Abstract
CYP130 is one of the 20 Mycobacterium tuberculosis cytochrome P450 enzymes, only two of which, CYP51 and CYP121, have so far been studied as individually expressed proteins. Herein we characterize a third heterologously expressed Mycobacterium tuberculosis cytochrome P450, CYP130, by UV-visible spectroscopy, isothermal titration calorimetry, and x-ray crystallography, including determination of the crystal structures of ligand-free and econazole-bound CYP130 at a resolution of 1.46 Å and 3.0 Å, respectively. Ligand-free CYP130 crystallizes in an ‘open’ conformation as a monomer, whereas the econazole-bound form crystallizes in a ‘closed’ conformation as a dimer. Conformational changes enabling the ‘open-closed’ transition involve repositioning of the BC loop and the F and G helices that envelop the inhibitor in the binding site and reshape the protein surface. Crystal structure analysis shows that the portion of the BC-loop relocates as much as 18 Å between the open and closed conformations. Binding of econazole to CYP130 involves a conformational change and is mediated by both a set of hydrophobic interactions with amino acid residues in the active site and coordination of the heme iron. CYP130 also binds miconazole with virtually the same binding affinity as econazole and clotrimazole and ketoconazole with somewhat lower affinities, which makes it a plausible target for this class of therapeutic drugs. Overall, binding of the azole inhibitors is a sequential two-step entropy-driven endothermic process. Binding of econazole and clotrimazole exhibits positive cooperativity that may reflect a propensity of CYP130 to associate into a dimeric structure.
The pathogenic bacterium Mycobacterium tuberculosis (Mtb)1 continues to be an enormous threat to human health. It is responsible for more deaths worldwide than any other infectious agent, and is the major cause of death for HIV-infected individuals in developing countries. An aggravating factor associated with the global resurgence of tuberculosis is the proliferation of strains resistant to isoniazid and rifampicin, the two major frontline antitubercular drugs. Therefore, new drug strategies are needed to combat the rising incidence of tuberculosis, especially the multidrug-resistant forms, and to shorten the duration of tuberculosis treatment (1).
It has been demonstrated that azole drugs such as econazole and clotrimazole, which inhibit the sterol 14α-demethylase CYP51 and were originally developed as fungal antibiotics (2), display inhibitory potential against the latent and multidrug-resistant forms of tuberculosis both in vitro and in tuberculosis infected mice (3–7). Furthermore, econazole exhibits synergistic activities with rifampicin and isoniazid against the multidrug-resistant Mtb strains (3). The 4.4 Mb Mtb genome encodes 20 different cyp genes (8), whose biological roles are not yet understood. To date, physiological roles have been proposed for CYP125 and CYP142 in cholesterol catabolism (9), and for CYP132 in fatty acid metabolism (10). A catalytic function, the demethylation of sterols, has been demonstrated for Mtb CYP51 (11) that, in the absence of a sterol biosynthetic pathway in Mtb, potentially links this enzyme to cholesterol-mediated Mtb entry into macrophages and its subsequent intracellular survival (12).
The cyp130 and cyp141 genes are missing from the virulent Mycobacterium bovis strain and from its avirulent counterpart, Mycobacterium bovis BCG, suggesting that they are not essential for Mtb growth, but may be relevant for Mtb virulence and infectivity towards the human host (13). The gene Rv1256c encoding Mtb CYP130 is possibly part of a functional operon along with the gene Rv1258c that encodes for a tetracycline/aminoglycoside-resistance (TAP)-like efflux pump. Both the Mycobacterium fortuitum TAP1 efflux pump and its Mtb Rv1258c homologue confer significant resistance to tetracycline and aminoglycosides, including streptomycin, a third major drug in antituberculosis treatment (14). Deletion of the Rv1258c gene from the M. bovis BCG chromosome increases the susceptibility of the organism to these two drugs, confirming involvement of the efflux pump in the intrinsic resistance of M. bovis and Mtb to tetracycline and streptomycin (15). Furthermore, a correlation has been established between expression of the Rv1258c gene and drug resistance in a clinical Mtb isolate resistant to the two major antitubercular drugs, rifampicin and isoniazid (16). However, no evidence yet exists of a functional link between CYP130 and Rv1258c.
The large number of distinct cytochrome P450 (P450) enzymes and the susceptibility of Mtb to azole agents that target such enzymes suggest important roles for them in Mtb physiology and, hence, their potential use as therapeutic targets. To date, only two Mtb P450 enzymes, CYP51 and CYP121, have been studied as individually expressed recombinant proteins. Both have been shown to tightly bind econazole, the agent of the azole class with the highest known antimycobacterial activity, as well as other azole and triazole drugs (17). The interactions of CYP51 and CYP121 with the azole inhibitors have been addressed by x-ray crystallography resulting in the determination of several crystal structures, including those of their complexes with the triazole antifungal agent fluconazole (18,19). Although econazole is so far the most potent antimycobacterial azole agent interacting in vitro with CYP51 and CYP121 (17), and herein with CYP130, no crystal structure of econazole bound in any P450 active site has ever been reported.
In the current work, we report determination of the x-ray crystal structures for ligand-free and econazole-bound Mtb CYP130. We have also examined the binding of azole drugs by UV-visible spectroscopy and isothermal titration calorimetry (ITC). Our data demonstrate that a conformational change in the protein is required for binding of econazole to CYP130 through a set of hydrophobic protein contacts and coordination to the heme iron. In addition to econazole, CYP130 binds a number of other antifungal agents with micromolar affinity, which makes it a plausible target for this class of therapeutic agents. Collectively, binding azoles to CYP130 is an endothermic entropy-driven complex process, which consists of two steps deducible from the titration calorimetry and exhibits spectrally detectable ligand-specific binding cooperativity that can be attributed to a potential for intramolecular or intermolecular protein-protein interactions inherent to CYP130.
EXPERIMENTAL PROCEDURES
Chemicals
Econazole, miconazole, clotrimazole, ketoconazole, glutaraldehyde, and other chemicals were purchased from Sigma-Aldrich unless otherwise specified. Crystallization screening kits were purchased from both Hampton Research and Qiagen.
Molecular Cloning of Rv1256c Encoding CYP130
Genomic DNA from M. tuberculosis H37Rv was obtained through the TB Vaccine Testing and Research Materials Contract at Colorado State University. The region of the Rv1256c gene encoding the putative cytochrome P450 CYP130 was amplified by PCR using Pfu Turbo DNA polymerase (Stratagene) and upstream 5'-CTCTGCTCCATATGACATCAGTAATGTCTCACG-3' and downstream 5'-AAGCTTTCATCTAGAGGATGTCACTCGGAACG-3' primers. The letters in bold in the upstream primer indicate an engineered Nde I restriction cloning site, including the initiation codon ATG. The underlined letters in the downstream primer indicate a Hind III restriction-cloning site. Amplification conditions were 94 °C for 5 min, 5 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 3 min followed by 25 cycles of 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 3 min. The PCR program was ended by a polymerization step at 72 °C for 25 min. To confirm the DNA sequence, the PCR fragment was first cloned into a pCR2.1 TOPO vector (Invitrogen) and then the Nde I-Hind III digested fragment was subcloned into a pCWori vector, which allows the expression of the recombinant protein with an N-terminal His6-tag (20).
