometry.55 The presence of four sets of ligands available to bind nickel in a square planar geometry in the apoHpNikR structure and the observation that nickel is bound to all four of these sets of ligands in a square planar geometry in the structures of holo-EcNikR and holo-PhNikR, led us to predict that holo-Ni(II)HpNikR would also house four nickel ions in four square planar sites. Surprisingly, we found that holo-Ni(II)HpNikR is a heterotetramer with two unique classes of nickel sites. Nickels Ni1 and Ni2 are found in square planar sites (Figure 2A), consistent with that seen in the Ec and Ph structures. Each nickel ion (Ni1 and Ni2) is ligated to His99, His101 and Cys107 from one MBD and His88 from a second MBD, as expected.
Figure 2.
(A) Representation of the square planar nickel site in holo-Ni(II)HpNikR (B) Representation of the 5-coordinate nickel site in holo-Ni(II)HpNikR. The electron density around the water molecules is shown; (C) Representation of the 6-coordinate nickel site in holo-Ni(II)HpNikR. The electron density around the water molecules is shown.
The remaining two nickel ions (Ni3 and Ni4) present in the holo-Ni(II)HpNikR structure are coordinated to three histidine residues (His 74 and His 101 from one MBD and His 88 from the second MBD) as well as 2 or 3 water molecules making a pentacoordinate, square pyramidal geometry in one case (Ni3) and an hexacoordinate, octahedral geometry (Ni4) in the other case (Figure 2B and 2C, respectively). This second site is analogous the site that was named “I” for intermediate in the nickel soaked structure, Ni1-HpNikr. 27 The second site has two ligands, His 101 and His 88, in common with the anticipated square planar geometry making it prohibitive for two nickel ions to simultaneously participate in the two coordination geometries at this second class of sites (Figure S1). Inspection of the apo-HpNikR structure indicates that the ligands that coordinate nickel at the 5/6 coordinate sites are pre-organized, which may facilitate binding in this geometry.
Hydrogen Bonding Interactions
One of the water molecules at each of the 5/6 coordinate sites is stabilized by a hydrogen-bonding interaction with Gln87 (Figure 2B and 2C), (also seen in the “I” site of the nickel soaked apo-HpNikR protein, Ni1-HpNikr)27 which DFT calculations (described below) posit as important for stabilizing the nickel-aquo bond. These interactions appear to lead to two of the Ni(II) binding sites assuming the unexpected 5 or 6 coordination geometries. Ligation in either of these geometries locates the nickel approximately 5 Å from its position if ligated in the anticipated square planar geometry. While this result was unanticipated the presence of different metal sites in families of metallotranscription factors is not without precedent. For example, Glasfeld and co-workers recently reported a novel metal site in ScaR, a member of the DtxR/MntR family of metalloregulators. 56
Hydrogen bonding between the occupied 5/6 coordinate sites and the occupied 4-coordinate sites in H. pylori NikR is limited. This contrasts E. coli NikR where an extensive hydrogen bonding network linking two individual square planar sites was mapped out by Drennan and co-workers. 22 The E. coli NikR hydrogen bonding network is composed of a Glu97, Gln75 and Tyr58 (Glu109, Gln87 and Val70 in HpNikR). Replacement of the tyrosine involved in EcNikR hydrogen bonding by valine in HpNikR disrupts the hydrogen bonding between the two sites.
Relationship between coordination chemistry and method used to generate crystals
The two previously reported structures of Ni(II)HpNikR were generated by soaking apo-Ni(II)HpNikR crystals with excess nickel sulfate or nickel chloride (20–100 millimolar).27 In these structures, nickel is found bound to multiple different sites including the 5/6 coordinate site, in addition to the square planar sites and neither of the structures has four 4-coordinate sites.27 Because the structures were generated by soaking nickel with apo-HpNikR that was already crystallized, it was thought that the structures did not represent the ground state associated with the ‘holo’ form of NikR. However, as we report here, holo-Ni(II)HpNikR does not contain four 4-coordinate sites but rather two 4-coordinate sites and two 5/6 coordinate sites, with the latter two sites lying approximately 5 Å from what would correspond to the 4-coordinate ligation geometry. Because this structure was obtained directly from holo-Ni(II)HpNikR, it is more likely representative of the conformation of the holo form of the protein. Thus, we propose that holo-Ni(II)HpNikR in the absence of DNA contains two distinct and stable classes of high affinity sites – the expected square planar sites and the unanticipated 5/6 coordinate sites.
