Abstract
Summary: Protein–protein interaction detection methods are applied on a daily basis by molecular biologists worldwide. After generating a set of potential interactions, biologists face the problem of highlighting the ones that are novel and collecting evidence with respect to literature and annotation. This task can be as tedious as searching for every predicted interaction in several interaction data repositories, or manually screening the scientific literature. To facilitate the task of evidence mining and novelty assessment of protein–protein interactions, we have developed a Cytoscape plugin that automatically mines publication references, database references, interaction detection method descriptions and pathway annotation for a user-supplied network of interactions. The basis for the annotation is ConsensusPathDB—a meta-database that integrates numerous protein–protein, signaling, metabolic and gene regulatory interaction repositories for currently three species: Homo sapiens, Saccharomyces cerevisiae and Mus musculus.
Availability: The ConsensusPathDB plugin for Cytoscape (version 2.7.0 or later) can be installed within Cytoscape on a major operating system (Windows, Mac OS, Unix/Linux) with Sun Java 1.5 or later installed through Cytoscape's Plugin manager (category ‘Network and Attribute I/O’). The plugin is freely available for download on the ConsensusPathDB web site (http://cpdb.molgen.mpg.de).
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: kamburov@molgen.mpg.de
1 INTRODUCTION
Due to the high explanatory power of protein–protein interactions for biological processes in health and disease (Ideker and Sharan, 2008), dedicated interaction detection methods like yeast-two-hybrid (Y2H) screening (Fields, 2005) and co-purification (Aebersold and Mann, 2003) are applied on a daily basis by molecular biologists worldwide and contribute to the completion of the map of protein–protein interactions for human and other species. An immediate task after generating a network of predicted interactions is to identify the ones that have not been published previously and to collect evidence for every single interaction from literature and annotation. This information is useful in order to estimate the performance of the interaction screen and to assess the contribution to the protein–protein interaction map of the species in question. To accomplish this task, biologists typically search their new data against every single protein–protein interaction repository like IntAct (Huntley et al., 2007) or MINT (Chatr-aryamontri et al., 2007). Even more tedious is the manual mining for interactions in scientific literature to collect the publication references and detection methods for the novel interaction list.
Cytoscape (Shannon et al., 2003) is a widely used, freely available software tool for visualization, manipulation and analysis of biomolecular interaction networks. To aid the process of interaction evidence mining, we have developed a plugin for Cytoscape that searches all interactions from the network of interest in the interaction space stored in ConsensusPathDB. ConsensusPathDB (Kamburov et al., 2009) is an interaction meta-database that integrates functional interaction repositories forming a heterogeneous interaction network which comprises protein–protein interactions, as well as signaling, metabolic and gene regulatory interactions. Currently, the database integrates 18 open-access repositories on human interactions and eight repositories for both yeast and mouse interactions and contains around 150 000 human, 195 000 yeast and 13 000 mouse distinct interactions (many of which are of non-binary nature, i.e. contain more than two interaction partners). In this article, we describe the functionality of the ConsensusPathDB plugin for Cytoscape and demonstrate its usage and performance.
2 DESCRIPTION
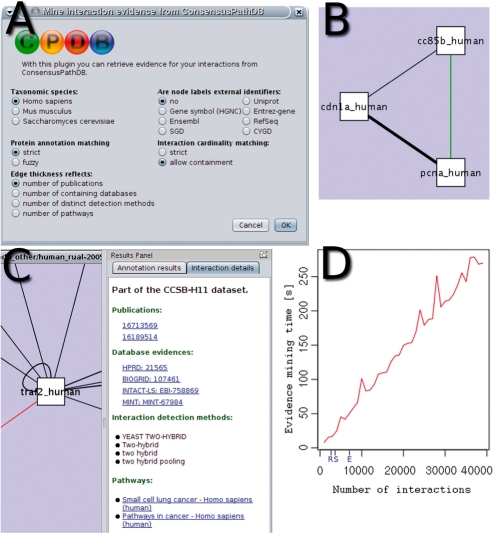
After installing the plugin, the user starts by loading the network of interest (denoted query network) represented by binary interactions in Cytoscape and launching the ConsensusPathDB plugin through Cytoscape's ‘Plugins’ menu (Fig. 1A). After setting a few parameters which we describe below, the user starts the evidence mining process. The plugin then communicates with the repository of ConsensusPathDB through a web service. Once the plugin sends the query network to the server, a search is executed on the server-side for all (or, optionally, just the selected) proteins and interactions from the query network in ConsensusPathDB through SQL queries. Proteins from the query network are matched to the data repository on the basis of accession numbers such as UniProt (The UniProt Consortium, 2010) or Ensembl (Flicek et al., 2010). Interactions from the query network are matched to the repository based on their participants.
