Abstract
Motivation: Selenoproteins are a group of proteins that contain selenocysteine (Sec), a rare amino acid inserted co-translationally into the protein chain. The Sec codon is UGA, which is normally a stop codon. In selenoproteins, UGA is recoded to Sec in presence of specific features on selenoprotein gene transcripts. Due to the dual role of the UGA codon, selenoprotein prediction and annotation are difficult tasks, and even known selenoproteins are often misannotated in genome databases.
Results: We present an homology-based in silico method to scan genomes for members of the known eukaryotic selenoprotein families: selenoprofiles. The core of the method is a set of manually curated highly reliable multiple sequence alignments of selenoprotein families, which are used as queries to scan genomic sequences. Results of the scan are processed through a number of steps, to produce highly accurate predictions of selenoprotein genes with little or no human intervention. Selenoprofiles is a valuable tool for bioinformatic characterization of eukaryotic selenoproteomes, and can complement genome annotation pipelines.
Availability and Implementation: Selenoprofiles is a python-built pipeline that internally runs psitblastn, exonerate, genewise, SECISearch and a number of custom-made scripts and programs. The program is available at http://big.crg.cat/services/selenoprofiles. The predictions presented in this article are available through DAS at http://genome.crg.cat:9000/das/Selenoprofiles_ensembl.
Contact: marco.mariotti@crg.es
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Selenoproteins are a rare class of proteins containing selenocysteine (Sec), an unusual amino acid which is a cysteine analog with selenium replacing sulfur. Specific machinery is needed for the recoding of the UGA codon (usually a stop codon) to Sec (Allmang et al., 2009; Hatfield et al., 2006; Xu et al., 2007). The main signal for UGA recoding is a RNA secondary structure element called SECIS (from SElenoCysteine Insertion Sequence) present in the 3′ UTR of eukaryotic selenoprotein gene transcripts (Copeland et al., 2001; Grundner-Culemann et al., 1999). Seleno-protein homologs (not containing Sec) have been found both as orthologs and paralogs. In most of them, a cysteine residue is aligned to Sec. There are currently 21 known families of selenoproteins in higher eukaryotes: Glutathione Peroxidases (GPx), Iodothyronine Deiodinase (DI), Selenoprotein 15 (Sel15 or 15kDa), Fish selenoprotein 15 (Fep15), SelM, SelH, SelI, SelJ, SelK, SelL, SelN, SelO, SelP, SelR, SelS, SelT, SelU, SelV, SelW, Thioredoxin Reductases (TR), SelenoPhosphate Synthetase (SPS). Some of these families may contain more than one member in a given genome (e.g. Homo sapiens contains 25 selenoproteins belonging to 17 families). All known selenoproteins contain just one Sec, with a few exceptions: SelP, SelN, some DI isoforms (Gromer et al., 2005), SelL (Shchedrina et al., 2007). In protists, selenoproteomes are variable, and recently some selenoprotein families limited to protist-specific lineages were identified (Cassago et al., 2006; Lobanov et al., 2006a, b; Novoselov et al., 2007; Obata and Shiraiwa, 2005). Some lineage-specific selenoprotein families have been identified in algae as well (Lobanov et al., 2009; Novoselov et al., 2002; Palenik et al., 2007). Selenoproteins' function is wide ranging, and still unknown for many families (Gromer et al., 2005; Lobanov et al., 2009).
During the last decade, several computational methods have been developed and used to identify novel selenoproteins (see Driscoll and Chavatte 2004 for a review; Jiang et al., 2010; Li et al., 2009; Zhang and Gladyshev, 2005). Most of these methods rely on the prediction of SECIS elements. A limitation of methods based on predicted SECISes is that they cannot identify selenoproteins with non-canonical SECIS elements, and they can be applied only to the species or taxonomic groups for which they were developed, since bacterial, archaeal and eukaryotic SECISes differ in their structure and also in their localization within the transcript (Krol, 2002). Also, SECIS prediction is problematic: while there is conservation of the secondary structure, the sequence is poorly conserved. Thus, genomic search for potential SECISes often lead to a large number of false positives (as well as, occasionally, some false negatives). Other strategies, not based on SECIS prediction, scan the target nucleotide sequence searching for ORFs with a conserved in frame UGA (Castellano et al., 2004; Jiang et al., 2010). These strategies also produces a large number of selenoprotein candidates in eukaryotic genomes. Like those based in SECIS searches, they require substantial manual curation. As a result, selenoprotein prediction is usually ignored in the standard genome annotation pipelines and selenoprotein genes are generally mispredicted, either by truncation of 3′ end of the gene (the UGA codon assumed to be the stop codon of the coding region), or by truncation of the 5′ end (the coding region assumed to start at the first AUG downstream of the UGA Sec codon), or by exclusion of the exon or the region containing the UGA/Sec codon. As the number of genome sequences available grows exponentially, automatic tools that produce high-quality genome annotations with minimal human intervention are essential. Here, we present a computational pipeline, which we name selenoprofiles, capable of producing reliable gene predictions for known eukaryotic selenoprotein families. Selenoprofiles can be used in conjunction with automatic gene annotation methods to predict otherwise misannotated selenoprotein genes in eukaryotic genomes. Importantly, selenoprofiles does not rely on the prediction of SECIS elements. Also, selenoprofiles does not rely on individual selenoprotein sequences to be used as initial queries, but on sequence profiles characteristic of each eukaryotic selenoprotein family. For each eukaryotic selenoprotein family, we have thus built an high-quality, manually curated multiple amino acid sequence alignment including all known orthologous and paralogous members of the family, and we derived a position-specific scoring matrix (PSSM) from it. Profiles derived from multiple sequence alignments implemented as PSSM, Markov models or other structures, capture more precisely the intrinsic variation within a protein family, and often lead to searches that are both more sensitive (thus allowing for the identification of distant relatives) and more specific (easing the identification of spurious hits) (Altschul et al., 1997). We show that selenoprofiles can be used with little or no human intervention to accurately identify known selenoproteins in eukaryotic genomes. Application of selenoprofiles to the publicly available reference annotation of metazoan genomes reveals hundreds of misannotated selenoprotein genes.