Expression of Native and Se-methionine Containing CYP130
Recombinant CYP130, both native and as the Se-methionine containing derivative, was expressed under the control of the tac promoter of the pCWori vector using the Escherichia coli DH5α cells. For the native protein, cells were grown at 37 °C with vigorous agitation (250 rpm) in 2.8-L flasks containing 1 L of terrific broth (TB) medium supplemented with 200 µg/ml of ampicillin until the OD600 reached 0.5–0.8. At that time IPTG (0.5 mM), δ-aminolevulinic acid (0.5 mM), FeCl3 (250 µM) and ampicillin (200 µg/ml) were added. The cells were incubated for an additional 36 h at 25 °C at reduced agitation (180 rpm). The cells were harvested by centrifugation at 5,000 × g for 20 min at 4 °C and were then kept frozen at −80 °C until used.
For the Se-methionine containing CYP130 derivative, the transformed cells were grown at 37 °C and 250 rpm in 2.8-L flasks containing 1 L of Luria-Bertani (LB) medium supplemented with 200 µg/ml ampicillin until the OD600 reached 0.8–1.0. Cells were harvested by centrifugation at 2,000 × g for 15 min at 18 °C, washed with 100 ml of SelenoMet Medium base (AthenaES, Baltimore, MD) according to the protocol provided by the manufacturer, and re-centrifuged. Re-centrifuged cells were resuspended, transferred into 1 L of fresh SelenoMet Medium base and incubated at 25 °C and 250 rpm for 2 h before IPTG (0.5 mM), δ-aminolevulinic acid (0.5 mM), ampicillin (200 µg/ml), and SelenoMet Nutrient Mix (AthenaES, Baltimore, MD) containing a mixture of all the amino acids except methionine, vitamins and seleno-methionine were added according to the protocol provided by the manufacturer. The cells were incubated for 36 h at 25 °C and 180 rpm and harvested by centrifugation as described above.
CYP130 Purification
Both native and Se-methionine containing CYP130 were purified to homogeneity by fast protein liquid chromatography (FPLC). Cells obtained from 6 L of culture were thawed on ice and resuspended in 200 ml of buffer A (50 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 0.1 mM EDTA, 20 mM imidazole, and 1 mM PMSF). The cell suspension was incubated on ice for 30 min after the addition of lysozyme (0.5 mg/ml) and DNAse I (0.1 mg/ml). The cells were lysed by sonication using a Branson sonicator (3 × 4-min bursts at 50% power, with 2 min cooling on ice between each burst). Cell debris was removed by centrifugation at 100,000 × g, for 1 h at 4 °C. The soluble extract was loaded onto a 20 ml His/PrepFF 16/60 column (Amersham-Biosciences) equilibrated with buffer A. The column was first washed with 100 ml of buffer A and then with 100 ml of buffer B (50 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, and 20 mM imidazole). The protein was eluted with 200 ml of a linear gradient (20–200 mM) of imidazole in buffer B. All the fractions containing P450 were pooled and the protein was further purified by flow-through chromatography on SP-sepharose Fast-Flow (Amersham Bioscience) and subsequent binding to Q-sepharose Fast-Flow (Amersham Bioscience). The protein was eluted with 200 ml of a linear gradient (0–250 mM) of NaCl in 50 mM Tris-HCl, pH 7.5 and 0.1 mM EDTA. The fractions were analyzed by SDS-PAGE, and those containing pure CYP130 were pooled and concentrated to at least 1 mM using an Amicon Ultra concentrating device (Millipore). The content of Se-methionine in the CYP130 Se-methionine derivative was assessed by trypsin digestion and analysis of the tryptic fragments by MALDI-TOF MS using a Q-STAR XL mass spectrometer (Applied Biosystems/MDS Sciex).
Optical Absorption Spectroscopy
UV-visible absorption spectra of the purified CYP130 were recorded on Cary UV-visible scanning spectophotometer (Varian) using 1-cm path length quartz cuvette at 23 °C in 50 mM potassium phosphate (KPi) buffer, pH 7.4, containing 0.1 mM EDTA. The ferric-nitrosyl species was obtained in anaerobic conditions by flushing pure NO gas (Matheson Tri Gas, CA) over the ferric protein solution previously flushed with argon for 20 min. Formation of the ferrous carbon monoxide complex was achieved by bubbling CO gas (Airgas, CA) into the ferric enzyme solution for approximately 30 s through a septum-sealed cuvette prior to the injection of 1 mM sodium dithionite using a gas tight syringe (Hamilton, Reno, NV). Difference spectra were generated by subtracting the spectrum of the ferrous deoxy form from that of its carbon monoxide complex. The concentration of P450 was determined from difference spectra using the extinction coefficient 91,000 M−1cm−1 (21).
Equilibrium Binding Assay
Binding of the antifungal azole agents econazole, miconazole, clotrimazole and ketoconazole to CYP130 was monitored by UV-visible spectroscopy at 23 °C in 50 mM KPi buffer, pH 7.4, containing 0.1 mM EDTA. Stock solutions of the inhibitors at concentrations of 1 and 10 mM were prepared in DMSO. Difference spectra were recorded following the addition of a series of 0.25–1.0 µl aliquots of inhibitor to the sample cuvette containing 1 ml of 2.5 µM CYP130 for a maximal volume of 10 µl. The same amounts of DMSO alone were added to the reference cuvette. Increasing concentrations of KCl were added when specified in Table 3. To determine the Kd values, titration data points were fitted to the rectangular hyperbola (Eq. 1) for ketoconazole, quadratic hyperbola (Eq. 2) for miconazole, and the Hill equation (Eq. 3) for both econazole and clotrimazole using the Kaleidagraph software (Synergy).
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
TABLE 3.
Effects of ionic strength on affinity and cooperativity of CYP130-inhibitor binding.
| KCl (mM) |
Kd (µM) |
Hill coefficient n |
|---|---|---|
| 0 | 1.93 ± 0.03 | 1.37 ± 0.02 |
| 50 | 0.90 ± 0.04 | - |
| 125 | 0.64 ± 0.04 | - |
| 250 | 0.79 ± 0.05 | - |
In all equations, Aobs is the absorption shift determined at any ligand concentration, Amax is the maximal absorption shift obtained at saturation, Kd, is the apparent dissociation constant for the inhibitor-enzyme complex, Et is the total enzyme concentration used, S is the ligand concentration, and n is a Hill coefficient, a measure of cooperativity.
