Abstract
The most common first-line, highly active anti-retroviral therapy (HAART) received by individuals infected with HIV-1 in Cameroon is the combination therapy Triomune, comprised of two nucleoside reverse transcriptase inhibitors (NRTI) and one non-NRTI (NNRTI). To examine the efficacy of these drugs in Cameroon, where diverse non-B HIV-1 subtypes and recombinant viruses predominate, the reverse transcriptase (RT) viral sequences in patient plasma were analyzed for the presence of mutations that confer drug resistance. Forty-nine HIV-1-positive individuals were randomly selected from those receiving care in HIV/AIDS outpatient clinics in the South-West and North-West Regions of Cameroon. Among the 28 patients receiving HAART, 39% (11/28) had resistance to NRTIs, and 46% (13/28) to NNRTIs after a median of 12 months from the start of therapy. Among those with drug-resistance mutations, there was a median of 14 months from the start of HAART, versus 9 months for those without; no difference was observed in the average viral load (10,997 copies/ml vs. 8,056 copies/ml). In contrast, drug-naïve individuals had a significantly higher average viral load (27,929 copies/ml) than those receiving HAART (9,527 copies/ml). Strikingly, among the 21 drug-naïve individuals, 24% harbored viruses with drug-resistance mutations, suggesting that HIV-1 drug-resistant variants are being transmitted in Cameroon. Given the high frequency of resistance mutations among those on first-line HAART, coupled with the high prevalence of HIV-1 variants with drug-resistance mutations among drug-naïve individuals, this study emphasizes the need for extensive monitoring of resistance mutations and the introduction of a second-line HAART strategy in Cameroon.
Keywords: drug-resistance mutations, HIV-1, NRTI, NNRTI, HAART, drug naïve
Introduction
Scaling up highly active anti-retroviral therapy (HAART) to achieve universal access is the current priority of the World Health Organization (WHO) and UNAIDS for the treatment of individuals infected with HIV-1 living in resource-constrained countries. In sub-Saharan Africa, where the greatest gain in HAART coverage was seen among the low- and middle-income countries surveyed in 2007, an estimated 2.1 million people received treatment, representing an increase of 54% [WHO, 2008]. Currently, 97% of individuals infected with HIV-1 in resource-constrained countries who are prescribed anti-retroviral (ARV) drugs are receiving a WHO-recommended first-line HAART [WHO, 2008]. In Africa, a generic, low-cost, fixed-dose combination (FDC) of two nucleoside reverse transcriptase inhibitors (NRTI) and one non-NRTI (NNRTI) is prescribed most frequently [Calmy et al., 2006; Laurent et al., 2008]. The safety and efficacy of a commonly prescribed FDC containing stavudine, lamivudine, and nevirapine (Triomune; Cipla, Mumbai, India) in HAART naïve and patients receiving HAART has shown positive results in resource-constrained countries [Laurent et al., 2008]. However, long-term use of thymidine analogue-based therapies are known to influence amino acid substitutions and deletions in viral reverse transcriptase (RT) and can confer drug resistance, particularly among those individuals who received previously mono- or dual- NRTI therapy or when an interruption in therapy has occurred [Gallant, 2007; Sungkanuparph et al., 2007; Marconi et al., 2008].
It is certain that the use of HAART, especially in resource-constrained countries where the logistics of medication disbursement and patient follow-up are not always optimal, will result in the emergence and transmission of drug-resistant HIV-1 variants which in turn will provide an obstacle to any successful HAART program. For this reason, the monitoring of HIV-1 transmitted drug resistance (TDR) in North America and Western Europe, where ARV therapy has been available for many years, remains an important aspect of clinical management of the disease [Hirsch et al., 2008; Little et al., 2008]. However, while access to HAART is becoming increasingly available in resource-constrained countries [Geretti, 2007], data on the emergence of drug-resistance mutations and transmission of viruses harboring drug-resistance mutations remains sparse for these countries. Therefore, in countries where clinical monitoring and drug-resistance testing is limited, it is critical for both the continued scale-up of first-line regimens of HAART and the successful introduction of effective second-line regimens that the emergence of drug-resistance mutations be monitored.
In the sub-Saharan African country of Cameroon, the Ministry of Public Health established the first HIV treatment center in March of 2001, and in May of 2007 announced that eligible individuals could receive ARV drugs free-of-charge through a national distribution program. Since then, no studies detailing the drug-resistance patterns of HIV-positive individuals who are on first-line HAART, nor of those who are drug naïve, have been performed. In the current cross-sectional study, RT drug-resistance mutations among patients receiving ARV drugs and those who were ARV drug naive, living in the North-West and South-West Regions of Cameroon, is reported. Many of these individuals are now receiving first-line HAART free-of-charge.
Materials And Methods
Study Participants
The samples analyzed in this study are part of a larger study aimed at analyzing the HIV-1 genetic diversity of infected individuals in Cameroon. Thus, 49 plasma samples were selected at random from HIV-1-positive individuals attending and/or receiving ARV therapy in clinics in the South-West and North-West Regions. The specimens used were obtained from these individuals in calendar years 2006 and 2007, and included drug-naïve individuals (n = 21) as well as individuals receiving first-line HAART (n = 28) due to a CD4 T-cell count of ≤200 cells/mm3. A detailed questionnaire, including the date of diagnosis of their HIV infection, mode of infection, treatment history, and demographic information was administered to each participant after signing an informed consent.