2 METHODS
2.1 Algorithm: the selenoprofiles pipeline
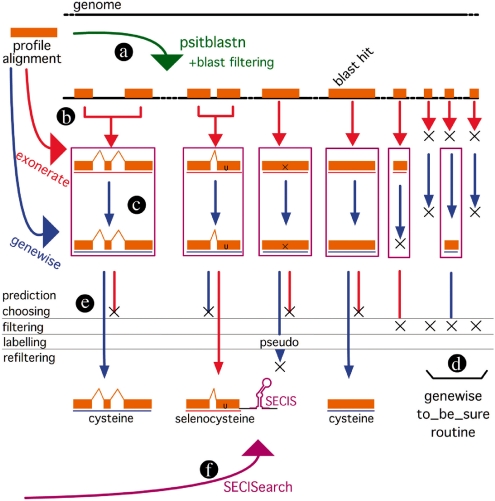
Selenoprofiles is a computational pipeline that, provided an alignment for a protein family, identifies all members of said family encoded in a target genome sequence. Selenoprofiles includes curated amino acid sequence alignments of all known eukaryotic selenoprotein families and selenoprotein factors. However, it can actually be used with alignments from any protein family. Technically, therefore, the pipeline is a general homology-based gene finder program with specific features that make it particularly suitable for selenoprotein identification. In selenoprofiles, the program psitblastn is used to identify matches in the target genome to the selenoprotein sequence alignments (Fig. 1a). These matches are then used, through two different splice alignment programs, exonerate (Slater and Birney, 2005, and Fig. 1b) and genewise (Birney et al., 2004, and Fig. 1c and d) to deduce the exonic structure of the candidate selenoprotein genes. The predictions of these two programs are analyzed to produce a final one (Fig. 1e). Finally, the program SECISearch (Kryukov et al., 1999) is used to identify suitable SECIS elements downstream of the coding region of the candidate selenoprotein genes (Fig. 1f). Through the entire pipeline, a number of steps are performed (detailed below) to filter out likely false positives and to keep the number of potential candidates under manageable levels. Next, we detail first the building of the selenoprotein profiles and then the different steps in the pipeline.
Fig. 1.
Schema of the selenoprofiles pipeline. Initially, a psitblastn search is run using a PSSM built from the profile alignment (a). The resulting genomic intervals are merged into ‘superexon’ intervals, and cyclic exonerate is run on each of them (b). Then, genewise is run both to refine exonerate predictions (c) and when exonerate failed recovering blast alignments (d—genewise ‘to be sure’ routine). The exonerate or the genewise prediction is chosen (e), and then results are filtered, labeled and then refiltered with family-specific filters. Lastly, SECISearch is used to detect potential SECIS elements downstream of the genes (f).
2.2 Multiple sequence alignments of protein families
Selenoprofiles includes amino acid sequence profiles for all known eukaryotic selenoproteins, as well as for all known selenoprotein-specific factors, that is, proteins involved in the synthesis of selenoproteins: SECIS binding protein 2 (SBP2), selenocysteine-specific elongation factor (eEFsec), O-phosphoseryl-tRNAsec kinase (PSTK), O-phosphoseryl-tRNAsec: selenocysteine synthase (SepSecS or just SecS), selenocysteine tRNA associated protein 43 (secp43) and SPSs (SPS1/SPS2). Searching for selenoprotein factors, in addition to the search for selenoproteins, is important because some of these factors appear to be good markers of selenoprotein encoding (Chapple and Guigó, 2008). While all selenoprotein factors (apart SPS2) are not selenoproteins themselves, and therefore their annotation does not suffer from the intrinsic limitations of selenoproteins, still the usage of selenoprofiles may result in a more accurate annotation than that produced by standard automatic annotation methods.
The seed sequences (one per family) to build the selenoprotein profiles were taken from SelenoDB (Castellano et al., 2008), a database of selenoproteins and selenoprotein factors. The human protein sequence was used when available. One exception was the SelK family, for which two distinct profiles were built, one utilizing the human sequence as seed and another utilizing the Drosophila sequence. This was necessary because this protein family is very divergent in insects. Also, the two selenoprotein families, SelV and SelW, were merged into a single profile alignment, since they share high sequence similarity (even though SelV possesses an additional N-terminal domain). Representative sequences from families not yet included in SelenoDB: SelJ, SelL, Fep15, were taken from the genomes where they were identified (see, respectively, Castellano et al., 2005; Novoselov et al., 2006; Shchedrina et al., 2007). For all families, the sequences used to build the profile were selected running the seed protein with either psiblast or blastp (Altschul et al., 1997) against nr (Sayers et al., 2010), with a very loose e-value filtering (max e-value = 1). The resulting sequences were aligned with the seed with t_coffee ver. 5.65 (Notredame et al., 2000). The alignment was then trimmed for redundancy with the t_coffee trim subroutine. Each alignment was then manually inspected and modified to remove spurious sequences or to add sequences that were missed during this process.