Comparison to Isothermal Titration Calorimetry Data
There is solution-based evidence to support the presence of two different high-affinity nickel binding sites in Ni(II)HpNikR. Ciurli, Scarlato and co-workers used Isothermal Titration Calorimetry (ITC) to measure nickel binding to apo-HpNikR and reported that two of the nickel ions bound with 10 nM affinity and the remaining two nickel ions bound with 100 nM affinity.57 We note that Zamble and co-workers used a competitive titration with EGTA and UV visible spectroscopy to measure nickel affinity for HpNikR and only observed one high affinity site (pM binding).14 Ciurli, Scarlato and co-workers argue that the EGTA approach yields inaccurate binding constants because EGTA can form polynuclear complexes with Ni(II) and suggest that the binding affinities determined from the direct titration with Ni(II) using ITC are more accurate.57
Role of pH in Nickel Binding Stoichiometry
The structures of EcNikR have all been reported at pH 7.5–8.5; whereas, the structure that we report here was determined at pH 5.6 To determine whether the pH affected the nickel sites in HpNikR, titrations of Ni(II) with apo-HpNikR at a range of pH were performed (ph 5.6, 6.5, 7.5, 8.5). The spectra all exhibited an absorbance maximum at 304 nm which saturated upon addition of 1 nickel ion/HpNikR (4 nickel ions/tetramer). An overlay of the titrations is shown in Figure S2. An overlay of the absorbance spectra at all four pH values as a function of extinction coefficient shows no differences in the spectra as a function of pH as shown in Figure S3. This suggests that the nickel sites are not affected by pH in HpNikR. Zamble and co-workers and Michaud-Soret and co-workers have also reported that they do not observe any changes in nickel stoichiometry and extinction coefficient of Ni(II)-HpNikR over a similar range of pH. 14,58,59 Interestingly, Michaud-Soret and co-workers do observe a pH dependence on nickel binding for E. coli NikR suggesting that E. coli NikR may be more sensitive to pH. 58
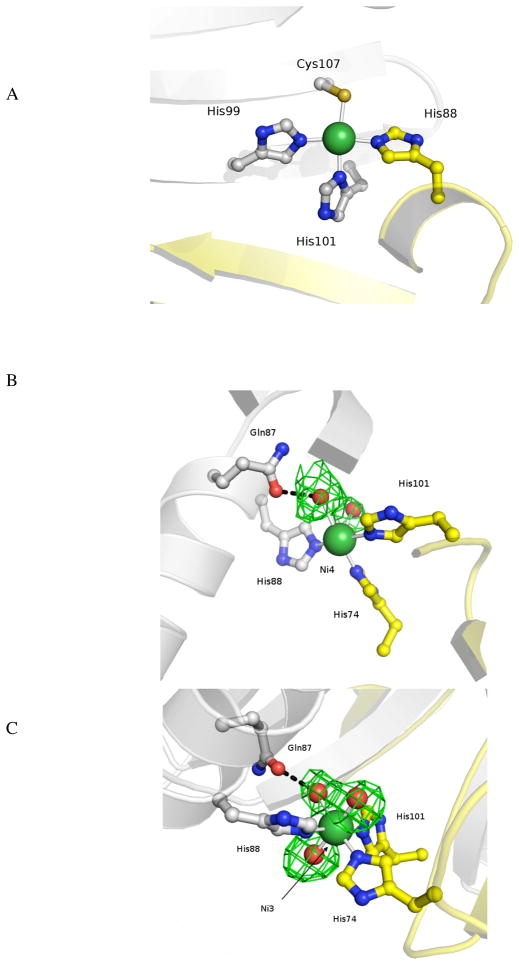
Role of Tryptophan (W54) in Locking the DBD
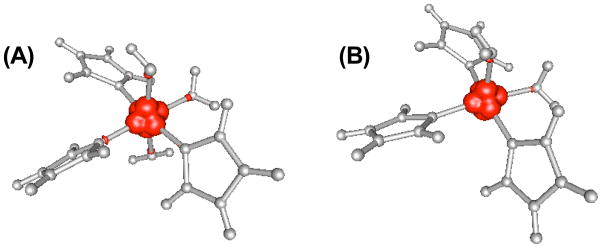
The two DNA binding domains are oriented in a ‘trans’ conformation relative to the MBD in the holo-Ni(II)-HpNikR structure (Figure 3A). Close inspection of the interface reveals that Trp54, which is part of the DBD, lies in a pocket that is stabilized by two lysine residues (Lys64 and Lys140) in the MBD (Figure 3A and B). Due to domain swapping in the holo-Ni(II)HpNikR structure both Trp54 residues from the DBD are located in pockets formed by Lys64/Lys140. We propose that the tryptophan serves to orient the DNA binding domain perpendicular to the MBD. We also speculate that the tryptophan may play a role in directing the DNA binding domain to the correct orientation to recognize its partner DNA. This tryptophan is not conserved in E. coli NikR and the structures of E. coli NikR all have the DNA binding domains in a linear conformation relative to the MBD.23 P. horikoshii NikR contains this tryptophan, but the protein’s DNA targets are not known limiting any comparison.26
Figure 3.