RNA Extraction, PCR, and Sequencing
Viral RNA extraction from the participant's plasma was performed using the QIAamp Viral RNA Mini kit (Qiagen, Inc., Valencia, CA) followed by reverse transcription using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. A 1,596-bp region of the pol gene, covering the RT (p51) and most of the RNase H (p15) was amplified from the extracted RNA using nested PCR. For the first round PCR, the forward primer RTPOL1F (5′-GTATTAGTAGGACCTACACCTGTC-3′; HXB2 location 2475–2498) and the reverse primer RTPOL1R (5′-ACCTTCCTGATTCCATTACTGAC-3′; HXB2 location 4203–4225) were used. For the nested PCR, forward primer RTPOL2F (5′-TAAAGCCAGGAATGGATGGCCC-3′; HXB2 location 2584–2605) and reverse primer RTPOL2R (5′-CCTCCAATCCCTTTGTGTGCTG-3′; HXB2 location 4159–4180) were used. The cycling conditions for both PCRs were an initial denaturation of 2 min at 94°C, followed by 35 cycles at 94, 50, and 72°C for 60, 60, 90 sec, respectively, and a final extension step of 7 min at 72°C. PCR products were then separated by agarose gel electrophoresis and amplicons of the expected size were excised and extracted from the gel using the QIAquick spin gel extraction kit (Qiagen, Inc.). Direct sequencing of the 5′ and 3′ ends of the purified PCR amplicons were performed with the two nested primers RTPOLF2 and RTPOLR2, and primer walking using the ABI Big Dye Terminator cycle sequencing chemistry on an ABI 3730XL instrument.
Phylogenetic Analysis
All sequences were automatically aligned with reference sequences of all known HIV-1 group M (sub-) subtypes (A1, A2, B, C, D, F1, F2, G, H, J, and K) and circulating recombinant forms (CRFs) from the Los Alamos HIV sequence database using CLUSTAL X with minor manual adjustments. Phylogenetic analyses were conducted using the MEGA version 3.1 software package [Kumar et al., 2004] with pairwise evolutionary distances estimated by using Kimura's two-parameter method. Phylogenetic trees were constructed by the neighbor-joining method and the reliability of the topologies was estimated by performing bootstrap analysis (1,000 replicates). Clustering of sequences with a bootstrap value of more than 70% was considered significant for subtyping.
Drug-Resistance Genotyping
The RT DNA sequences amplified from patients receiving HAART and those who were ARV drug naive were analyzed for potential drug-resistance mutations using the Stanford University HIV database genotypic-resistance interpretation algorithm (http://hivdb.stanford.edu/index.html). This program identifies documented drug-resistance mutations in user-entered sequences and infers the level of resistance to NRTIs, NNRTIs, and protease inhibitors (PIs). In addition, the drug-resistance results were cross-checked with the International AIDS Society-USA consensus mutation figures [Johnson et al., 2008] and only major mutations conferring a several-fold reduction in viral sensitivity to all current, FDA approved, ARVs (including those prescribed to the study individuals) were included in the analysis.
RTI Genotypic Drug-Resistance Mutation Analysis
The RT sequences from the 49 HIV-1-positive individuals were analyzed for mutations associated with NRTI and NNRTI drug resistance using the HIValg program available on the Stanford University HIV Drug-Resistance Database website [Rhee et al., 2003]. Mutations in the study sequences were defined as differences from the consensus B reference sequence and were further characterized as NRTI-resistance mutations, NNRTI-resistance mutations, or other mutations which consists primarily of accessory mutations (those which have little to no effect on drug susceptibility in of themselves) and polymorphisms. In addition, the HIValg program scores each mutation, using three different algorithms (ANRS, Agence Nationale de Recherches sur le SIDA; HIVDB, Standford University HIV Drug-Resistance Database; Rega Institute) in order to compare predicted levels of associated drug resistance. For each sequence analyzed, the drug-resistance interpretation was compared for consistency and only the results given by the HIVDB algorithm, whereby drug resistance was predicted to be “high-level” are presented here unless otherwise indicated (Table I).
TABLE I.
Genotypic Resistance Mutations That Emerge Among HIV-1 Non-B Subtype Infected Patients on Anti-Retroviral Therapy
| Patient ID | Age | Sex | Current ART | Months from start of ART | Viral load copies/μl | Genotypic-resistance mutations | High-level drug resistancea | RT subtype | ||
|---|---|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | NRTI | NNRTI | |||||||
| 06ARC001 | 47 | F | Trio | 14 | 5,484 | M184V | K103N | 3TC, FTC | NVP, DLV, EFV | CRF02_AG |
| 06ARC007b | 36 | F | Duovir + stocrinc | 8 | <75 | A62V, K65R, F116Y, Q151M, M184I | V108I, Y181C | 3TC, FTC, ABC, d4T, ddI | NVP, DLV | CRF02_AG |
| 06ARC009 | 35 | F | 3TC+d4T | 9 | <75 | None | V179E | — | — | CRF02_AG |
| 06ARC036b | 54 | M | Duovir + stocrin | 39 | 7,156 | M184V, T215Y | K103N, V179E, Y181C, G190A | 3TC, FTC | NVP, DLV, EFV, ETR | CRF02_AG |
| 06ARC071 | 34 | F | Trio | 12 | 9,524 | M184V | K103N | 3TC, FTC | NVP, DLV, EFV | CRF02_AG |
| 06BDHS005 | 50 | M | Trio | 22 | 20,899 | A62V, V75I, F77L, F116Y, Q151M, M184V | Y188L | 3TC, ABC, AZT, d4T, ddI | NVP, DLV, EFV | CRF11_cpx |
| 07BDHS040 | 33 | F | Trio | 27 | <75 | M184V | Y181C | 3TC, FTC | NVP, DLV | D |