2.3 Finding matches to the selenoprotein profiles in the target genomes
In selenoprofiles, the multiple sequence alignments in input are compared with the sequence of the target genome using psitblastn, a member of the psiblast family of programs. This program is an extension of tblastn, that uses a protein PSSM to search nucleotide sequences translated in all six frames. While the psiblast programs are generally used to search iteratively a database and build an increasingly accurate profile, in this pipeline the profile is given as input, so a single search is performed against the target genome. Selenoprofiles utilizes psitblastn from the ncbi blastall package, version 2.2.22. The results of the search are filtered using the program alignthingie.pl (C.E.Chapple, personal communication). Three types of blast hits pass the filter: those in which the Sec position is aligned to a UGA codon, those hits in which it is aligned to a cysteine-coding codon, and all other hits whose e-value is below a certain threshold.
2.4 Inferring the exonic structure of the selenoprotein candidate genes
For each selenoprotein alignment, the output of the step above is a set of hits in the genomic sequence (genomic intervals), roughly corresponding to the exons of candidate selenoprotein genes (Fig. 1a). Each such genomic interval is used to initiate an iterative exonerate alignment that would ideally recover the entire exonic structure of the candidate selenoprotein gene. This initial structure may be subsequently refined through the usage of genewise, another splice alignment tool. Before running exonerate, the genomic intervals likely to correspond to exons of the same gene are merged in ‘superexon’ genomic intervals, to minimize subsequent computation (Fig. 1b). For two hits to be merged, one must align a region of the profile that is downstream of the region aligned in the other one, and also be located downstream along the genome sequence within a given distance.
2.4.1 Cyclic_exonerate
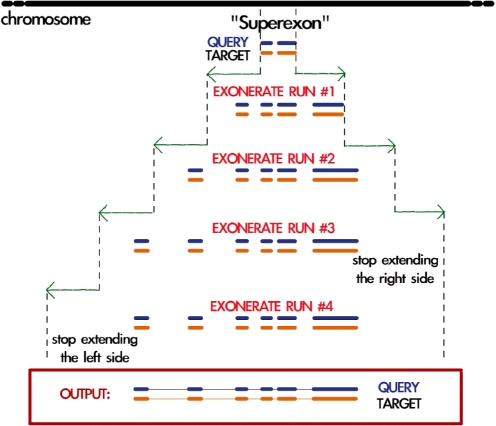
Exonerate is a generic tool for pairwise sequence comparison. Selenoprofiles utilizes exonerate version 2.2.0, in protein-to-genome mode, that aligns a single protein sequence (the query) to a nucleotide sequence (the target or subject), incorporating prediction of splice sites. Selenoprofiles runs exonerate in a peculiar way, hereafter described as the cyclic_exonerate routine (Fig. 2). We use this procedure to ensure that the whole gene structure of a candidate is found, without the need to use as subject the whole target chromosome and neither making a priori assumptions on the gene width. This method initially runs exonerate using as target the genomic interval defined in a blast hit (or in a ‘superexon’). It then runs exonerate again on the same interval extended at both ends, and compares the two alignments produced. In case that the second run of exonerate extends the coding sequence with respect to the first, then additional runs will be performed, as long as extending the genomic interval results in an extended gene structure prediction. If the extension parameter is chosen larger than the largest expected intron, the whole gene structure of the target should be detected.
Fig. 2.
Schema illustrating the cyclic exonerate routine. The program is run on a genomic interval initially defined by a blast hit (or a set of merged blast hits—‘superexon’), which is extended at each cycle. After each exonerate run (except the first one), the resulting prediction is compared with the previous one and the program decides whether to perform another run or not. Just before running exonerate (not displayed), the current alignment is mapped to the profile alignment and the query protein which is most similar to the target sequence is chosen. Although in the shown example, exonerate is run four times, cyclic exonerate runs on average 3.03 cycles (on well-assembled genomes such as the ones used for testing).
Exonerate can only take as protein query a single sequence—and not an entire alignment or a profile. At each run of exonerate, cyclic_exonerate thus maps the current query–target alignment into the profile alignment, and selects as a query the sequence in the profile which is the most similar to the predicted protein sequence. In selenoprofiles, only a subset of the sequences of the profile are allowed to be chosen as exonerate/genewise queries, since the profile may contain also incomplete sequences. Cyclic_exonerate launches exonerate with a custom scoring matrix (derived from BLOSUM62), which is favoring the extension of the alignment over Sec-encoding UGA codons: in the query protein selected from the profile, the position(s) aligned to Sec are replaced with a flag character (*). The custom scoring matrix contains positive values corresponding to the alignments of this character with * (any stop codon, Score 8), and with cysteine (Score 4), as well as with arginine (Score 2) and threonine (Score 1), since these amino acids have been found aligned to Sec in some eukaryotic selenoprotein families. The alignment of * with any other amino acid is scored with −4.