Binding stoichiometry by Job’s titration
The stoichiometry for the binding of inhibitors to CYP130 was determined by the method of continuous variation known as a Job’s titration (22) using UV-visible spectroscopy and a sub-micro quartz cuvette designed for 0.1- to 1.55-ml sample (Starna, Atascadero, CA). Experiments were carried out at 23 °C in 50 mM KPi, pH 7.4, containing 0.1 mM EDTA and 0.5% DMSO. 125 µl of 15 µM CYP130 were placed into the optical cell, and the inhibitor solution of the same concentration was added gradually, until the volume of the mixture reached 1.525 ml. The sum of the concentrations of the reactants was therefore kept constant and equal to 15 µM.
Crystallization and Data Collection
Purified CYP130 diluted to a concentration of 0.2 mM was subjected to automated screening of crystallization conditions using a nanoliter drop setter Mosquito (TTP LabTech). Both ligand-free and econazole-bound CYP130 crystallized from the different sets of crystallization conditions, which were further optimized to generate crystals of diffraction quality. Ligand-free crystals grew from 1.6 M ammonium sulfate, 0.1 M Na citrate, pH 5.2, and 2% isopropanol and diffracted in the monoclinic space group C2 (Table 1) to a resolution of 1.46 Å. The asymmetric unit contained one protein molecule and 40% solvent. Econazole-bound crystals grew from 1.4 M ammonium sulfate, 0.1 M MES, pH 6.25, 40 mM NaF, and 2 mM econazole. Crystals belonged in the space group P3(2)21 and diffracted to a resolution of 3.0 Å (Table 1). Despite a large unit cell, there were only two molecules in the asymmetric unit, both related by non-crystallographic two-fold symmetry. Thus, high solvent content (78%) and peculiarities of the molecule packing probably account for a low resolution of these crystals. Data were collected at 100–110 K at beamline 8.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory, USA. The images were integrated, and the intensities merged by using HKL2000 software suite (23). Anomalous data were collected at two wavelengths using a Se-methionine derivatized crystal (Table 1).
Table 1.
Data collection and refinement statistics
| PDB ID | Ligand-free 2UUQ | ECO-bound 2UVN | Se-Met | |
|---|---|---|---|---|
| Data Collection | ||||
| Resolution, Å | 1.52 | 3.0 | 1.75 | 1.75 |
| Wavelength, Å | 0.97926 | 1.11588 | 0.97970 | 0.96863 |
| Space group | C2 | P3(2)21 | C2 | C2 |
| Cell dimensions a, b, c, Å a, b, g, ° |
160.0, 53.9, 43.7 90.0, 96.5, 90.0 |
131.7, 131.7, 229.4 90.0, 90.0, 120.0 |
160.3, 53.9,43.8 90.0, 96.6, 90.0 |
160.3, 53.9,43.8 90.0, 96.6, 90.0 |
| Molecules in an asymmetric unit | 1 | 2 | 1 | 1 |
| Solvent content, % | 40 | 78 | 40 | 40 |
| Rsyma,b, % | 4.7 (31.3) | 8.1 (51.2) | 4.4 (45.8) | 4.4 (43.6) |
| I/s | 21.8 (2.0) | 29.9 (4.2) | 26.0 (2.1) | 26.2 (2.3) |
| Unique reflections | 52466 | 45935 | 71697 | 70994 |
| Completeness, % | 91.6 (56.9) | 100.0 (100.0) | 97.5 (92.4) | 97.7 (94.0) |
| Redundancy | 4.1 (2.2) | 9.8 (9.6) | 3.9 (3.5) | 3.9 (3.6) |
| Phasing | ||||
| Resolution range, Å | 43.5-2.2 | |||
| Number of used sites | 9 | |||
| Phasing power | 2.5 | |||
| Figure of merit | 0.64 | |||
| After density modification | 0.95 | |||
| Refinement | ||||
| Reflections used | 44721 | 41548 | ||
| Rcryst/Rfreec, % | 18.7/23.1 | 20.0/23.4 | ||
| No. atoms | ||||
| Protein | 3079 | 6120 | ||
| Heme | 43 | 86 | ||
| Ligand | N/A | 48 | ||
| Water/Ion | 289/9 | 77/34 | ||
| Wilson plot B-values, Å2 | 18.6 | N/A | ||
| Mean B-factors, Å2 | 22.1 | 64.6 | ||
| Protein | 21.4 | A: 64.4/ B: 65.4 | ||
| Heme | 12.5 | A: 48.2/B: 60.5 | ||
| Ligand | N/A | A: 37.7/B: 53.4 | ||
| Water | 31.2 | 43.7 | ||
| r.m.s. diviations | ||||
| Bond length, Å | 0.010 | 0.008 | ||
| Bond angles, ° | 1.28 | 1.30 | ||
| Ramachandrand, % | A:88.9/10.8/0.3/0.0 | A: 84.2/14.0/1.2/0.6 B: 83.8/14.0/1.5/0.6 |
||
Numbers in parentheses correspond to the highest resolution shell.
Rsym = Σ̣| Ii − <I> | / ΣIi, where Ii is the intensity of the ith observation, and <I> is the mean intensity of reflection.
Rcryst = Σ ‖Fo|−|Fc‖/Σ̣|Fo|, calculated with the working reflection set. Rfree is the same as Rcryst but calculated with the reserved reflection set.
Program PROCHECK [Laskowski, 1993 #302], portions of the protein residues in most favored/additional allowed/generously allowed/disallowed regions.
Structure Determination and Refinement
The crystal structure of the ligand-free CYP130 was determined to 1.46 Å resolution using the multiwavelength anomalous dispersion (MAD) protocol implemented in the CNS software suite (24) and a two-wavelength data set (Table 1). Electron density was traced using the ARP/wARP program (25). Model refinement was performed by alternation of automated model building with COOT (26,27) and refinement with the REFMAC5 (28) programs. The structure of the econazole-bound form was determined by molecular replacement using the program MOLREP (29) and the ligand-free CYP130 structure (PDB ID 2UUQ) as a search model (Table 1). The structure was further refined by alternation of manual model building using program O (30) and refinement using CNS (24). The quality of the final structures was assessed with the program PROCHECK (31) and the Ramachandran statistics are shown in Table 1.
Isothermal Titration Calorimetry
Experiments were performed using a VP-ITC calorimeter equipped with the control and data acquisition and analysis software ORIGIN 7 (MicroCal Inc., Northampton, MA). Solutions of the protein and inhibitors were prepared in 50 mM KPi, pH 7.4, containing 0.1 mM EDTA and 0.5% DMSO. Due to the low solubility of the azole inhibitors in aqueous solutions, the experiments were carried out in the reverse mode. The inhibitor solution (25 µM) was placed in the calorimetric cell and titrated with the CYP130 (400 µM) in the titration syringe. First injection (1 µl, omitted from the analysis) was followed by 30 injections of 4 µl with 4 min intervals. The titration syringe was continuously stirred at 305 rpm and the temperature of the calorimetric cell was maintained at 25 °C. Injecting the protein into the buffer alone was also carried out as a reference titration, and the resulting heat of dilution was subtracted from the protein-inhibitor titration.