Representation of the (A) protein backbone and the pocket formed at the interface of the MBD and DBD. (B) Trp54 from the DBD is stabilized by a stacking interaction with Lys64 and Lys140.
DFT Calculations of the Nickel Sites
Density functional calculations were performed on the nickel sites of holo-Ni(II)HpNikR using models of the active site to understand the geometries observed in the crystal structures. Ni(II) can form complexes with coordination numbers ranging from 3 to 6.55,60 Qualitative rationalization of the geometrical preferences of these complexes can be made with the help of the molecular orbital (MO) interaction diagram in Figure S4. Square-planar coordination is a consequence of the d8 configuration, since the D4h ligand field causes one of the d orbitals, b1g (x2−y2), to be high in energy while another d orbital, a1g (z2) lowers its energy and approaches the non-bonding manifold, eg+b2g (xy, xz, yz). Eight electrons can occupy the b2g, eg and a1g orbitals thus originating diamagnetic complexes. This is illustrated in the rightmost part of Figure S4. Ni(II) can also form 5 and 6 coordination complexes as illustrated in the central and left and panels of Figure S4, respectively. If the separation between the eg and t2g orbitals is small, the energy gained from forming a triplet state from additional ligands compensates orbital destabilization penalties, leading to 6-coordinate paramagnetic complexes. In 5-coordinated C4v systems the (x2−y2) and (z2) orbitals are no longer degenerate; however due to the electron-electron repulsion contribution the triplet state will become the ground state, such that the complex is also paramagnetic. Accordingly, both the 5- and 6-coordinate geometries were treated as paramagnetic species in the QM calculations; attempts to optimize both complexes as singlets lead to dissociation of the axial ligands.
Description of geometries and electronic structure
The computed DFT geometries are summarized in Table 2 and illustrated in Figures 4 and S3. Figure 4 shows details of the coordination around the Ni centers. Relevant hydrogen bond interactions involving Ni ligands and neighboring groups are included while atoms comprising the backbone atoms included in the model are omitted for clarity. These atoms are included in Figure S5, which also compares the x-ray experimental and the optimized model cluster structures. A ribbon representation of the local protein backbone is included to help to understand how the model clusters were defined. In all cases the coordination is well maintained, though shifts in the coordination spheres, presumably associated with the omission of much of the protein, as described below, are evident. While differences in the structures do exist, as elaborated below, the ability of the DFT calculations to yield coordination geometries consistent with those observed experimentally further validates our crystallographic results.
Figure 4.
Optimized coordination geometries with hexa, penta and tetra coordination extracted from the large cluster optimized structures. (A) is the 6-coordinated large cluster (Ni3); (B) is the 5-coordinated large cluster (Ni4); (C) and (D) are the 4-coordinated large clusters, Ni2 and Ni1 respectively. Numbers are used throughout the text for referencing relevant atoms.
Optimization of the 6-coordinate large cluster resulted in geometries that differ from the experimental structure (Table 2). Hexa coordination around Ni was only possible when the complexes were optimized in their corresponding triplet spin-states. The Ni-O bond distances are consistently longer in the optimized model complex compared to the experimental data and no trends among the Ni-O distances are observed in either structure. The differences between the computed and experimental structures can be attributed to the omission of electronic effects of neighboring moieties on the water ligands associated with interactions not accounted for in the computational model including (1) the omission of side chains and (2) the Ni(II) sites being located on the protein surface. A more complete study of these effects is beyond the scope of the present work. On the other hand the Ni-N distances show better agreement, and the trend among the Ni-N distances observed experimentally are reproduced in the computational model. This better agreement may result, in part, from the geometrical constraints imposed by the protein. Other structural features are similar for the calculated and experimental geometries in both the 5 and 6-coordinate complexes. For example, the Oax-Ni-Oax,2 angle deviates from 180°, being approximately 172 and 175°, for the computed and experimental structures.
Table 2.