When multiple predictions are present in a exonerate output, only the main prediction (defined as the highest scoring among those overlapping the original input ‘superexon’) is reported by selenoprofiles. Often, however, exonerate fails to join predictions which actually belong to the same gene, because no canonical splice sites are found or because a region of the query sequence that would bridge the predictions is not found in the target sequence. Therefore, selenoprofiles uses secondary exonerate predictions to extend the main one: such predictions must align a region of the profile that is downstream (upstream) of the region aligned in the main one, and they should also be located downstream (upstream) in the genome sequence. That is, co-linearity needs to be maintained.
It is possible that more than one exon per gene initiates the exonerate cycle. In most of these cases, the procedure just described converges, leading to the choice of the same query protein and therefore to identical gene structure predictions. In a few cases, the procedure does not converge and slightly different gene structures are predicted. Exonerate predictions are processed to produce a unique gene structure per genomic loci: identical predictions are considered just once, and predictions which are completely included within the boundaries of another are discarded. Rarely, partially overlapping predictions, not including each other, are produced by this procedure. These will be output separately at the end of the pipeline. Note that there may be multiple non-overlapping exonerate predictions for a given selenoprotein profile, which could correspond to different members of the selenoprotein family. Selenoprofiles attempts next to refine the exonerate predictions through genewise.
2.4.2 Genewise
Genewise is a program belonging to the Wise2 package that performs a protein to DNA splice alignment (analogously to exonerate). Selenoprofiles utilizes genewise from Wise version 2.2.3. Generally, genewise is used to refine the gene boundaries of predictions already produced by exonerate (Fig. 1c). Sometimes, however, the exonerate routine seeded by a psitblastn hit (or by a ‘superexon’) produces no output. We also use genewise in these cases to produce a prediction on the genomic interval outlined by blast and extended by 10 000 bp on each side (genewise ‘to be sure’—see Fig. 1d). As with exonerate, the query sequence to be used is chosen from the profile alignment maximizing the sequence similarity to the predicted peptide sequence in the target. Genewise is run just once, so the query sequence in the standard routine is always the last one used by exonerate. When no exonerate output is available, the query sequence is chosen maximizing the similarity to the target peptide sequence predicted by psitblastn. Genewise can accept as query also a profile (not only a single sequence), in the form of a hidden Markov model (HMM). Nonetheless, selenoprofiles implements the use of genewise only with single protein queries, to keep the time of computation acceptable in genome-wide searches. As with exonerate, genewise is run with a custom scoring matrix favoring the alignment of Sec with UGA codons, with cysteine codons, or, with a lower score, with arginine or threonine codons. The query sequence chosen from the profile is replaced with a flag character (in this case U) in the positions that are aligned to Sec in the profile. In the case of genewise, though, it is possible to customize the program behavior to favor only the alignment of the U with UGA codons (not with other stop codons): this is accomplished by providing a different codon table to genewise, in which UGA codes for U.
2.4.3 Final prediction
At this point, selenoprofiles compares the genewise prediction with the prediction by exonerate, and chooses only one of them (Fig. 1e). In our experience, using the two programs instead of just one of them improves both the performance and the stability of the pipeline (Supplementary Section S3). Since the scores of the two programs are not comparable, selenoprofiles chooses the prediction with the longest protein sequence, unless it is likely to correspond to a pseudogene (i.e. it contains frameshifts or non-Sec coding stop codons), or it does not include a residue aligned to the Sec position(s) of the profile. In this case, the shorter prediction is chosen provided that it does not verify these two conditions. In our analysis, the genewise and exonerate predictions are identical in 27% of the cases. When they are different, selenoprofiles chooses the genewise prediction over the original prediction by exonerate in 68% of the cases. The final predictions are then filtered (see next section).
2.5 Filtering, labeling and outputting
Gene predictions are filtered so that only predictions spanning at least a given fraction of the profile alignment (40%) or longer than a given threshold (60 amino acids) are reported. All gene predictions that pass this filtering step are output, producing sequence files (in fasta format) and gene coordinate files [in General Feature Format (GFF), see http://www.sanger.ac.uk/resources/software/gff/spec.html]. Each gene prediction is labeled according to the codon that aligns to Sec in the profile. If a UGA codon is occurring at this position, the gene is labeled as ‘selenocysteine’. If another codon is occurring, the label takes the name of the correspondent amino acid (which is cysteine most of the times). There are some other possible labels, detailed in the caption of Figure 3.