| 07BDHS077 | 42 | F | Trio | >42 | 29,154 | M184V, T215Y | V106A, F227L | 3TC, ABC, AZT, d4T, ddI | NVP, DLV, EFV | CRF02_AG |
| 06LPH17HT | 32 | M | Trio | 13 | 3,774 | None | G190A | — | NVP | CRF02_AG |
| 06LPH20SC | 42 | M | Trio | 20 | 37,754 | F116Y, Q151M, M184V | V108I, Y181C, F227L | 3TC, d4T | NVP | CRF02_AG |
| 07LPH142 | 27 | F | Trio | 18 | 5,185 | M184V, T215Y | Y181C | 3TC, FTC | NVP | CRF02_AG |
| 07LPH149 | 42 | M | Trio | 9 | 23,533 | M184V | V106A, F227L | 3TC, FTC | NVP | D |
| 07LPH02NJM | 48 | M | Trio | 12 | 504 | M184V | K103S, G190A | 3TC | NVP | CRF02_AG |
| 06ARC003 | 29 | M | Trio | 7 | 428 | None | None | — | — | CRF02_AG |
| 06ARC011 | 29 | F | Trio | 6 | <75 | None | None | — | — | CRF02_AG |
| 06ARC053 | 34 | F | 3TC + NVP | <1 | 68,242 | None | None | — | — | CRF02_AG |
| 06ARC055 | 38 | F | Trio | 5 | <75 | None | None | — | — | D |
| 06ARC058 | 38 | M | 3TC + NVP | 1 | 183 | None | None | — | — | CRF02_AG |
| 06ARC075 | 23 | M | Trio | 2 | <75 | None | None | — | — | CRF02_AG |
| 06BDHS004 | 37 | M | Trio | 17 | <75 | None | None | — | — | CRF02_AG |
| 06BDHS006 | 44 | M | Trio | 15 | <75 | None | None | — | — | CRF02_AG |
| 06BDHS007 | 32 | F | Trio | 21 | 138 | None | None | — | — | CRF02_AG |
| 06LPH16SL | 58 | F | Trio | 17 | 34,939 | None | None | — | — | CRF02_AG |
| 07LPH01SG | 26 | M | Stocrin | <1 | 604 | None | None | — | — | CRF02_AG |
| 07LPH110 | 33 | F | Trio | 11 | 121 | None | None | — | — | CRF02_AG |
| 07LPH112 | 36 | F | Trio | 15 | 77 | None | None | — | — | CRF02_AG |
| 07LPH128 | 41 | M | Trio | 9 | ND | None | None | — | — | CRF02_AG |
| 07LPH162 | 39 | F | Trio | 11 | ND | None | None | — | — | CRF02_AG |
ART, antiretroviral therapy; RT, reverse transcriptase; Trio, Triomune (3TC/d4T/NVP); ND, not done.
Stanford University HIV Drug-Resistance Database, interpretation of NRTI and NNRTI-resistance mutations (susceptible-, low-, and intermediate-level of drug resistance not shown).
06ARC007 and 06ARC036 first HAART regimen was Trio for 3 months and 4 months, respectively.
Duovir + Stocrin (3TC+AZT+EFV).
Viral Load Determination
The viral load of each study individual was determined by the Versant HIV RNA 3.0 Assay (bDNA; Siemens, IL) using the same plasma specimen used for the sequencing of RT, and was performed as recommended by the manufacturer.
Results
Characteristics of Study Individuals
All patients receiving HAART were provided with the drugs free-of-charge from the Ministry of Public Health when CD4 T-cell-counts of ≤200 cells/mm3 or a WHO clinical stage 4 was determined. Among the treated individuals (n = 28), the average number of months on HAART prior to inclusion in the study was 13.7 (median = 12 months), and 79% were receiving the FDC Triomune (3TC/d4T/NVP). The remaining six individuals were prescribed either a combination of lamivudine and stavudine (3TC/d4T; n = 1), lamivudine and nevirapine (3TC/NVP; n = 2), Duovir and Stocrin (3TC/AZT and EFV; n = 2), or Stocrin alone (EFV; n = 1). The median age of all study individuals (including those receiving HAART and HAART naïve) was 34 years (range: 18–58) and 59% were women (Tables I and II). The average viral load for the patients receiving HAART with drug-resistance mutations (n = 13) was 10,997 copies/ml compared to 8,056 copies/ml for those without (n = 13; 2 not tested). Among the drug-naïve individuals (n = 21), the average viral load was 27,929 copies/ml (Table II). The percentage of individuals with undetectable viral loads (<75 copies/ml) for patients receiving HAART with drug-resistance mutations, patients receiving HAART without mutations, and drug-naïve individuals were 23%, 38%, and 10%, respectively.
TABLE II.
Prevalence of Genotypic-Resistance Mutations Among Drug-Naïve HIV-1 Non-B Subtype Infected Patients
| Patient ID | Age | Sex | Viral load copies/ml | Genotypic-resistance mutations | High-level drug resistancea | RT subtype | ||
|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | NRTI | NNRTI | |||||
| 06ARC004 | 35 | M | 8,358 | None | None | — | — | CRF02_AG |
| 06ARC010 | 30 | F | 14,715 | None | None | — | — | CRF02_AG |
| 06ARC013 | 31 | F | 1,823 | None | None | — | — | U |
| 06ARC076 | 33 | F | 3,649 | K65R | K101E | — | — | F2 |
| 06BDHS019 | 28 | F | <75 | None | None | — | — | U |
| 06BDHS020 | 30 | F | 93 | None | None | — | — | CRF02_AG |
| 06BDHS023 | 38 | F | 293 | None | None | — | — | CRF13_cpx |
| 06BDHS024 | 27 | F | 1,400 | M184V | V1081, Y181C | 3TC, FTC | NVP, DLV | F2 |
| 06BDHS028 | 31 | M | 511 | M184V | K103N | 3TC, FTC | NVP, DLV, EFV | CRF02_AG |
| 06BDHS035 | 29 | F | 1,604 | None | None | — | — | CRF02_AG |
| 07BDHS053 | 28 | M | 38,442 | None | None | — | — | A |
| 07BDHS054 | 29 | M | 185 | None | G190A | — | — | CRF02_AG |
| 07BDHS055 | 27 | F | 3,368 | None | None | — | — | CRF02_AG |
| 06LPH01OJ | 36 | M | 4,401 | None | None | — | — | U |
| 06LPH02MG | 35 | F | 141,198 | None | None | — | — | CRF02_AG |
| 06LPH03VJ | 52 | M | 25,517 | None | None | — | — | CRF02_AG |
| 06LPH05DE | 48 | F | 15,135 | None | None | — | — | CRF02_AG |
| 06LPH11TT | 27 | F | <75 | None | None | — | — | A |
| 06LPH21ZC | 29 | M | 1,629 | None | None | — | — | CRF02_AG |
| 06LPH22NH | 31 | F | 292,627 | None | None | — | — | CRF02_AG |
| 06LPH19CM | 18 | F | 31,557 | None | None | — | — | A |
RT, reverse transcriptase.