Selected bond distances and angles for the DFT optimized complexes and experimental x-ray structure. Bond distances are in Å and bond angles in degrees.
| 6-Coordinated (Ni3) | 5-Coordinated (Ni4) | |||
|---|---|---|---|---|
| Model complex | Exper. | Model complex | Exper. | |
| Ni-Oeq | 2.12 | 2.1 | 2.11 | 2.1 |
| Ni-Oax | 2.14 | 2.1 | 2.07 | 2.1 |
| Ni-Oax,2 | 2.22 | 2.1 | NA | NA |
| Ni-N1 | 2.14 | 2.1 | 2.08 | 2.1 |
| Ni-N2 | 2.12 | 2.1 | 2.15 | 2.1 |
| Ni-N3 | 2.13 | 2.1 | 2.01 | 2.1 |
| Oeq-O1 | 2.59 | 2.5 | 2.64 | 3.2 |
| Oeq-O2 | NA | NA | 2.81 | 2.6 |
| O1-O3 | NA | NA | 2.53 | 3.3 |
| Oax- Ni- Oax,2 | 171.69 | 175.05 | NA | NA |
| Oax- Ni- Oeq | 92.29 (79.84) | 82 (93) | 95.03 | 71 |
| Oax- Ni- N1 | 86.72 (89.42) | 94 (87) | 96.05 | 79 |
| Oax- Ni- N2 | 95.84 (92.02) | 89 (96) | 92.78 | 102 |
| Oax- Ni- N3 | 88.40 (94.35) | 92 (87 | 95.82 | 91 |
| 4-Coordinated (Ni1) | 4-Coordinated (Ni2) | |||
| Model complex | Exper. | Model complex | Exper. | |
| Ni-S | 2.25 | 2.3 | 2.26 | 2.3 |
| Ni-N1 | 1.91 | 2.1 | 1.97 | 2.1 |
| Ni-N2 | 2.00 | 2.1 | 2.00 | 2.1 |
| Ni-N3 | 1.95 | 2.1 | 1.92 | 2.1 |
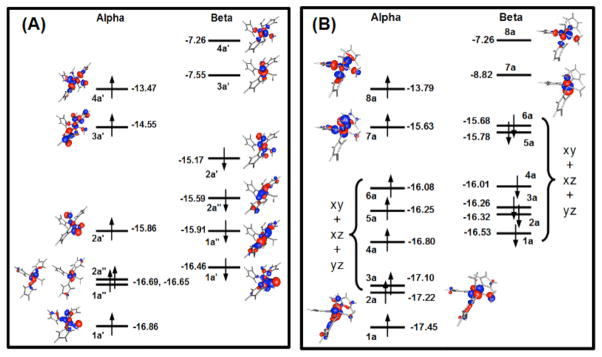
Frontier orbitals that include metallic character for the optimized small model complexes for the 5- and 6-coordinate geometries are shown in Figure 5. For the 6-coordinate complex the frontier orbitals are from the optimized small complex where a plane of symmetry was maintained to mimic the large cluster. The computed frontier orbitals closely resemble the qualitative MO interpretation for a six-coordinate nickel site (Figure S4). Some orbital mixing is visible, particularly for the orbitals (eg. 1a′ and 2a′) corresponding to the t2g set of an ideal complex with s-donor ligands only. The mixing occurs in both the alpha and beta sets, and involves mostly the xz orbital of Ni and a p orbital of the equatorial water oxygen. Orbitals 1a′ and 2a′ are the resulting bonding and anti-bonding combinations. Orbitals mix more in the beta set but the pattern is similar to the alpha set. The 6-coordinated Ni(II) complexes are stable, despite the d8 configuration, because the splitting of the eg and t2g orbitals is relatively small; the energy difference between the 3a′ and 4a′ (eg) orbitals in Figure 5A and the highest energy orbital of the t2g set, 2a′, being 1.3 eV. Inspection of the plot of the spin density for the 6-coordinated complex in Figure 6A confirms single occupation of the x2−y2 and z2 orbitals.
Figure 5.
Illustration of the frontier orbitals of the hexa (A) and penta-coordinated (B) small complexes of Ni. A mirror plane was used during the optimization of the hexa-coordinated model and no-symmetry was used in the optimization of the penta-coordinated model.
Figure 6.
Spin-density of the small model complexes: (A) is for the 6-coordinated complex and (B) is for the 5-coordinated complex.
The minimized structure of the 5-coordinated large cluster shows good agreement between the computed and crystal structures around Ni(II) (Table 2). The Ni-N and Ni-O bond separations follow the same trend in both structures, the biggest difference being 0.04 Å. In the QM model there are significant out-of-plane distortions as measured by the O, Ni and the equatorial ligand angles (Table 2), with those distortions being more pronounced in the experimental structure. The positioning of the oxygen atoms participation in the hydrogen bonding network with the equatorial water differs significantly in the two structures.