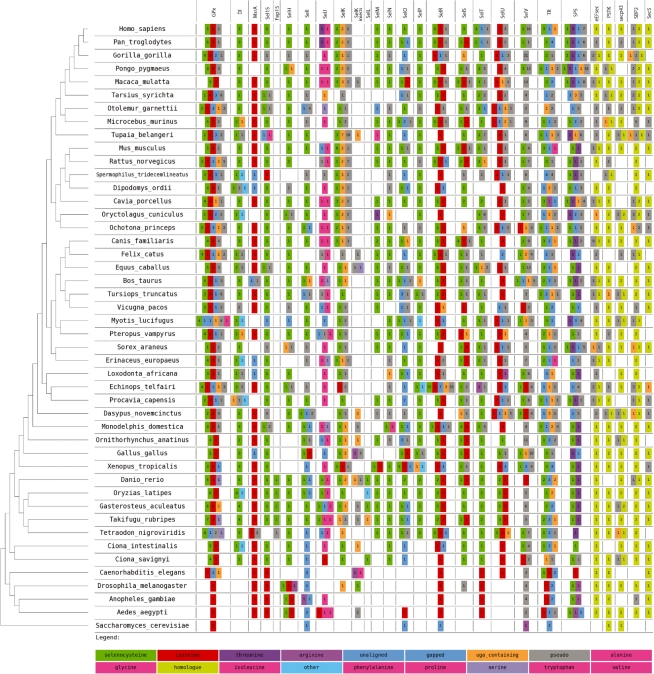
Fig. 3.
Graphical summary of non-redundant selenoprofiles predictions on all Ensembl genomes. The summary have been obtained with the program selenoprofiles_drawer. For each species, the numbers in the colored boxes indicate how many hits were found for each protein family (column) and label (box color). A color-to-label legend is located at the bottom: selenoproteins are in green, cysteine homologs in red and so on. Rare labels (such as ‘glutamine’, ‘tryptophan’ and ‘glutamic acid’) are all indicated with the pink color and cannot be differentiated in the picture. Hits labeled as ‘pseudo’ contain frameshifts or stop codons other than UGA (these were included in this figure although they are filtered out by selenoprofiles). The label ‘uga_containing’ is used when the only in-frame stop codon(s) are UGAs (not aligned to any Sec). This is useful since the scoring scheme rarely allows the alignment over a non-Sec encoding UGA. When no profile Sec position is aligned, the hit is labeled as ‘gapped’ in case the prediction aligns regions in the profile both upstream and downstream of the Sec position, ‘unaligned’ otherwise. The label ‘other’ is only for selenoprotein families with more than one Sec, when none of them are aligned to a UGA. The selenoprotein machinery families (not containing Sec) are on the right of the figure. The non-pseudo, non-uga_containing predictions for these families are labeled as ‘homologue’. A phylogenetic tree serves to indicate the evolutionary position of the investigated species (T.Gabaldón, personal communication). In the tree, three unresolved nodes were given an arbitrary topology for visualization purposes. This image can be downloaded at http://genome.crg.es/datasets/selenoprofiles2010/results_ensembl52.png.
2.6 SECISearch
Finally, selenoprofiles utilizes SECISearch version 2.0 (Kryukov et al., 1999), as adapted in (Chapple et al., 2009), to search for potential SECIS elements in the genomic region downstream from the predicted selenoprotein genes (Fig. 1f). By default, a region of 3000 bp is scanned. Initially, selenoprofiles attemps to find SECIS element matching the standard pattern, which fits both forms of eukaryotic SECISes (Krol, 2002). If no SECISes are found matching this pattern, SECISearch is run with two increasingly degenerate SECIS patterns (all patterns are reported in Supplementary Material, Section S1). It is possible that more than one SECIS is found in this way. It is also possible that no SECIS is found at all. Nevertheless, selenoprofiles does not drop a prediction for lacking a SECIS prediction. We believe that in most cases the occurrence of a UGA aligned to a Sec position of a known selenoprotein family is a very strong evidence for selenoprotein function. The lack of a detectable SECIS in the genomic region downstream of a real selenoprotein gene can be due to unusual features of the SECIS, and also to poor quality in the genome assembly, or to the presence of long and/or many introns in the 3′ UTR of the candidate.
2.7 Refiltering
Some profiles report false hits, either because the protein alignment for the family features poor sequence information (causing spurious hits along the genome), or because the family shares a certain degree of similarity with members of some other non-selenoprotein families (causing the profile to identify these genes). Through our experience with specific protein families, we have learnt to recognize these cases, and we have thus implemented a number of filters to identify, label and remove them. Filters are specific of each selenoprotein family. As an example, the refiltering for the SelV family is as follows. This family is characterized by a long, unstructured N-terminal domain showing very poor conservation, and a conserved C-terminal region. The N-terminal region sometimes causes this protein profile to produce many spurious hits in the genome. Through the refiltering, we ignore the hits that align only in this unstructured N-terminal region.
SelV: result_obj.label! = ‘pseudo’ and
result_obj.boundaries_in_profile()[1] >=300
2.8 Implementation
Selenoprofiles has been implemented in python. Selenoprofiles contains a number of profile alignments and scripts, including a program for graphical output: selenoprofiles_drawer.pl (Fig. 3). A Perl program (get_annotation.pl) is used when searching genomes with annotations in Ensembl. This program interrogates online the Ensembl database utilizing the Perl Core API, and retrieve the most similar annotation in Ensembl to each selenoprofile prediction. The database releases for all species considered in this article are reported in Supplementary Table S2. The code and manual of selenoprofiles is available at http://big.crg.cat/services/selenoprofiles. Selenoprofiles scanned the human genome for all the 27 implemented families in 1100 min (∼18 h) in a computer equipped with two double-core Intel(R) Xeon(TM) processors (2.80 Hz) and 4 GB of RAM. About 46% of the time was spent on the SelV family alone.