Glutaraldehyde Cross-linking and Gel-electrophoresis Analysis
CYP130 and the reference P450 enzymes dissolved at 20 µM concentration in 50 mM MES, pH 6.5, containing KCl (when specified) at concentrations ranging from 50 to 300 mM, were mixed with freshly prepared glutaraldehyde (1%) to final concentrations of 0.05 to 0.1%. After 15 min incubation the samples were loaded either directly onto 20% pre-cast polyacrylamide gel equilibrated with the native buffer or (after quenching excess of glutaraldehyde with 1/10 (v/v) of 1 M Tris-glycine buffer, pH 7.5 followed by 5 min incubation at 95 °C in the presence of SDS) onto 12.5% polyacrylamide gel equilibrated with SDS-containing buffer and run at 15 °C using a FastSystem apparatus (General Electric) according to the standard protocols until bromophenol blue dye reached the gel bottom. The protein bands were stained with Coomassie Blue.
RESULTS
Expression and Purification of CYP130
CYP130 is the third Mtb P450 expressed and purified to homogeneity in a soluble recombinant form, yielding 60 mg of native protein and 30 mg of Se-methionine derivative from 1 L of bacterial culture. In the Se-methionine derivative, 54% of the methionine was substituted by Se-methionine, as judged by mass spectrometric analysis (not shown).
Spectroscopic Characterization of CYP130
UV-visible absorption spectroscopy was used for initial characterization of purified CYP130. CYP130 displayed the spectral properties typical for a ferric P450 with the heme iron in a low-spin state, exhibiting a Soret γ band at 418 nm and α and β bands at 567 and 535 nm, respectively (Fig. S1A). Bubbling of NO into the ferric CYP130 under anaerobic conditions resulted in the formation of a stable ferric nitrosyl adduct with a Soret band at 434 nm (Fig. S1A). Coordination of the imidazole of econazole to the CYP130 ferric heme iron caused a typical Type II red shift of the Soret band to ~424–425 nm, reflecting replacement of the heme distal water ligand by the Nε1 atom of the azole moiety (Fig. S1A). However, the econazole-induced red shift in CYP130 was temperature dependent (Fig. S1B), progressively and reversibly shifting from 421.7 nm at 15 °C to 424.2 nm at 40 °C, suggesting an equilibrium between low-spin heme iron complexes involving either direct iron-nitrogen ligation or indirect coordination mediated by the water molecule, as observed elsewhere for the CYP121-fluconazole interactions (19).
One-electron reduction of the iron by sodium dithionite followed by binding of CO shifted the Soret band to 447 nm, as expected for conversion of the ferric CYP130 to its ferrous-CO complex (Fig. S1A).
Binding of Antifungal Azole Inhibitors
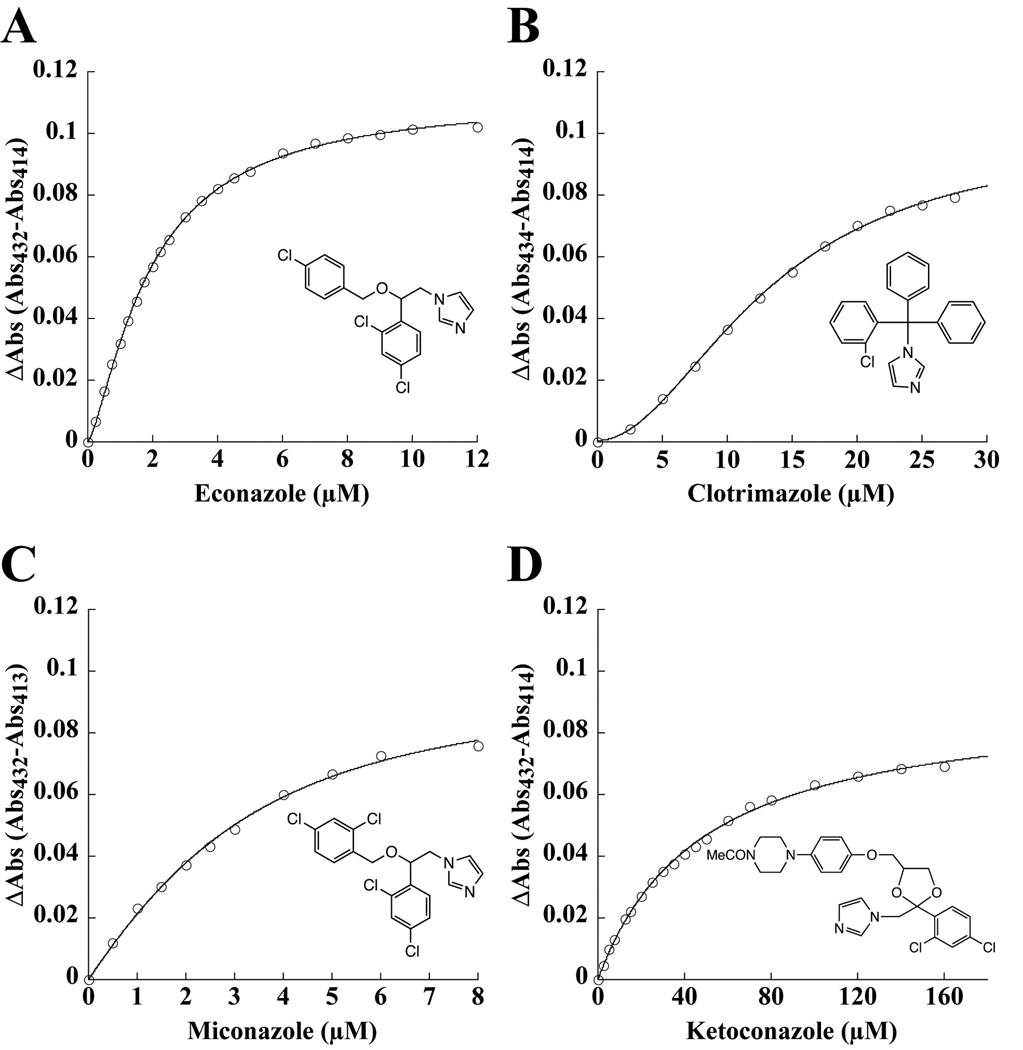
Binding of the azole antifungal drugs econazole, miconazole, clotrimazole, and ketoconazole (Fig. 1) to CYP130 was monitored via the type II shift of the heme Soret band caused by coordination of the inhibitors to the heme iron atom. The Kd values for the inhibitors were obtained from the spectral titration curves (Fig. S2) and are summarized in Table 2. For comparison, the Kd values for CYP121 obtained elsewhere and for CYP51 determined herein are also listed. The sigmoid titration plots obtained for both econazole (Fig. 1A) and clotrimazole (Fig. 1B) were best fitted to the Hill equation (Eq. 3) with coefficients of 1.37 and 1.93, respectively, indicating the presence of binding cooperativity. The titration curves for ketoconazole (Fig. 1C) and miconazole (Fig. 1D) were fitted with the rectangular (Eq. 1) and the quadratic (Eq. 2) hyperbolas, respectively. Collectively, the binding affinities of all the inhibitors are about an order of magnitude lower for CYP130 than for CYP121. Miconazole, clotrimazole, and ketoconazole also bind to CYP51 somewhat more tightly than to CYP130, whereas econazole has about the same binding affinities for both CYP130 and CYP51.