Fig. 1.
(A) The splash screen of the plugin showing the different parameters; (B) the ConsensusPathDB visual style where reproduced interactions are weighted by evidence and novel interactions are highlighted in green; (C) newly imported attributes of a selected interaction are shown in the ‘Interaction details’ tab of Cytoscape's results panel; (D) evidence mining time plot for networks of different size with default parameters (here, all query interactions were present in ConsensusPathDB such that the mining process took maximal time). The sizes of the networks predicted using large-scale interaction screening by Rual et al. (2005) (R), Stelzl et al. (2005) (S) and Ewing et al. (2006) (E) are marked on the x-axis.
The performance of the interaction matching depends critically on how well proteins in the query network are annotated with accession numbers. In the case that accession numbers are not available, the user is prompted to specify whether the node labels represent accession numbers of a certain type. The interaction matching performance is influenced by two parameters, ‘protein annotation matching’ (strict/fuzzy) and ‘interaction cardinality matching’ (strict/allow containment). Strict protein annotation matching denotes that a protein from the query network and a protein from the database are considered identical only if all identifiers of a type match. Fuzzy matching means that the identifiers of the query protein may form a sub-set of the identifiers of the database counterpart or vice versa. Fuzzy matching is useful, e.g. when proteins on the one side are compared with protein families on the other side. The ‘interaction cardinality matching’ parameter specifies whether the binary interactions from the query network should be matched only with binary interactions from the database network (strict matching) or whether they may be matched to complex interactions, i.e. interactions of more than two proteins that contain the binary interactions. More details about protein and interaction mapping can be found in the Supplementary Material to this paper.
After matching proteins and interactions, the web service server sends annotation attributes for matched query interactions in the form of publication references (Pubmed identifiers), interaction detection methods, database references (such as IntAct and MINT) and pathway annotations (i.e. pathways that contain both participants of a protein–protein interaction) to the client plugin. The plugin creates a custom visual style in Cytoscape where the thickness of interaction edges reflects (optionally) the number of publications, number of containing interaction databases, number of distinct detection methods, or number of containing pathways for the protein interaction (Fig. 1B). Interactions that are not found in the repository, and thus represent potential novel interactions, are highlighted in green. In the results tab of Cytoscape, an interaction mapping summary is displayed together with a legend. The interaction attributes that have been retrieved from ConsensusPathDB can be viewed for selected interactions under the ‘Interaction details’ tab of the results panel (Fig. 1C). If applicable, this information is provided as web links to the primary data and can be viewed in a web browser.
Figure 1D shows the performance of the plugin implementation with respect to the mining of interaction annotation for different network sizes. Results show that even for large networks evidence mining executes in minutes, for example ∼2 min for a network with 20 000 nodes. It should be noted, however, that the Internet connection speed of the client influences the overall speed of interaction matching.
Funding: Max Planck Society (IMPRS-CBSC); Cytoscape Project (GM070743); European Union's project APO-SYS (HEALTH-F4-2007-200767); BMBF MedSys project PREDICT (0315428A).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2006;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 2005;272:5391–5399. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Sharan R. Protein networks and disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, et al. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.