3 RESULTS
3.1 Evaluation of selenoprofiles
We have tested the performance of selenoprofiles on the genomes of Homo sapiens (25 selenoproteins and 5 selenoprotein factors), Drosophila melanogaster (3 selenoproteins and 5 selenoprotein factors) and Saccharomyces cerevisiae (no selenoproteins and no selenoprotein factors), since these genomes are well annotated in Ensembl and have all entries in SelenoDB. We ran selenoprofiles removing pre-emptively all sequences belonging to the tested species from the profiles alignments. In addiction to the families already mentioned, we included the methionine sulfoxide reductase A (MsrA) family as well, since this family is included in SelenoDB (although it was found as selenoprotein only in Chlamydomonas reinhardtii; Novoselov et al., 2002). Overall, selenoprofiles found 27 out of the 28 selenoprotein genes, 10 out of 10 selenoprotein factor genes, and 26 out of 28 annotated selenoprotein homologs. The three genes missed by selenoprofiles are Drosophila SelK2, and human SelW1 and SelW2.
SelK2 is a cysteine homolog of SelK, and is located adjacent to it on the fly genome, confounding selenoprofiles. The human SelW proteins (the selenoprotein SelW1 and the cysteine homolog SelW2) have an exon structure made of very short exons which produces, in the psitblastn search, e-values that are higher than the threshold. The sequences are correctly predicted when searching the ncbi human ESTs database with selenoprofiles (data not shown).
For selenoproteins (meaning in this case all predictions labeled ‘selenocysteine’), selenoprofiles produced no false positives in the yeast and Drosophila genomes (Supplementary Table S3). In the human genome, five selenoprotein genes that were not present among Ensembl or SelenoDB annotations were predicted—these are very likely to be false positives (Supplementary Section S5). Regarding the selenoprotein machinery, four false positives in total were predicted by selenoprofiles in the three genomes (Supplementary Section S5). For non-Sec homologs of selenoproteins, more false positives were predicted (Supplementary Table S3). Their number depends mostly on the protein family considered (i.e. on the effectiveness of the refiltering steps specific to that family).
In addition to assessing whether selenoprofiles were able to identify the selenoprotein and machinery genes in complete genomic sequences, we also evaluated the quality of the exonic structure inferred by selenoprofiles for these genes. Predicted and annotated gene structures were compared and the usual measures of sensitivity and specificity at gene, exon and nucleotide level (Burset and Guigó, 1996) were computed using the script evaluation.pl (E.Eyras, personal communication). The details of the procedure and the results appear in Supplementary Table S3. Overall, accuracy values are comparable (or even higher) with those obtained through the most accurate automatic gene annotation pipelines: for selenoproteins, both the average sensitivity and the average specificity at the nucleotide level are >90%.
3.2 Using selenoprofiles to identify selenoproteins in eukaryotic genomes
To further assess both selenoprofiles and the current status of selenoprotein annotation in eukaryotic genomes, we ran selenoprofiles on all 46 currently available Ensembl genomes (all eukaryotes). Eight hundred and thirty-seven selenoprotein genes, 925 non-Sec homologs and 236 selenoprotein factors were found. A summary of the results is given in Figure 3. The figure, produced by the program selenoprofiles_drawer, lists the selenoprotein families found in the analyzed genomes and the number of genes in each family, indicating whether these are selenoproteins, cysteine homologs or contain other amino acids at the Sec position. Consistent with our assessment in the human, fly and yeast genomes, results indicate that, while selenoprofiles finds most of the known selenoprotein genes, it also misses some of them. This is due in part to limitations of the profiles, but mostly to the quality of the genome sequence.
For example, the mosquitoes Aedes aegypti and Anopheles gambiae are known to possess the selenoprotein SelK (Chapple and Guigó, 2008), but their protein sequence is quite distant from both Drosophila SelK (used to seed the SelK_insect profile) and vertebrate SelK (with human SelK used to seed the SelK profile). Consequently, the annotated SelK is missed in these two genomes by both SelK profiles searches. Other genes are missed in the psitblastn search because of the e-value of the alignment is above the threshold. In other cases, selenoproteins are not found because of incompleteness in the genome sequence. Thus, no SPS2 is predicted by selenoprofiles in Gallus gallus genome, but this gene can be easily found searching the EST data available at ncbi for this organism (data not shown). Other cases of genes that we expect to be present, but are missed by selenoprofiles correspond to predictions labeled as pseudogenes, because of frameshift(s) or inframe stop codons. This happens with selenoprotein families as well with machinery proteins (e.g. SecS in Microcebus murinus and PSTK in Rattus norvegicus). Since all Ensembl species (apart from S.cerevisiae) possess selenoproteins and therefore must have the necessary machinery, we believe this suggests the occurrence of sequencing errors in the genomes. Many genomes included in Ensembl are characterized by low coverage, and this is known to heavily affect the inferences on gene presence in such species (Milinkovitch et al., 2010). Out of the 837 selenoproteins predicted by selenoprofiles, 658 of them contain a putative SECIS elements. We find a correspondent gene annotation in Ensembl for 604 of them. In 66 cases, the gene was correctly annotated as a selenoprotein. Given the low false positive rate of selenoprofiles, most of the 771 remaining cases are likely to correspond to misannotations. For the 233 cases in which no correspondent Ensembl annotation was found, we believe that the in-frame UGA confounded the Ensembl annotation pipeline to the point that no annotation at all was produced. Among the 538 remaining cases, we observed a few recurrent patterns of misannotation: in 154 (28.6%) cases, the annotated coding region in Ensembl ends exactly at the Sec-UGA site (mostly for families with a C-terminal Sec), while in 100 (18.6%) cases starts downstream of it (for families with a N-terminal/central Sec). In 231 (42.9%) cases, there is a deletion in the annotated coding region compared with the selenoprofiles prediction that includes the Sec-UGA codon. Often the deletion eliminates only this codon through the annotation of a 3 bp intron. The 53 (9.9%) remaining cases do not fall in any of the previous categories. A list of the misannotated genes for each category is provided as Supplementary Material. Selenoprofiles predictions on all Ensembl genomes can be accessed through DAS at http://genome.crg.cat:9000/das/Selenoprofiles_ensembl.