Stanford University HIV Drug-Resistance Database, interpretation of NRTI and NNRTI-resistance mutations (susceptible-, low-, and intermediate-level of drug resistance not shown).
NRTI-Resistance Mutations Among Patients Receiving HAART
Of the 28 study individuals who were on treatment, 11 (39%) harbored viruses that had at least one genotypic NRTI-resistance mutation conferring drug resistance (Table I). The viral RT sequences from all of these individuals contained the M184VI mutation, which is known to confer up to 300-fold reduced susceptibility to lamivudine (3TC) and a high level of resistance to emtricitabine (FTC) (http://hivdb.stanford.edu/index.html). Additionally, the thymidine analog mutation (TAM) T215Y, which confers high-level drug resistance to stavudine (d4T) and zidovudine (AZT), was found in the viruses infecting individuals 06ARC036, 07BDHS077, and 07LPH142 (Table I). The multi-NRTI-resistance mutation, Q151M, was found in the three patients receiving HAART; 06ARC007, 06BDHS005, and 06LPH20SC, of which only 06BDHS005 had the additional accessory mutations V75I, F77L, and F116Y. The combination of mutations found in individual 06BDHS005 form the Q151M mutation complex, which confers a greater level of resistance to abacavir (ABC), didanosine (DDI), d4T, and AZT. Moreover, the Q151M complex reduces 3TC susceptibility by 10-fold [Shafer et al., 1995]. Lacking this complex, individuals 06ARC007 and 06LPH20SC were predicted to have unaffected 3TC susceptibility yet a high level of resistance to ABC, DDI, d4T, and AZT (Table I).
The mutation K65R was detected in viruses from two individuals, 06ARC007 and 06ARC076. The virus infecting patient 06ARC076 contained no other NRTI-resistance mutations and was predicted to have intermediate resistance to all NRTIs (data not shown) except AZT, for which the K65R mutation has been shown to increase susceptibility [Delaugerre et al., 2005; Parikh et al., 2006; White et al., 2006]. Conversely, four additional NRTI-resistance mutations, A62V, F116Y, Q151M, and M184VI were identified for the virus infecting individual 06ARC007, indicating a high level of resistance to 3TC, FTC, ABC, d4T, and DDI (Table I). Overall analysis of NRTI-resistance mutations among the patients receiving HAART (n = 28) revealed a total of eight different mutations, including M184VI (39%) and T215Y (11%), conferring high-level resistance to 3TC and d4T, respectively. Both aforementioned ARVs are components of Triomune, the most frequently prescribed therapy in Cameroon. Of the remaining six NRTI mutations, five are considered multi-resistance mutations, including: F116Y (11%), Q151M (11%), A62V (7%), V75I (4%), and F77L (4%). The last mutation, K65R, was found in the virus from one individual (4%), conferring high-level resistance to ABC, DDI, and TDF.
NNRTI-Resistance Mutations Among Patients Receiving HAART
A total of 13 (46%) of the 28 patients receiving HAART were infected with viruses that harbored at least one NNRTI-resistance mutation, of which 11 concomitantly had at least one NRTI-resistance mutation (Table I). The most frequently observed NNRTI-resistance mutations were K103NS and Y181C, representing 14% and 18% of the viruses infecting patients receiving HAART in this study, respectively. Both of these mutations confer high-level resistance to nevirapine (NVP) and delaviridine (DLV), while additional resistance to either efavirenz (EFV) or etravirine (ETR) has been attributed to the mutation K103NS and Y181C, respectively. Also contributing to a high level of resistance to NVP, the G190A mutation was identified in viruses infecting three individuals, 06ARC036, 06LPH02NJM, and 06LPH17HT. Notably, patient 07LPH17HT was the only individual whose virus was predicted to have high-level resistance mutation to an NNRTI but lacked any NRTI-resistance mutation (Table I). Conferring high-level resistance to both NVP and DLV, the NNRTI-drug-resistance mutation V106A was identified in viruses infecting individuals 07BDHS077 and 07LPH149. The remaining four resistance mutations, conferring a lesser degree of resistance, V108I, V179E, Y188L, and F227L, were found among the patients receiving HAART, at frequencies of 7%, 7%, 4%, and 11%, respectively. The remaining 15 (54%) out of 28 patients receiving HAART had no major genotypic mutations (Table I); however, it was observed that these individuals had been on treatment for fewer months than those who had mutations, as the median months from the start of HAART was 10 months for this group compared to 14 months for the individuals harboring mutations.