Interpretation of the electronic structure of the 5-coordinated model (Figure 5B) closely follows the arguments used in the case of 6-coordinated complexes. Interestingly, symmetry could not be used during optimization of the small model complex. All attempts resulted in a 4-coordinated complex of pseudo-tetrahedral geometry and loss of one of the water ligands. The consequence of the lack of symmetry is more orbital mixing. In Figure 5B, only MOs with a clear metallic character are shown. Increasing in energy are z2 and x2−y2 in both the alpha and beta sets and xz, xy in the alpha set alone. All others are heavily mixed and cannot be easily assigned. Interesting is the nature of the two topmost orbitals of the alpha set, 7a and 8a. They are derived from the z2 and x2−y2 orbitals of Ni and correspond to the 3a′ and 4a′ of the 6-coordinated model. It is relevant to note that orbital 7a of the square base pyramid complex has a lower energy than the equivalent orbital of the octahedral complex, 3a′, their respective energies being −15.63 and −14.55 eV. The energy of the x2−y2 derived orbital is almost the same, −13.79 and −13.47 eV for the 5 and 6-coordinated complexes. This agrees with the qualitative MO picture of Figure S4. The spin density shown in Figure 6B confirms single occupation of the x2−y2 and z2 orbitals.
Geometry optimizations of both 4-coordinated Ni centers were performed by enforcing a singlet ground state. The optimized geometries maintained the square-planar geometry around both Ni sites (Figure 4C and 4D). Distortion of the optimized clusters relative to the experimental geometries is smaller than with the 5 and 6-coordinated centers (Figure S5). The geometrical parameters shown in Table 2 are very similar for both Ni centers. Compared to the higher coordination complexes, Ni-N bonds are considerably shorter. There is also a clear trend with Ni-N2 (opposite to the Ni-S bond) which is considerably longer than Ni-N1 and Ni-N3.
Mechanistic Implications
HpNikR recognizes multiple DNA sequences. Half of the target DNA sequences are recognized with high affinity (Kd = nM) and the other half with low affinity (Kd = μM);16 and a key, unanswered question is how does HpNikR distinguish between its target promoter sequences? We hypothesize that the 5/6 coordinate site plays a role in DNA discrimination. Molecular dynamics studies of HpNiKR support a model in which the DBD are highly flexible until DNA is introduced.61 We speculate that the presence of both 4-coordinate and 5/6-coordinate sites promote conformational diversity and contribute to recognition of multiple DNA sequences. Another possible role for the 5/6 coordinate site is to link nickel occupancy to DNA binding. In this scenario, the overall conformation of HpNikR may be altered as a function of nickel:protein stoichiometry. As a result, differential DNA recognition occurs as a function of nickel occupancy. Such a role would correlate well with in vivo studies that have demonstrated that the function of HpNikR is related to nickel availability. 20 A final scenario involves the nickel ions switching between the 4 coordinate and 5/6 coordinate sites upon DNA binding. The thermodynamic cost of this switching would serve to control the protein/DNA interaction. Inspection of the crystal structure identifies two histidine residues (His 75 and His 105) located between the two sites which may serve to traffic nickel between the two sites. The exact role of the 5/6 coordinate site in DNA recognition remains to be determined; however, we propose that this site is an important player in the complex mechanism of DNA recognition by HpNikR.
Comparison to other NikR structures
A key structural difference between HpNikR, EcNikR and PhNikR is that the nickel binding domains are asymmetric in HpNikR and symmetric in both EcNikR and PhNikR. The structures were attained under different pH conditions (EcNikR and PhNikR at pH 7.5–8.5, HpNikR at pH 5.6). By comparing all three structures, we can identify certain residues that may explain why holo-Ni(II)HpNikR binds nickel at a 5/6 coordinate site in the absence of DNA. Among the five residues involved in coordinating nickel in the two distinct classes of binding sites, four belong to the central β-sheet sandwich of the NikR fold while one, His74, is located on a loop between a helix and a β strand. We identified the conformational change that would occur if nickel were to move from a 5/6 coordinate to a 4 coordinate geometry by aligning the MBD of HpNikR with its homologues. Pairs of MBDs forming the interface containing both nickel binding sites were superimposed based on the Cα atoms using PyMOL, resulting in overall RMSD of 1.4/1.9 Å relative to EcNikR and PhNikR, respectively. Interestingly, the corresponding RMSD values are reduced to 0.7/0.9 Å when only the β-sheets are included in superposition, while the RMSD increases dramatically (4.2/3.1 Å) when only the α-helices are aligned. In addition, alignment of the EcNikR and PhNikR MBDs yields RMSD values of 0.5 Å for the β-sheets and 1.0 Å for the α-helices, showing the greater similarity of these homologs versus HpNikR. This analysis suggests that the conformational changes required to accommodate the all square planar sites in HpNikR could be described as positional shift of α-helices.