4 DISCUSSION
In spite of significant advances, gene annotation of newly sequenced genomes remains a challenging task. While manual curation is still essential to produce high-quality gene and transcript annotations (Guigó et al., 2006), automatic genome annotation pipelines produce increasingly accurate gene sets (Harrow et al., 2009), in particular for well-characterized protein coding families and when other well-annotated evolutionary close genomes exist. Due to their peculiar recoding of the standard genetic code, selenoproteins constitute the most notable exception; even for well-annotated genomes, they are often mispredicted. Indeed, as we have shown through the analysis described here, most eukaryotic selenoproteins are misannotated in the available reference gene sets. Since misannotation invariably involves the deletion of the region in the protein sequence including the Sec-UGA—key to proper family assignation—misprediction in the case of selenoproteins have the additional negative effect, beyond simply protein truncation, of impairing proper functional characterization.
Proper annotation of selenoprotein genes—even those belonging to well-characterized protein families—requires substantial human intervention. Indeed, due to the degeneration of the sequence of the SECIS element, and to the complex evolutionary history of selenoprotein genes, with frequent gene duplications and family expansions, pseudogenizations, and the yet not completely understood evolutionary dynamics of Cys to Sec interconversion (Castellano et al., 2009), detection of sequence homology is, in general, not sufficient for correct selenoprotein identification. In fact, the correct annotation of the two dozen (at the most) selenoprotein genes corresponding to known selenoprotein families, which may be encoded in a newly sequenced eukaryotic genome takes, in our experience, 2–3 weeks of full-time work of an experienced scientist. He/she has to browse through a maze of multiple sequence alignments and SECIS predictions, making often ad hoc decisions, which generally involve running additional, more sophisticated alignment programs and post-processing their output. In selenoprofiles, we have attempted to encapsulate the experience that we have accumulated during the years in manual identification of selenoproteins. Selenoprofiles includes standard sequence similarity search and sequence alignment programs together with custom made post-processing scripts and a number of rules that direct the overall flow of the process. The core of selenoprofiles is a set of very high-quality multiple sequence alignments for the different selenoprotein families and subfamilies. Given that we know a priori which positions in a profile alignment are allowed to bear a selenocysteine, selenoprofiles favors the alignment to UGA codons only if these are aligned to one such position. Therefore, an important feature of each profile alignment is the position or positions that contain Sec, and one of the major determinants of the efficiency of the selenoprofiles pipeline are the species and the subfamilies represented in the profile. Selenoprofiles automatically selects the best sequence to be used as query from the profile. Consequently, if the profile contains at least one sequence that is very similar to the protein coded by the gene that is predicting, the prediction will be accurate. But if the most similar sequence in the profile differs from the real protein encoded in the investigated genome in the presence or absence of some domains, or if there is poor conservation between the two sequences at some regions (often at one or both ends), then the prediction may be inaccurate. Input profile alignments for selenoprofiles should, therefore, be as consistent, complete and representative as possible. In this regard, as new genomes are being analyzed, we keep updating selenoprofiles, and we are working in a procedure to substantially automate this updating.
While selenoprofiles does not completely eliminate the need for manual intervention, it dramatically reduces it. We estimate that, after running selenoprofiles on a (newly sequenced) genome, an experienced scientist will need, in general, only a few hours to produce a high-quality annotation of the selenoprotein genes corresponding to known families in the genome. But, given its low false positive rate, even the default output of selenoprofiles will generally be a much superior annotation of selenoprotein genes than that produced by automatic annotation pipelines—including the most sophisticated ones. In this regard, we believe that selenoprofiles would be a useful complement of such pipelines, and we are working on a method to automatically correct the misannotated selenoproteins taking into account the selenoprofiles output. Using directly this output may not be an option, since sophisticated annotation pipelines rely on transcript information (such as ESTs and cDNA sequences), as well as genomic sequence conservation across species, and the overall gene structure delineated using this information is likely to be superior to the one delineated by selenoprofiles, with the exception of the region including the Sec-UGA. Therefore, a better strategy will be to conciliate the selenoprofiles prediction with the annotated gene, giving predominance to the selenoprofiles prediction in the region (exon) containing the Sec-UGA, but to the annotated prediction in the rest of the gene/transcript.