Fig. 1. Binding of antifungal azole drugs to CYP130.
The concentration dependence of econazole (A), clotrimazole (B), miconazole (C) and ketoconazole (D) binding deduced from the difference absorption changes obtained from the titration of CYP130 (2.5 µM) with increasing concentrations of the inhibitor. The structure of the inhibitor is shown in each panel.
TABLE 2.
The values of dissociation constants (Kd, µM) and Hill coefficients (n, if applicable) for the four azole antifungal inhibitors and the three structurally-characterized Mtb CYP enzymes.
Cooperativity of P450-ligand Binding
Binding cooperativity was observed for two (econazole and clotrimazole) of the four ligands used in this study. Two potential sources of CYP130 binding cooperativity can be considered: (i) multiple site cooperativity, in which two or more ligands bind simultaneously to the same protein molecule, and (ii) multimer cooperativity, where protein-protein interactions are promoted by the binding of a ligand to a molecule of the protein. Multiple site cooperativity has been demonstrated by a variety of experimental techniques for a number of microsomal (CYP3A4, CYP1A2, CYP2C9) and bacterial (EryF) P450 enzymes (32,33). Simultaneous binding of two ligands within the P450 active site has been validated by x-ray structures for EryF (34), CYP3A4 (35), and CYP158A2 (36). Although multimer binding cooperativity has not, to our knowledge, been reported for P450 systems, substrate-dependent P450-P450 interactions have been shown to significantly influence individual functions of drug-metabolizing CYP2 and CYP3A4 enzymes (37–43), presumably by altering the rate of association and/or affinity for the P450-reductase. These changes could reflect ligand-dependent dimerization or oligomerization of the P450 enzymes (44). Among the reported P450 structures, the majority of mammalian CYP2 enzymes have been crystallized in dimeric (CYP2C9, CYP2C8, CYP2B4, CYP2R1, CYP3A4) or higher (CYP2A6, CYP2A13, CYP3A4) oligomerization states. Among the bacterial proteins, dimers with two-fold non-crystallographic symmetry have been detected for a P450 enzyme from Thermus thermophilus (PDB ID code: 1WIY) and for CYP154C1 from Streptomyces coelicolor (1GWI) (45).
Stoichiometry of CYP130-inhibitor Binding
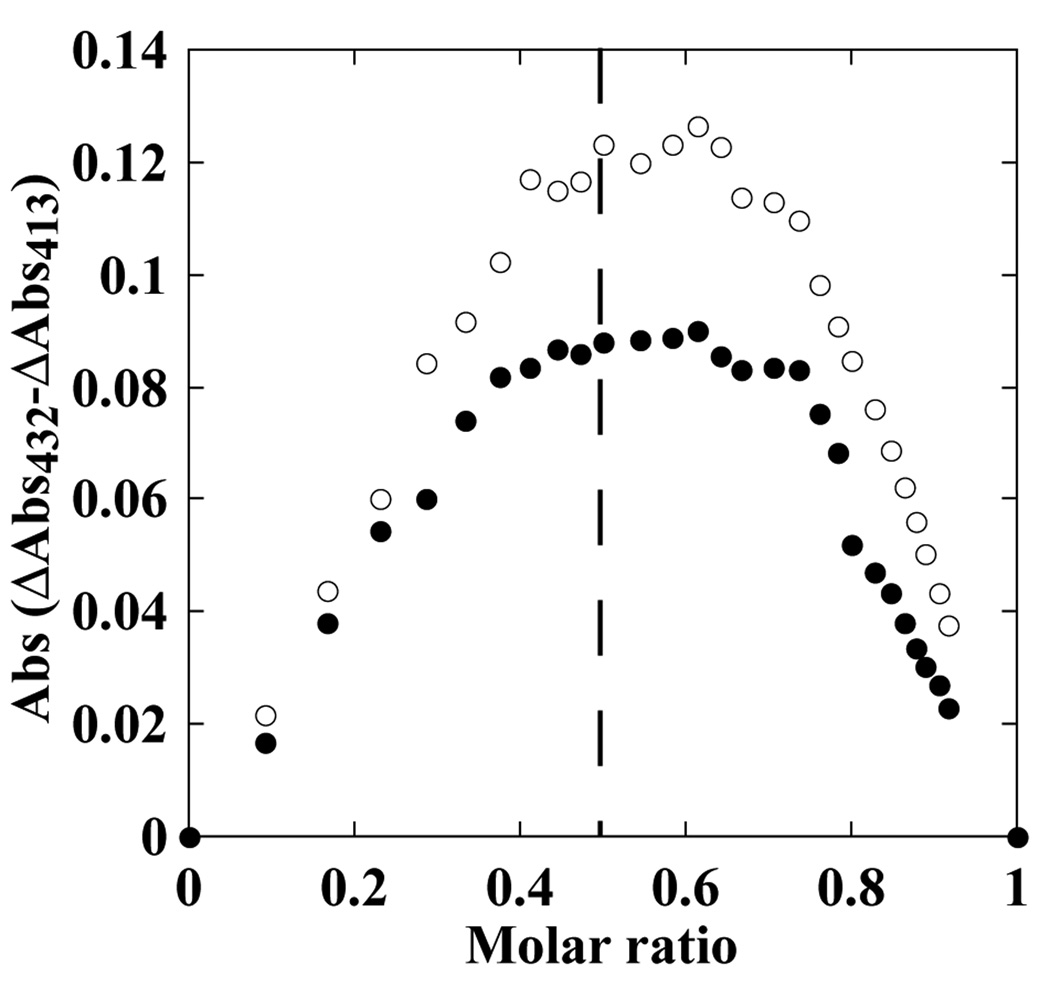
To address the possibility that binding cooperativity may arise from the binding of multiple inhibitor molecules in the CYP130 active site, the stoichiometry for the CYP130/econazole and CYP130/miconazole complexes was determined using the Job’s titration method (22), which is based on mixing of the reactants in such a way that their molar ratio varies, while the total molar concentration remains constant. The data for both econazole and miconazole were fit to a binary complex mechanism with the maximum of the bell-shaped plot located close to the center corresponding to a molar fraction (defined as [CYP130]/([CYP130]+[inhibitor])) of ~0.55, indicating virtually a 1:1 enzyme:inhibitor ratio (Fig. 2). This result suggests that the binding cooperativity observed for econazole is unlikely due to simultaneous binding of two molecules in the active site, which agrees with the x-ray structure data. This conclusion is consistent with the fact that the structurally related miconazole, which binds CYP130 with the same stoichiometry, does not exhibit binding cooperativity.