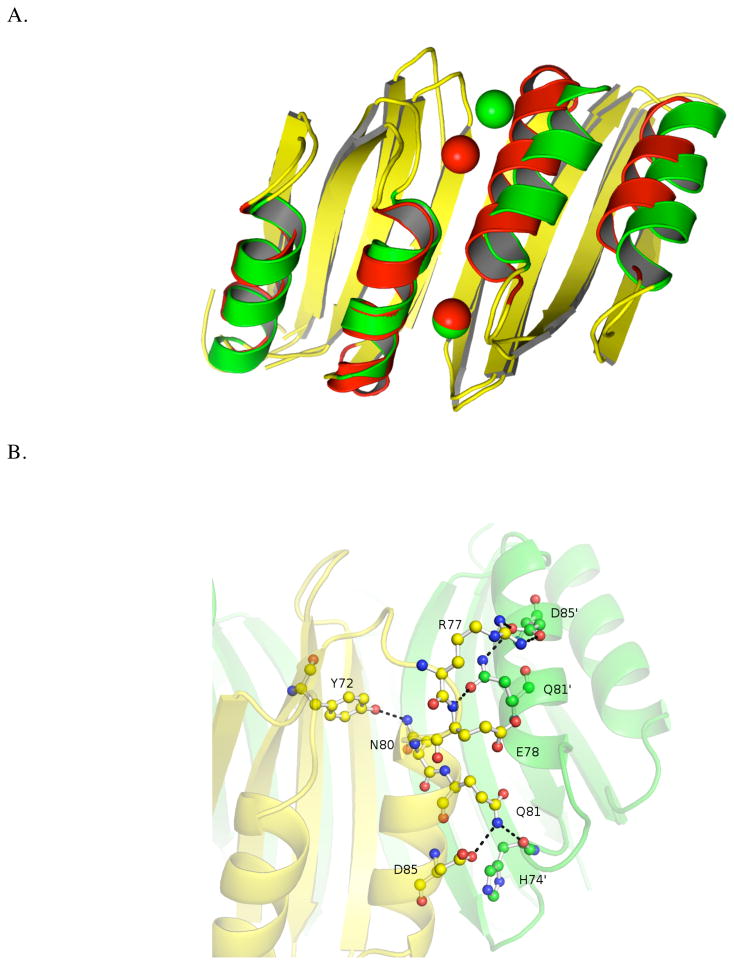
Figure 7A shows the alignment of HpNikR and EcNikR based on only the β-sheet Cα atoms. To shift the Ni(II) at the 5/6 coordinate geometry to a 4 coordinate geometry, the second α-helix of the HpNikR MBD would need to shift such that there would be steric clash at the N-terminus of the helix. Sequence alignment indicates that HpNikR contains an NQ-motif (Asn80-Gln81) in this region that is absent in the other homologues. These two residues form the basis of the extended hydrogen bonding network (Figure 7B) which appears to be crucial in determining the conformation of the loop that contains His74. On the “4-coordinate side” (lower region of Figure 7B), the side chain of Gln81 makes a hydrogen bond to the backbone of His74′, potentially making it more difficult for that residue to ligate the nickel ion. On the “5/6 coordinate side” (upper region of Figure 8b), Asn80 makes a hydrogen bond to Tyr72 which leads to a shift in the position of Gln81′, such that it no longer interacts with His74, but rather with the backbone of Glu78 and stabilizes the salt bridge between Arg77 and Asp85. We propose that these hydrogen-bonding interactions help stabilize the 5/6 coordinate geometries relative to the 4-coordinate geometry by allowing His74 to assume the proper orientation necessary to ligate the nickel ion. In both EcNikR and NikR, the NQ motif is replaced by less polar residues and an analogous hydrogen bonding network is absent (Figures S6 and S7).
Figure 7.
(A) MBD of holo-Ni(II)HpNikR (alpha helices are colored green) overlaid with MBD of holo-Ni(II)EcNikR (α-helices are colored red). Beta sheets from both homologs are colored yellow. Structural alignment was based on the Cα atoms in the β-sheets. (B) The extended hydrogen bonding network at the N-terminus of neighboring alpha helices of the MBD of holo-HpNikR. Amino acid side chains are shown as ball and stick. Dashed lines indicate the hydrogen bonds.
Recently, Michaud-Soret and co-workers reported the structural characterization of several mutants of HpNikR to assess the role of the external site observed in their nickel-soaked apo-HpNikR structures.62 This site is unoccupied in our holo-Ni(II)HpNikR structure. The mutant designed to address the role of the external site is a double histidine mutant H74G/H75G. This mutant also disrupts the 5/6 coordinate site so it is not specific for the external site. As with wt-HpNikR, the published structure was generated by soaking crystals of apo-H74G/H75G with excess nickel. In the nickel bound structure of the mutant, only two square planar sites occupied by nickel ions are present which adds further support to the idea that holo-Ni(II)HpNikR only has two square planar nickel sites.