NRTI- and NNRTI-Resistance Mutations Among Drug-Naive Individuals
Of the 21 drug-naïve individuals studied, 5 (24%) were infected with viruses that harbored one or more genotypic drug-resistance mutations, 2 of whom had mutations to both the NRTI and NNRTI classes of ARVs (Table II). Of interest, the viruses infecting the two individuals 06BDHS024 and 06BDHS028 harbored the M184VI mutation, conferring a high level of resistance to 3TC and FTC. Additionally, the NNRTI-resistance mutations K103NS and Y181C were identified in viruses from patients 06BDHS024 and 06BDHS028, respectively. These NNRTI-resistance mutations both confer high-level resistance to NVP and DLV, while mutation K103NS is also predicted to confer high-level resistance to EFV. In addition to the mutations described above that confer high-level resistance, two additional NNRTI-resistance mutations, V179E and G190A, were found in the viruses infecting individuals, 06ARC013 and 07BDHS054, respectively. These mutations are predictive of low- or intermediate-level drug resistance. Taken together, the presence of the NRTI- and NNRTI-resistance mutations in the viruses infecting the drug-naïve individuals suggests that these variants were transmitted, possibly from patients on HAART.
Analysis of HIV-1 Subtypes and Drug-Resistance Mutations
Phylogenetic analysis of the RT gene revealed a predominance of the CRF02_AG variant and a broad HIV-1 genetic diversity as follows: 73% CRF02_AG (n = 36), 6% D, 6% A, 4% F2, 2% CRF11_cpx, 2% CRF13_cpx, and 6% U (unclassifiable) (Tables I and II; Fig. 1).
Fig. 1.
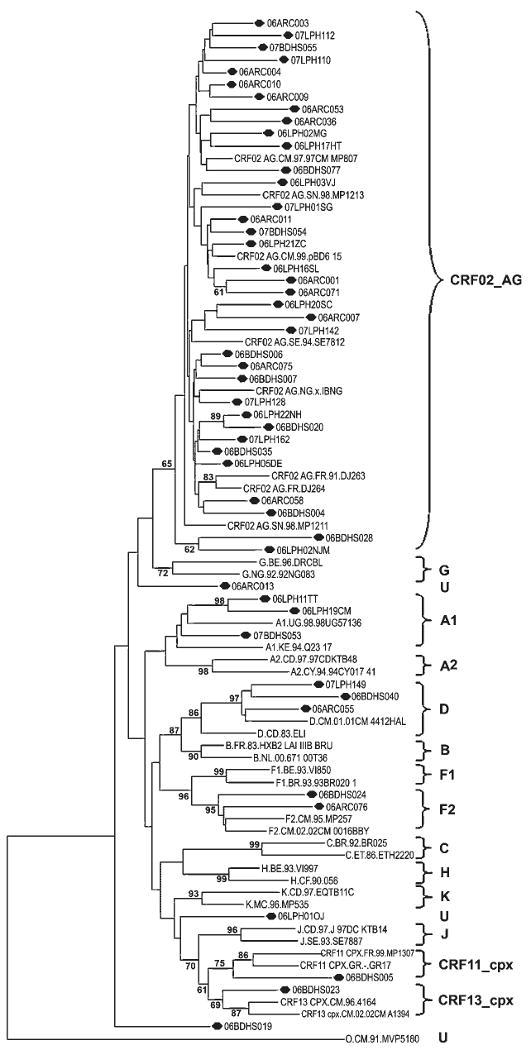
Phylogenetic tree of reverse transcriptase DNA sequences. The sequences analyzed are shown in bold italics. U, unclassifiable subtype. HIV-1 group M reference strains for subtypes A1, A2, B, C, D, F1, F2, G, H, J, K, CRF02_AG, CRF11_cpx, and CRF13_cpx were used to construct the tree using the neighbor-joining method of TREECON, Kimura's two-parameter method, mid-point rooting, and 1,000 bootstrap replicates.
First, a comparison of potential differences in the resistance mutations between patients receiving HAART who were infected with CRF02_AG (n = 24) or non-CRF02_AG (n = 4) viruses was performed (Table III). Overall, the genotypic-resistance mutations were comparable, notably the NRTI-resistance mutation M184VI was found with the most frequency in both groups of individuals. No single NNRTI-resistance mutation appeared to be more prevalent in either group. Of interest, some NRTI- and NNRTI-resistance mutations were found exclusively in one group of patients (those infected with CRF02_AG viruses vs. those infected with non-CRF02_AG viruses), however, given the small number of non-CRF02_AG viruses examined, more samples need to be tested in order to determine whether a differential pattern exists.
TABLE III.
HIV-1 Drug-Resistance Mutations by Viral Subtype (CRF02_AG vs. Non-CRF02_AG) Among HAART-Experienced Individuals
| Cameroon1 | Stanford University HIV Drug-Resistance DB2 | |||
|---|---|---|---|---|
| ARV class | Mutation | CRF02_AG (%), n = 24 | Non-CRF02_AG (%), n = 4 | CRF02_AG (%), n = 139 |
| NRTI | A62V | 1 (4) | 1 (25) | 1 (1) |
| K65R | 1 (4) | 0 (0) | 4 (3) | |
| V75I | 0 (0) | 1 (25) | 2 (1) | |
| F77L | 0 (0) | 1 (25) | 1 (1) | |
| F116Y | 2 (8) | 0 (0) | 2 (1) | |
| Q151M | 2 (8) | 1 (17) | 3 (2) | |
| M184V | 8 (33) | 3 (75) | 42 (30) | |
| T215Y | 3 (13) | 0 (0) | 10 (7) | |
| NNRTI | K103NS | 4 (17) | 1 (0) | 40 (29) |
| V106A | 1 (4) | 1 (25) | 2 (1) | |
| V108I | 2 (8) | 0 (0) | 6 (4) | |
| V179E | 2 (8) | 0 (0) | 3 (2) | |
| Y181C | 4 (17) | 1 (25) | 10 (7) | |
| Y188L | 0 (0) | 1 (25) | 5 (4) | |
| G190A | 3 (13) | 0 (0) | 4 (3) | |
| F227L | 2 (8) | 1 (25) | 1 (1) | |
The number and percentage of RTI-resistance mutations based on the analysis of
28 HAART-experienced individuals from Cameroon infected with RT subtype CRF02_AG or non-CRF02_AG (D or CRF11_cpx) were compared to
CRF02_AG-infected individuals on a HAART of two NRTIs and one NNRTI that were compiled using the “literature prevalence of mutations in submitted sequences” of the HIVseq program available at the Stanford University HIV Drug-Resistance Database website.