Fig. 2. Stoichiometry of CYP130-inhibitor binding.
The bell-shaped Job plots at a total protein and inhibitor concentration of 15 µM display a maximum close to a mole fraction of 0.55, the value that corresponds to a 1:1 ratio for the binding to CYP130 of econazole (open circles) and miconazole (closed circles).
Protein-protein Interactions and Binding Cooperativity
To examine a possible role for protein-protein interactions in the binding cooperativity of econazole and clotrimazole, a series of binding experiments was conducted in the presence of increasing concentrations of KCl (Table 3). The binding cooperativity of econazole was abolished by 50 mM KCl. In the case of clotrimazole, the influence of ionic strength could not be explored due to protein aggregation at even the lowest concentration of KCl, an effect similar to that observed for the CYP3A4/ketoconazole complex in the presence of apolar solvents and elevated ionic strength (35). These data support the inference that the binding cooperativity of econazole may arise from protein-protein interactions.
Overall Structure of CYP130
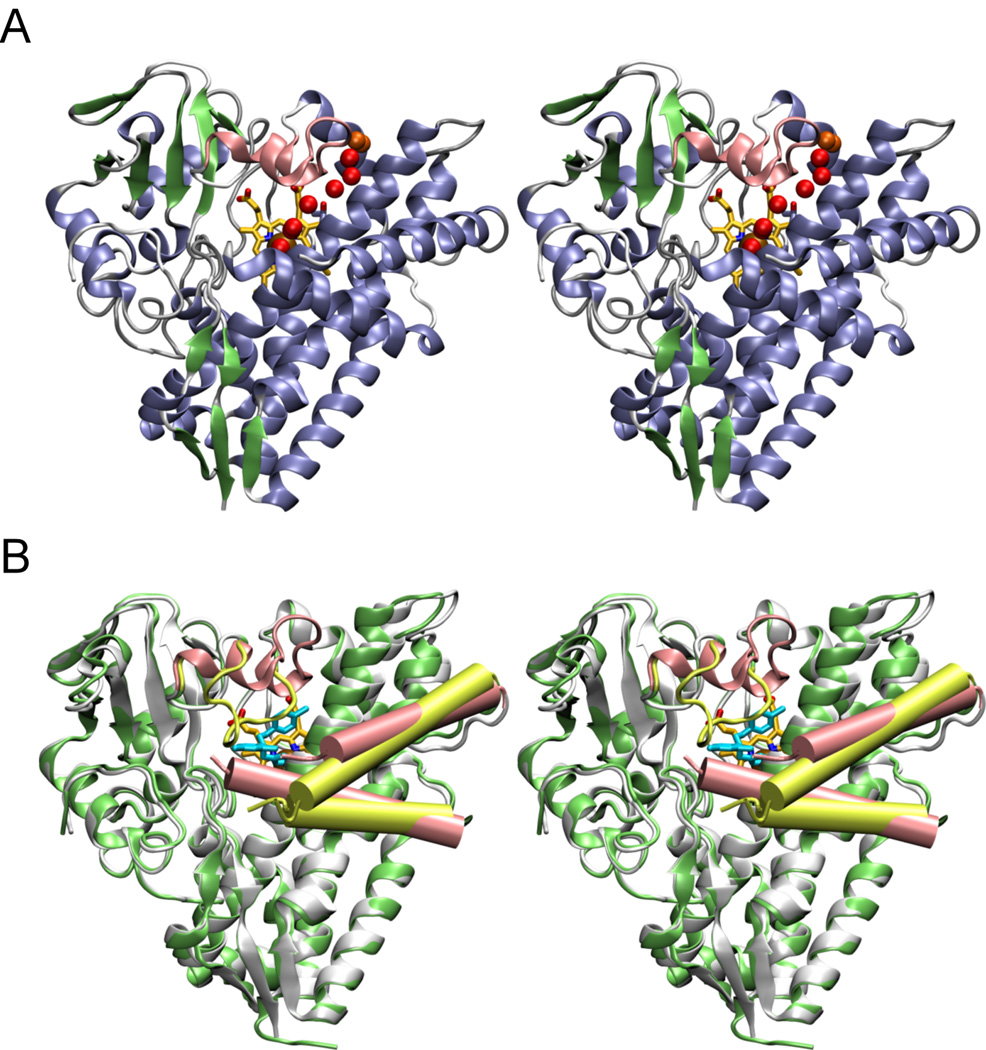
The interaction of CYP130 with econazole was addressed by x-ray crystallography. Crystal structures of CYP130 were determined for the ligand-free and econazole-bound forms. CYP130 has the characteristic fold common to all structurally defined cytochromes P450 but exhibits conformational and oligomerization differences between the ligand-free and -bound forms. Ligand-free CYP130 crystallized as a monomer in a relatively ‘open’ conformation (Fig. 3A), which is largely achieved by an extended conformation of the BC-loop unmasking a route for substrate access. In the dimeric econazole-bound form, the BC-region (residues 80–91, colored pink in Fig. 3B, 4A) looses secondary structure and relocates as much as 18 Å to generate multiple contacts with econazole, primarily with its two chlorinated phenyl moieties. At the same time, the F helix looses one helical turn while the G helix gains one helical turn, causing a drift of the connecting loop in the direction of the N-terminus along the primary sequence. Thus, the FG-loop in CYP130 is rather short (note that four amino acids are missing from the electron density in both the open and closed CYP130 forms (Fig. 3B, 4A)) and serves only as a turn between two helices. Conformational mobility of the BC- and the FG-regions has been previously observed in other structurally defined P450 enzymes and may serve to enable substrate access/product release to/from the active site.
Fig. 3. Stereo views of ligand-free and econazole-bound CYP130.
A, the ligand-free CYP130 is in a ribbon representation colored according to the secondary structure elements: helices are in blue, β-sheets in green, and loops and turns in grey. The BC-loop region (highlighted in pink) is well-structured having two short helices αB' and αB". A hydrogen-bonding network of water molecules linking the stability of the distal water ligand to the I-helix N-terminus is marked by the red spheres. In orange are shown water molecules having contacts with the bulk solvent. Residue T239 in the N-terminal portion of the I-helix is shown in sticks. B, superimposition of the ligand-free (gray) and econazole-bound (lime green) forms. The BC-loop region containing residues 80–91 which relocates up to 18 Å to a position where they interact with the econazole is shown in pink in the ligand-free and in yellow-green in the econazole-bound forms. The F and G helices are shown as pink cylinders in the ligand-free and yellow-green cylinders in econazole bound forms. The G helix is on top. Econazole is in cyan and heme is in yellow. Images were generated using VMD software (51) unless specified otherwise.