The H74G/H75G mutant exhibits decreased DNA binding affinity compared to wild type and has no in vivo activity.62 Based upon these results it was concluded that the external site is required for DNA binding because the His74G/His75G mutation has disrupted this site. However, His74 is one of the ligands in the 5/6 coordinate site in addition to participating in the external site and the mutation may disrupt the 5/6 coordinate site as well as the external site. Thus, an alternate interpretation is that the decrease in DNA binding coupled with lack of in vivo activity observed for the mutant may be due to disruption of the 5/6 coordinate site, an interpretation with is consistent with our hypothesis that the 5/6 coordinate site is essential for HpNikR function. Further studies are required to address these issues.
Conclusion
Here, we report the first crystal structure of holo-Ni(II)HpNikR. This structure reveals that holo-Ni(II)HpNikR contains two distinct classes of high affinity nickel sites: two 5/6 coordinate sites and two 4 coordinate sites. DFT calculations show that the 5/6 coordinate sites are stabilized by key amino acid side chains from the protein. Spectroscopic analysis of nickel binding to HpNikR at a range of pH values suggest that occupation of the nickel sites is pH independent. Based on these observations we hypothesize that the 5/6 coordinate site plays a role in HpNikR’s ability to specifically and selectively bind to a series of promoter sequences.
Supplementary Material
Acknowledgments
We would like to thank the Dept. of Pharmaceutical Sciences at the University of Maryland Baltimore and NIH GM051501 to ADM for financial support for this research.
Footnotes
Supporting Information Available. Additional DFT and structural figures, pH dependent Ni titrations and complete reference 39. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Marshall BJ, Warren JR. Lancet. 1984;1:1311–5. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. Annu Rev Med. 1992;43:135–45. doi: 10.1146/annurev.me.43.020192.001031. [DOI] [PubMed] [Google Scholar]
- 3.Sepulveda AR, Coelho LG. Helicobacter. 2002;7(Suppl 1):37–42. doi: 10.1046/j.1523-5378.7.s1.6.x. [DOI] [PubMed] [Google Scholar]
- 4.Kusters JG, van Vliet AH, Kuipers EJ. Clin Microbiol Rev. 2006;19:449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loughlin MF. Expert Opin Ther Targets. 2003;7:725–35. doi: 10.1517/14728222.7.6.725. [DOI] [PubMed] [Google Scholar]
- 6.van Vliet AH, Ernst FD, Kusters JG. Trends Microbiol. 2004;12:489–94. doi: 10.1016/j.tim.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 7.van Vliet AH, Kuipers EJ, Stoof J, Poppelaars SW, Kusters JG. Infect Immun. 2004;72:766–73. doi: 10.1128/IAI.72.2.766-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Zamble DB. Chem Reviews. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 10.van Vliet AH, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, Penn CW, Kuipers EJ, Kusters JG. Infect Immun. 2002;70:2846–52. doi: 10.1128/IAI.70.6.2846-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benanti EL, Chivers PT. J Biol Chem. 2007;282:20365–75. doi: 10.1074/jbc.M702982200. [DOI] [PubMed] [Google Scholar]
- 12.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. J Bacteriol. 2005;187:7703–15. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dosanjh NS, Hammerbacher NA, Michel SL. Biochemistry. 2007;46:2520–9. doi: 10.1021/bi062092w. [DOI] [PubMed] [Google Scholar]
- 14.Abraham LO, Li Y, Zamble DB. J Inorg Biochem. 2006;100:1005–14. doi: 10.1016/j.jinorgbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Mol Microbiol. 2003;49:947–63. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 16.Dosanjh NS, West AL, Michel SL. Biochemistry. 2009;48:527–36. doi: 10.1021/bi801481j. [DOI] [PubMed] [Google Scholar]
- 17.Dosanjh NS, Michel SL. Curr Opin Chem Biol. 2006;10:123–30. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Chivers PT, Sauer RT. Protein Sci. 1999;8:2494–500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. J Bacteriol. 1999;181:670–4. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AH. Infect Immun. 2005;73:7252–8. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chivers PT, Sauer RT. Chem Biol. 2002;9:1141–8. doi: 10.1016/s1074-5521(02)00241-7. [DOI] [PubMed] [Google Scholar]