A comparison was also made of the RTI-resistance mutations among CRF02_AG-infected patients receiving HAART from the current study (n = 24) with a panel available in the “HIVseq: Literature Prevalence of Mutations in Submitted Sequences” program at the Stanford University HIV Drug-Resistance Database website was performed (Table III). A total of 139 sequences from CRF02_AG-infected individuals with an ARV history that included administration of both NRTIs and NNRTIs were examined. The CRF02_AG sequences compiled from the database revealed a similar NRTI-resistance mutation profile to those in the current study, whereby A62V, K65R F116Y, Q151M, and T215Y were all observed at a low frequency while M184VI was the most prevalent. Comparison of the NNRTI-resistance mutations revealed mutationY188L was absent among the CRF02_AG-infected individuals in the present study, yet occurred at a low frequency among the CRF02_AG-infected individuals in the database. Unlike the NRTI-resistance mutations, no single NNRTI-resistance mutation appeared to be more prevalent among the study individuals. However, this finding was not observed with the CRF02_AG sequences from the database, whereby the NNRTI-resistance mutation, K103NS was seen with the greatest frequency (29%). In addition, the NNRTI-resistance mutation Y181C was reported in 7% of the database CRF02_AG infections; while among the CRF02_AG-infected study individuals, a slightly higher percentage of 17% was observed.
Discussion
The current study reveals a high level of drug resistance among patients receiving HAART and drug-naïve individuals in Cameroon, demonstrated by 46% of the study individuals who were on HAART for a median period of 12 months harbored genotypic drug-resistance mutations. This finding is similar to that reported in other sub-Saharan African countries including Gabon (58%) [Vergne et al., 2002], Cote d'Ivoire (57%) [Adje et al., 2001], and Uganda (52%) [Richard et al., 2004]. In particular, 39% of the patients receiving HAART in this study were predicted to have resistance to an NRTI, and 46% to an NNRTI. Of note is that the 13 out of 28 patients receiving HAART with drug-resistance mutations were on treatment for a median of 14 months, while the remaining 15 individuals whose viruses did not harbor drug-resistance mutations had been on treatment for a median of only 9 months, a finding which is consistent with what is known about prolonged ARV pressure on HIV-1 and the frequency of drug-resistance mutations. This therefore suggests that the individuals who do not show evidence of drug-resistance mutations are likely to develop resistance within another year. Therefore, monitoring of such patients for the emergence of drug resistance is a necessity to better manage their disease course.
Studies conducted in Yaoundé, Cameroon from 2002 to 2004 [Laurent et al., 2006], found a lower level of prevalence of ARV resistance, whereby 16.4% of the participants had genotypic drug-resistance mutations after a median of 10 months, including 12.5% resistance to NRTIs, 10.2% to NNRTIs, and 2.3% to PIs [Laurent et al., 2006], even though treatment and laboratory expenses were paid by the patient—an obstacle to drug adherence that often leads to a higher rate of drug-resistance mutations. In both studies conducted in Cameroon, the median of months from the start of HAART prior to sampling was comparable (10 months vs. 12 months). However, important differences in treatment were observed, including the administration of PIs, and a lower number of individuals on suboptimal ART regimens in the Yaoundé study (2% vs. 10% dual-therapy; 0% vs. 3% mono-therapy). Several factors could account for the differences in the rate of mutations including prescription practices, drug availability, support programs, and the training of physicians in both Yaoundé and in the less urban settings of Cameroon.
The resistance mutation observed most frequently among the patients receiving HAART in this study was M184V/I, present in all the patients with NRTI-resistance mutations, conferring a high-level resistance to 3TC, and was the drug taken by 96% of the study individuals. The M184V/I mutation also confers resistance to emtricitabine (FTC), which potentially abrogates the use of this drug for salvage therapy. The remaining NRTI-resistance mutations among patients receiving HAART in the current study include K65R, Q151M, and T215Y representing resistance to all the remaining NRTIs currently approved by the FDA. In fact, 77% of the individuals in this study with drug-resistance mutations are infected with viruses that are resistant to two or more NRTIs. The mean viral load of 10,997 copies/ml for these individuals reflects the great number of drug-resistance mutations present and suggests the need for salvage therapy. Of note, three individuals, 06ARC007, 06ARC009, and 07BDHS040 had undetectable viral loads at the time of collection. Individuals 06ARC007 and 06ARC009 were previously taking Triomune for 3 months and 4 months, respectively, then switched therapy 5 months prior to sample collection.
In the present study among patients receiving HAART, the majority of mutations conferred NNRTI resistance; however, no single mutation predominated, in contrast to the NRTI-resistance mutations. The most predominant NNRTI-resistance mutations were Y181C (38%), which alone confers high-level resistance to NVP, DLV, and ETR, and mutation K103NS identified in 31% (4/13) individuals with drug-resistance mutations. Irrespective of NVP resistance, 25% of all patients receiving HAART and 9.5% of all drug-naïve individuals had mutations that confer high-level resistance to at least one of the remaining NNRTIs approved by the FDA, including DLV, EFV, and ETR. Therefore, the mutations identified here serve as clear markers for the emergence of drug-resistant variants in this population and point to potential alternative second-line treatment options with drugs that would be effective in controlling disease progression in such individuals.