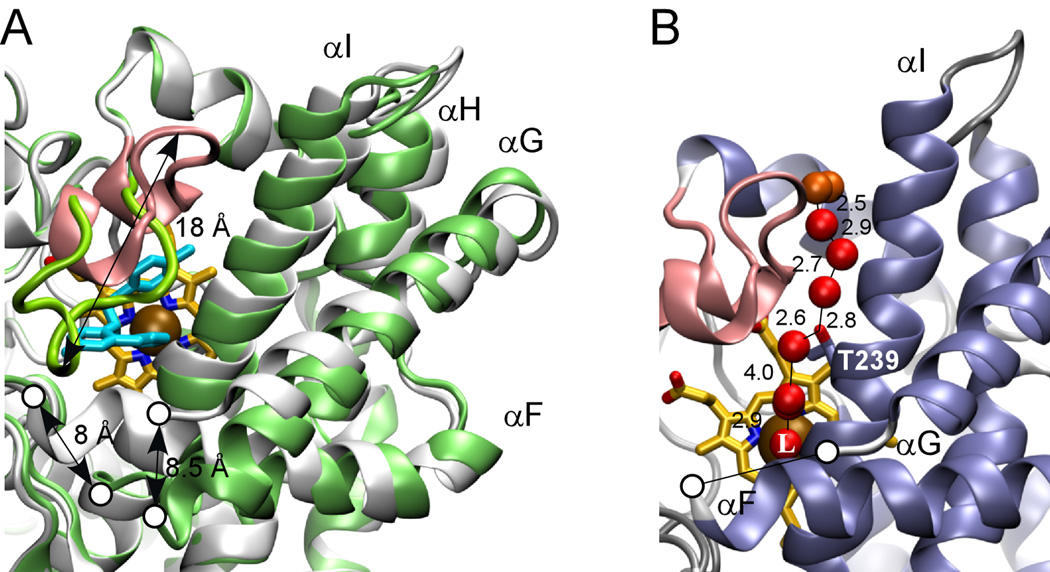
Fig. 4. CYP130 active site.
A, major conformational differences between the ligand-free (gray) and econazole–bound (green) states. The BC-region is in pink, heme is in yellow and econazole is in cyan. Gaps in the protein chain between the F and G helices due to the missing electron density are marked with the open circles. Relocation distances for selected structural elements are given in Angstroms. B, H-bonding network. The fragment of the ligand-free crystal structure showing the water molecules (red spheres) that stabilize the distal water in CYP130. Water molecules having contacts with the bulk solvent are colored in orange. Distances between oxygen atom centers are in Angstroms. T239 is shown in sticks. The iron axial water ligand is labeled with a capital L.
Dimerization of CYP130 in the Crystal
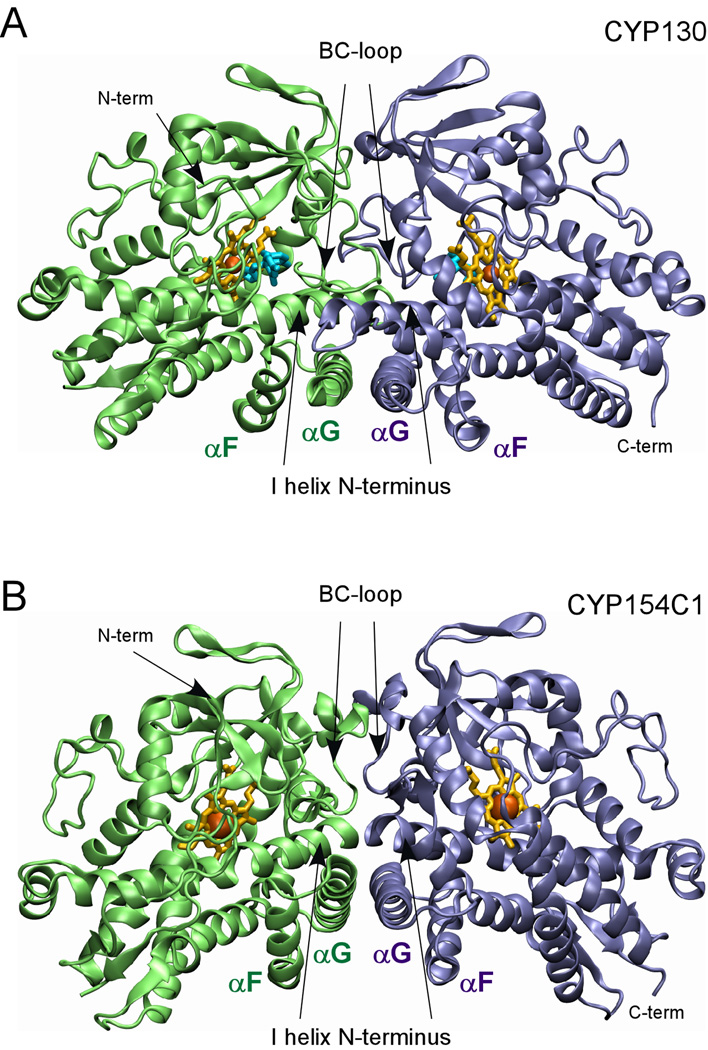
We were unable to crystallize the CYP130/econazole complex under conditions that favor crystallization of the open ligand-free form. Instead, a longer incubation under a different set of conditions was required to generate econazole-bound crystals that have a different morphology, unit cell dimensions, and diffract in a different space group (Table 1). The high solvent content in these crystals (78%), indicating loose packing, partially explains the low resolution of the diffraction data. Analysis of the econazole-bound structure and the crystal symmetry revealed that the crystal lattice is largely stabilized by (i) formation of a CYP130 dimer having two-fold rotation symmetry along the non-crystallographic axis which generates a dimerization interface utilizing about 2000 Å2 (12.5%) of the surface of each monomer, and (ii) a two-fold crystallographic symmetry generating a dimer of dimers with 1280 Å2 of total interface (Fig. S3). The non-crystallographic dimerization interface involves the most conformationally mobile P450 regions: the BC-loop, the F and G helices, and the N-terminal portion of the I-helix (Fig. 5). The interface is stabilized by partial overlap (about four helical turns) between the I-helix N-termini and complete overlap between the G helices packed in anti-parallel orientations (Fig. 5A). Together with the F helices, they constitute two layers of anti-parallel α-helices crossing each other at an angle of ~60°. A similar dimerization pattern, although with a smaller (600 Å2, 3.5% of the monomer surface) dimerization interface, is observed in ligand-free CYP154C1 (45) (Fig. 5B). It is worth mentioning that the BC- and FG-regions are also involved in the dimerization of a bacterial P450 from T. thermophilus (PDB ID code 1WIY).