- 22.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Nat Struct Biol. 2003;10:794–9. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 23.Schreiter ER, Wang SC, Zamble DB, Drennan CL. Proc Natl Acad Sci U S A. 2006;103:13676–81. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albright TA, Burdett JK, Whangbo M-H. Orbital Interactions in Chemistry. Wiley; New York: 1985. [Google Scholar]
- 25.Phillips CM, Schreiter ER, Guo Y, Wang SC, Zamble DB, Drennan CL. Biochemistry. 2008;47:1938–46. doi: 10.1021/bi702006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chivers PT, Tahirov TH. J Mol Biol. 2005;348:597–607. doi: 10.1016/j.jmb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Dian C, Schauer K, Kapp U, McSweeney SM, Labigne A, Terradot L. J Mol Biol. 2006;361:715–30. doi: 10.1016/j.jmb.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Macromolec Cryst, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey S. Acta Cryst. 1994;D50:760–763. [Google Scholar]
- 31.Potterton E, Briggs P, Turkenburg M, Dodson E. Acta Cryst. 2003;D59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. Acta Cryst. 2004;D60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 35.Painter J, Merritt EA. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, Isupov MN, Murshudov GN. Acta Cryst. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 37.Winn MD, Murshudov GN, Papiz MZ. Macromolec Cryst, Pt D. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 38.Kendall RA, Aprà E, Bernholdt DE, Bylaska EJ, Dupuis M, Fann GI, Harrison RJ, Ju J, Nichols JA, Nieplocha J, Straatsma TP, Windus TL, Wong AT. Computer Phys Comm. 2000;128:260–283. [Google Scholar]
- 39.Bylaska EJ, et al. NW Chem, A Computational Chemistry Package for Parallel Computers, Version 5.1.1. Pacific Northwest National Laboratory; Richland, Washington 99352–0999, USA: 2007. [Google Scholar]
- 40.Hohenberg P, Kohn W. Physical Review. 1964;136:B864. [Google Scholar]
- 41.Kohn W, Sham LJ. Physical Review. 1965;140:A1133. [Google Scholar]
- 42.Parr RG, Yang W. Density-Functional Theory of Atoms and Molecules. Oxford University Press; New York: 1989. [Google Scholar]
- 43.Becke AD. J Chem Phys. 1993;98:1372–1378. [Google Scholar]
- 44.Kim K, Jordan KD. J Phys Chem. 1994;98:10089–10094. [Google Scholar]
- 45.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. J Phys Chem. 1994;98:11623–11627. [Google Scholar]
- 46.Hay PJ, Wadt WR. J Chem Phys. 1985;82:299. [Google Scholar]
- 47.Binkley JS, Pople JA, Hehre WJ. J Am Chem Soc. 1980;102:939–947. [Google Scholar]
- 48.Ditchfield R, Hehre WJ, Pople JA. J Chem Physics. 1971;54:724–728. [Google Scholar]
- 49.Hariharan PC, Pople JA. Theoretical Chemistry Accounts: Theory, Computation, and Modeling (Theoretica Chimica Acta) 1973;28:213–222. [Google Scholar]
- 50.Hehre WJ, Ditchfield R, Pople JA. J Chem Phys. 1972;56:2257–2261. [Google Scholar]
- 51.Barnes C. J App Cryst. 1997;30:568. [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 53.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, Mondragon A. Science. 2003;301:1383–7. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 54.Schreiter ER, Drennan CL. Nature Reviews Microbiology. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 55.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. 5. John Wiley and Sons; New York: 1988. [Google Scholar]
- 56.Stoll KE, Draper WE, Kliegman JI, Golynskiy MV, Brew-Appiah RA, Phillips RK, Brown HK, Breyer WA, Jakubovics NS, Jenkinson HF, Brennan RG, Cohen SM, Glasfeld A. Biochemistry. 2009;48:10308–20. doi: 10.1021/bi900980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zambelli B, Bellucci M, Danielli A, Scarlato V, Ciurli S. Chem Commun (Camb) 2007:3649–51. doi: 10.1039/b706025d. [DOI] [PubMed] [Google Scholar]
- 58.Fauquant C, Diederix RE, Rodrigue A, Dian C, Kapp U, Terradot L, Mandrand-Berthelot MA, Michaud-Soret I. Biochimie. 2006;88:1693–705. doi: 10.1016/j.biochi.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Zamble D. Biochemistry. 2009. [Google Scholar]
- 60.Greenwood NN, Earnshaw A. Chemistry of the Elements. 2. Butterworth-Heinemann; 1997. [Google Scholar]
- 61.Sindhikara DJ, Roitberg AE, Merz KM., Jr Biochemistry. 2009;48:12024–33. doi: 10.1021/bi9013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bahlawane C, Dian C, Muller C, Round A, Fauquant C, Schauer K, de Reuse H, Terradot L, Michaud-Soret I. Nucleic Acids Res. doi: 10.1093/nar/gkp1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss MS. J Appl Cryst. 2001;34:130–135. [Google Scholar]
- 64.Weiss MS, Hilgenfeld R. J App Cryst. 1997;30:203–205. [Google Scholar]
- 65.MSW J Appl Cryst. 2001;34:130–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.