Subtype analysis of the RT gene revealed a predominance of CRF02_AG viruses (73%; 36/49) among the study individuals with 86% (24/28) of the patients receiving HAART harboring this variant. A comparison of the drug-resistance mutations among the CRF02_AG and non-CRF02_AG variants revealed subtle differences in the frequency of specific mutations suggesting that there could be subtype-specific differences, which may confer resistance to different medications. For example, among NRTI-resistance mutations, V75I and F77L confer resistance to ABC, DDI, D4T, and ZDV and were present in the non-CRF02_AG strains but were absent in the CRF02_AG strains. Differences in mutational patterns that cause resistance to NNRTIs were also identified among the CRF02_AG and non-CRF02_AG strains. Though the number of non-CRF02_AG strains studied is small, others have reported similar findings. Soares et al. [2007], reported subtype differences in drug-resistance mutations among subtype B and C sequences of Brazilian HIV-1-infected patients on HAART. It was also striking to note differences in the mutational patterns between the Cameroon CRF02_AG and those available in the Stanford University HIV drug-resistance database, whereby the K103NS mutation that confers resistance to NNRTIs was present in 29% of the sequences from the database compared to 17% present among the Cameroonian sequences. Taken together, studies that examine subtype differences and the development drug-resistance mutations are urgently needed in the Central African region in particular, a region of the world that harbors the highest HIV-1 viral diversity and where first-line HAART use is increasing. Such studies are underway and will be reported elsewhere.
Data regarding the transmission of drug-resistance mutations in the current study found five (24%) drug-naïve individuals who harbored viruses with genotypic drug-resistance mutations to NRTIs and/or NNRTIs. Earlier studies conducted in Cameroon found the rate of drug-resistance mutation transmission among drug-naïve individuals ranging between 4.9% and 9.8% [Koizumi et al., 2006; Vergne et al., 2006; Vessiere et al., 2006; Ndembi et al., 2008]. The present analysis reveals an increase in the rate of transmission of viral variants with drug-resistance mutations, which could be due in part to the increasing availability of ARV therapy in the community and/or poor adherence to treatment. In addition, studies conducted in other African countries have found a lower frequency of RT drug-resistance mutations among drug-naive individuals including Mali (11.5%) [Derache et al., 2008], South Africa (3.6%) [Jacobs et al., 2008], and Tanzania (3% NRTI, 4% NNRTI) [Nyombi et al., 2008]. Taken together, this study reveals a high prevalence of drug-resistance mutations among patients receiving HAART (46%) and drug-naïve (24%) individuals in Cameroon. From a public health standpoint, it is critical to improve efforts to monitor the emergence of such mutations, to initiate a second-line HAART that includes a PI, and to monitor drug-naïve individuals to prevent the spread of resistant variants.
Nucleotide Sequence Accession Numbers
The DNA sequences of the HIV-1 RT region of Pol that were determined in this study were submitted to GenBank under the following accession numbers: GQ344955–GQ345003.
Acknowledgments
The authors are grateful to the individuals who have donated their specimens for these studies. The authors wish to acknowledge the continued support of the Ministry of Public Health, Cameroon. The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
References
- Adje C, Cheingsong R, Roels TH, Maurice C, Djomand G, Verbiest W, Hertogs K, Larder B, Monga B, Peeters M, Eholie S, Bissagene E, Coulibaly M, Respess R, Wiktor SZ, Chorba T, Nkengasong JN. High prevalence of genotypic and phenotypic HIV-1 drug-resistant strains among patients receiving antiretroviral therapy in Abidjan, Cote d'Ivoire. J Acquir Immune Defic Syndr. 2001;26:501–506. doi: 10.1097/00126334-200104150-00018. [DOI] [PubMed] [Google Scholar]
- Calmy A, Pinoges L, Szumilin E, Zachariah R, Ford N, Ferradini L. Generic fixed-dose combination antiretroviral treatment in resource-poor settings: Multicentric observational cohort. AIDS. 2006;20:1163–1169. doi: 10.1097/01.aids.0000226957.79847.d6. [DOI] [PubMed] [Google Scholar]
- Delaugerre C, Peytavin G, Dominguez S, Marcelin AG, Duvivier C, Gourlain K, Amellal B, Legrand M, Raffi F, Costagliola D, Katlama C, Calvez V. Virological and pharmacological factors associated with virological response to salvage therapy after an 8-week of treatment interruption in a context of very advanced HIV disease (GigHAART ANRS 097) J Med Virol. 2005;77:345–350. doi: 10.1002/jmv.20462. [DOI] [PubMed] [Google Scholar]
- Derache A, Maiga AI, Traore O, Akonde A, Cisse M, Jarrousse B, Koita V, Diarra B, Carcelain G, Barin F, Pizzocolo C, Pizarro L, Katlama C, Calvez V, Marcelin AG. Evolution of genetic diversity and drug resistance mutations in HIV-1 among untreated patients from Mali between 2005 and 2006. J Antimicrob Chemother. 2008;62:456–463. doi: 10.1093/jac/dkn234. [DOI] [PubMed] [Google Scholar]
- Gallant JE. Drug resistance after failure of initial antiretroviral therapy in resource-limited countries. Clin Infect Dis. 2007;44:453–455. doi: 10.1086/510752. [DOI] [PubMed] [Google Scholar]
- Geretti AM. Epidemiology of antiretroviral drug resistance in drug-naive persons. Curr Opin Infect Dis. 2007;20:22–32. doi: 10.1097/QCO.0b013e328013caff. [DOI] [PubMed] [Google Scholar]
- Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, Yeni PG, Jacobsen DM, Richman DD. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- Jacobs GB, Laten A, van Rensburg EJ, Bodem J, Weissbrich B, Rethwilm A, Preiser W, Engelbrecht S. Phylogenetic diversity and low level antiretroviral resistance mutations in HIV type 1 treatment-naive patients from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24:1009–1012. doi: 10.1089/aid.2008.0028. [DOI] [PubMed] [Google Scholar]
- Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug-resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- Koizumi Y, Ndembi N, Miyashita M, Lwembe R, Kageyama S, Mbanya D, Kaptue L, Numazaki K, Fujiyama Y, Ichimura H. Emergence of antiretroviral therapy resistance-associated primary mutations among drug-naive HIV-1-infected individuals in rural western Cameroon. J Acquir Immune Defic Syndr. 2006;43:15–22. doi: 10.1097/01.qai.0000226793.16216.55. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laurent C, Kouanfack C, Vergne L, Tardy M, Zekeng L, Noumsi N, Butel C, Bourgeois A, Mpoudi-Ngole E, Koulla-Shiro S, Peeters M, Delaporte E. Antiretroviral drug resistance and routine therapy, Cameroon. Emerg Infect Dis. 2006;12:1001–1004. doi: 10.3201/eid1206.050860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent C, Bourgeois A, Mpoudi-Ngole E, Ciaffi L, Kouanfack C, Mougnutou R, Nkoue N, Calmy A, Koulla-Shiro S, Delaporte E. Tolerability and effectiveness of first-line regimens combining nevirapine and lamivudine plus zidovudine or stavudine in Cameroon. AIDS Res Hum Retroviruses. 2008;24:393–399. doi: 10.1089/aid.2007.0219. [DOI] [PubMed] [Google Scholar]
- Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi VC, Sunpath H, Lu Z, Gordon M, Koranteng-Apeagyei K, Hampton J, Carpenter S, Giddy J, Ross D, Holst H, Losina E, Walker BD, Kuritzkes DR. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008;46:1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndembi N, Abraha A, Pilch H, Ichimura H, Mbanya D, Kaptue L, Salata R, Arts EJ. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: Evidence of major drug-resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol. 2008;46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyombi BM, Holm-Hansen C, Kristiansen KI, Bjune G, Muller F. Prevalence of reverse transcriptase and protease mutations associated with antiretroviral drug resistance among drug-naive HIV-1 infected pregnant women in Kagera and Kilimanjaro regions, Tanzania. AIDS Res Ther. 2008;5:13. doi: 10.1186/1742-6405-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh UM, Bacheler L, Koontz D, Mellors JW. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J Virol. 2006;80:4971–4977. doi: 10.1128/JVI.80.10.4971-4977.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard N, Juntilla M, Abraha A, Demers K, Paxinos E, Galovich J, Petropoulos C, Whalen CC, Kyeyune F, Atwine D, Kityo C, Mugyenyi P, Arts EJ. High prevalence of antiretroviral resistance in treated Ugandans infected with non-subtype B human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2004;20:355–364. doi: 10.1089/088922204323048104. [DOI] [PubMed] [Google Scholar]
- Shafer RW, Iversen AK, Winters MA, Aguiniga E, Katzenstein DA, Merigan TC. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. The AIDS Clinical Trials Group 143 Virology Team. J Infect Dis. 1995;172:70–78. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- Soares EA, Santos AF, Sousa TM, Sprinz E, Martinez AM, Silveira J, Tanuri A, Soares MA. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PLoS ONE. 2007;2:e730. doi: 10.1371/journal.pone.0000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Piyavong B, Chumpathat N, Chantratita W. Options for a second-line antiretroviral regimen for HIV type 1-infected patients whose initial regimen of a fixed-dose combination of stavudine, lamivudine, and nevirapine fails. Clin Infect Dis. 2007;44:447–452. doi: 10.1086/510745. [DOI] [PubMed] [Google Scholar]
- Vergne L, Malonga-Mouellet G, Mistoul I, Mavoungou R, Mansaray H, Peeters M, Delaporte E. Resistance to antiretroviral treatment in Gabon: Need for implementation of guidelines on antiretroviral therapy use and HIV-1 drug resistance monitoring in developing countries. J Acquir Immune Defic Syndr. 2002;29:165–168. doi: 10.1097/00042560-200202010-00009. [DOI] [PubMed] [Google Scholar]
- Vergne L, Diagbouga S, Kouanfack C, Aghokeng A, Butel C, Laurent C, Noumssi N, Tardy M, Sawadogo A, Drabo J, Hien H, Zekeng L, Delaporte E, Peeters M. HIV-1 drug-resistance mutations among newly diagnosed patients before scaling-up programmes in Burkina Faso and Cameroon. Antivir Ther. 2006;11:575–579. [PubMed] [Google Scholar]
- Vessiere A, Nerrienet E, Kfutwah A, Menu E, Tejiokem M, Pinson-Recordon P, Barre-Sinoussi F, Fleury H, Ayouba A. HIV-1 pol gene polymorphism and antiretroviral resistance mutations in drug-naive pregnant women in Yaounde, Cameroon. J Acquir Immune Defic Syndr. 2006;42:256–258. doi: 10.1097/01.qai.0000209909.20373.3b. [DOI] [PubMed] [Google Scholar]
- White KL, Chen JM, Feng JY, Margot NA, Ly JK, Ray AS, Macarthur HL, McDermott MJ, Swaminathan S, Miller MD. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir Ther. 2006;11:155–163. doi: 10.1177/135965350601100209. [DOI] [PubMed] [Google Scholar]
- WHO. Towards Universal Access: Scaling Up Priority HIV/AIDS Interventions in the Health Sector 2008 [Google Scholar]