One limitation of selenoprofiles is that it predicts, with a few exceptions only one transcript per gene. Nonetheless, if alternative splicing forms (Sec/non-Sec) exist for a gene, the pipeline is likely to pick the Sec containing transcript, or one of them, due to the scoring scheme used. If selenoprofiles is used on transcribed sequences (such as ESTs, cDNAs, or RNA sequences) instead of genomic sequences, it could potentially produce predictions for multiple splicing isoforms of selenoprotein genes. While we have developed and tested selenoprofiles to annotate eukaryotic selenoproteomes, the strategy that we have employed can be easily ported to prokaryotic genomes as well. This requires the building and curation of the corresponding profiles, the usage of the bacterial and archaeal SECIS patterns, and the modification of some of the selenoprofiles rules.
5 CONCLUSIONS
Selenoprofiles is an homology-based method to produce accurate predictions of known selenoprotein families, and can be used in conjunction with automatic annotation pipelines. Running selenoprofiles on all available eukaryotic genomes reveals hundreds of misannotated selenoprotein genes. Selenoprofiles predictions constitute the largest available collection of eukaryotic selenoproteins, and are in this regard, an invaluable resource for selenoprotein research.
Supplementary Material
ACKNOWLEDGEMENTS
We thanks Toni Gabaldón for providing a phylogenetic tree of all species present in Ensembl. Thanks also to Eduardo Eyras for the script evaluation.pl to test the performances of selenoprofiles. Finally, a special thanks to Charles E. Chapple for his script alignthingie.pl and for endless support.
Funding: Spanish Ministerio de Educacion y Ciencia (grant BIO2006-03380); National Institutes of Health/National Human Genome Research Institute (grant 1U54HG004555).
Conflict of Interest: none declared.
REFERENCES
- Allmang C, et al. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim. Biophys. Acta (BBA) Gen. Subj. 2009;1790:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Altschul S, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, et al. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Guigó R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- Cassago A, et al. Identification of Leishmania selenoproteins and SECIS element. Mol. Biochem. Parasitol. 2006;149:128–134. doi: 10.1016/j.molbiopara.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Castellano S, et al. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, et al. Diversity and functional plasticity of eukaryotic selenoproteins: identification and characterization of the SelJ family. Proc. Natl Acad. Sci. USA. 2005;102:16188. doi: 10.1073/pnas.0505146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, et al. SelenoDB 1.0: a database of selenoprotein genes, proteins and SECIS elements. Nucleic Acids Res. 2008;36:D332–D338. doi: 10.1093/nar/gkm731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, et al. Low exchangeability of selenocysteine, the 21st amino acid, in vertebrate proteins. Mol. Biol. Evol. 2009;26:2031. doi: 10.1093/molbev/msp109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CE, Guigó R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE. 2008;8 doi: 10.1371/journal.pone.0002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CE, et al. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics. 2009;25:674–675. doi: 10.1093/bioinformatics/btp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland P, et al. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell Biol. 2001;21:1491. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Chavatte L. Finding needles in a haystack. In silico identification of eukaryotic selenoprotein genes. EMBO Rep. 2004;5:140–141. doi: 10.1038/sj.embor.7400080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromer S, et al. Human selenoproteins at a glance. Cell Mol. Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner-Culemann E, et al. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA. 1999;5:625–635. doi: 10.1017/s1355838299981542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R, et al. EGASP: the human ENCODE genome annotation assessment project. Genome Biol. 2006;7(Suppl. 1):S2.1–S231. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, et al. Identifying protein-coding genes in genomic sequences. Genome Biol. 2009;10:201. doi: 10.1186/gb-2009-10-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D, et al. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- Jiang L, et al. In silico identification of the sea squirt selenoproteome. BMC Genomics. 2010;11:289. doi: 10.1186/1471-2164-11-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- Kryukov G, et al. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 1999;274:33888. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- Li M, et al. A method for identification of selenoprotein genes in archaeal genomes. Genomics Proteomics Bioinformatics. 2009;7:62–70. doi: 10.1016/S1672-0229(08)60034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, et al. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov A, et al. Selenium metabolism in Trypanosoma: characterization of selenoproteomes and identification of a Kinetoplastida-specific selenoprotein. Nucleic Acids Res. 2006a;34:4012. doi: 10.1093/nar/gkl541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov A, et al. The plasmodium selenoproteome. Nucleic Acids Res. 2006b;34:496. doi: 10.1093/nar/gkj450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinkovitch MC, et al. 2x genomes - depth does matter. Genome Biol. 2010;11:R16. doi: 10.1186/gb-2010-11-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, et al. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Novoselov S, et al. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov S, et al. Identification and characterization of Fep15, a new selenocysteine-containing member of the Sep15 protein family. Biochem. J. 2006;394:575. doi: 10.1042/BJ20051569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov S, et al. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc. Natl Acad. Sci. USA. 2007;104:7857–7862. doi: 10.1073/pnas.0610683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Shiraiwa Y. A novel eukaryotic selenoprotein in the haptophyte alga Emiliania huxleyi. J. Biol. Chem. 2005;280:18462. doi: 10.1074/jbc.M501517200. [DOI] [PubMed] [Google Scholar]
- Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchedrina V, et al. Identification and characterization of a selenoprotein family containing a diselenide bond in a redox motif. Proc. Natl Acad. Sci. USA. 2007;104:13919. doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G.SC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 2007;404:115. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.