Fig. 5. Dimerization interface.
The dimerization interfaces of (A) CYP130 (2000 Å3) and (B) CYP154C1 (610 Å3) formed largely via interactions between the G helices in anti-parallel orientations, overlapping N-termini of the I helices, and multiple contacts in the BC-loop regions are shown. The monomers are colored in green and blue, heme is in yellow and econazole in (A) is in cyan.
Dimerization of CYP130 in Solution
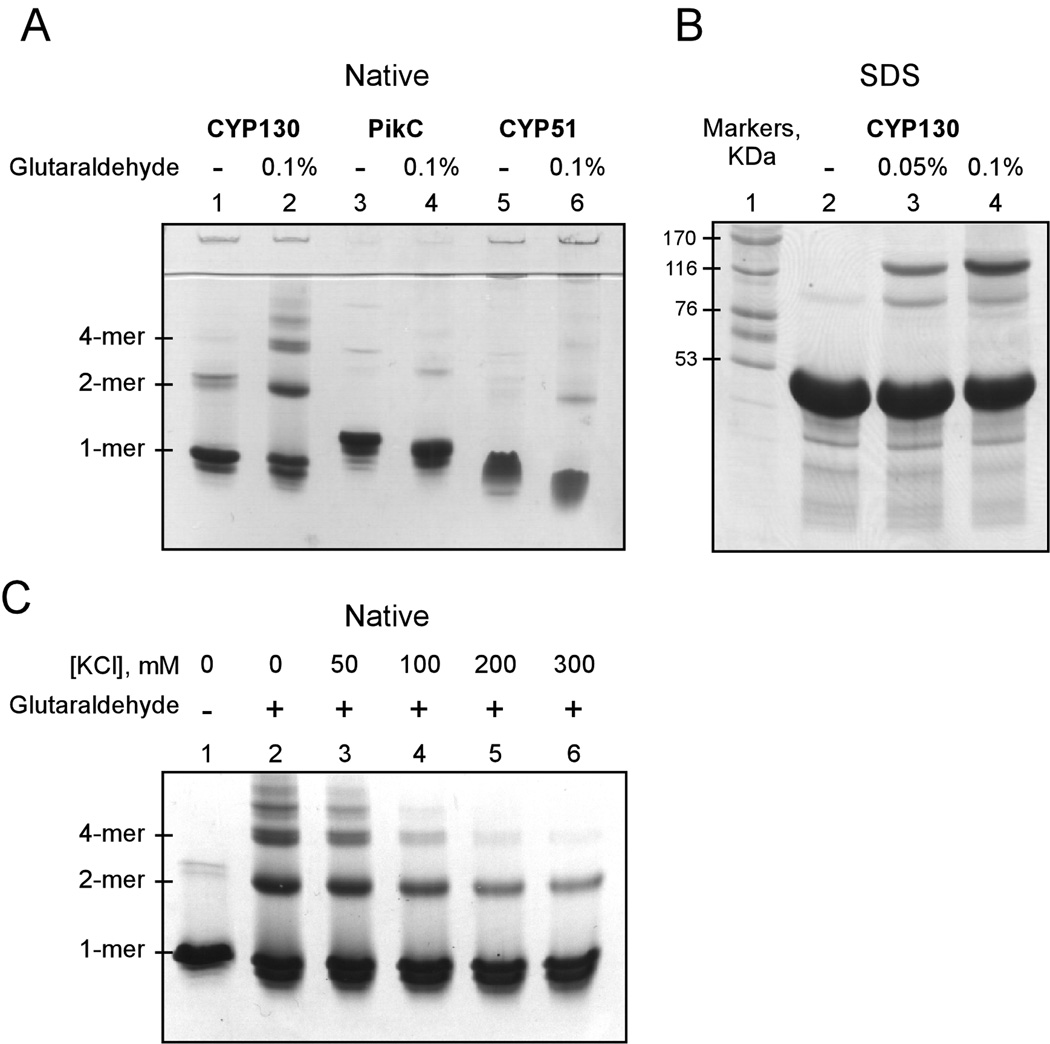
The CYP130 dimer in the crystal is stabilized via a number of hydrophobic and H-bonding interactions, while electrostatic interactions are involved in stabilizing the crystallographic tetrameric interface. The stability of the dimer, if formed, is not sufficiently high to detect it by equilibrium techniques such as gel filtration chromatography or native gel electrophoresis at protein concentrations up to 100 µM. However, CYP130 oligomerization in solution was detectable by chemical cross-linking using glutaraldehyde (46). A substantial fraction of CYP130 was found in dimeric/tetrameric forms at 20 µM concentration, whereas only marginal oligomerization was detected for two other soluble bacterial P450 enzymes, Mtb CYP51 and PikC from Streptomyces venezuelae, examined as controls (Fig. 6 A, B). No significant effect of inhibitors at up to a 500 µM concentration was observed, with the exception of a slightly reduced content of the higher molecular weight aggregates for clotrimazole and ketoconazole (not shown).
Fig. 6. Glutaraldehyde cross-linking.
Analysis of the CYP130 cross-linked products by native- (A, C) and SDS- (B) gel electrophoresis. A, cross-linking performed at a protein concentration of 20 µM is shown. Two soluble P450 enzymes, CYP51 from Mtb and PikC from S. venezuelae, were used as controls. B, the molecular weight of the cross-linked CYP130 products was confirmed by the SDS-gel electrophoresis. C, effect of ionic strength on stability of the CYP130 aggregates is shown. As the KCl concentration increases from 0 to 300 mM, the dimer product stabilized by glutaraldehyde cross-linking persists but the formation of tetramers and higher oligomers is suppressed.
When cross-linking was carried out with increasing KCl concentrations ranging up to 300 mM, the dimer product persisted unabated but the formation of tetramers and higher oligomers was suppressed at the higher salt concentrations (Fig. 6 C). This is consistent with the observation that dimer formation involves specific hydrophobic and H-bonding interactions, whereas higher oligomers are formed by relatively non-specific ionic ones. A small fraction of the dimer may be formed by such non-specific interactions, but the majority of the dimer does not involve ionic contacts and persists in the presence of higher salt concentrations. Collectively, the crystallographic and chemical cross-linking data suggest that the oligomerization of CYP130 seen in the crystal can also occur in solution even in the absence of a ligand, with the closed form being susceptible to dimerization. If, as expected, CYP130 exists in an equilibrium between the open and closed forms that is shifted toward the closed form by econazole binding, the accumulation of the cross-linked products in the absence of azole ligand is readily explained by irreversible removal of the closed form from the equilibrium mixture by the cross-linking reaction.
Active Site of CYP130
In the ligand-free form, the CYP130 heme iron is hexa-coordinated with a water molecule tightly bound as the distal axial ligand (bond distance 2.2 Å). This axial water is additionally stabilized via an H-bond to the carbonyl group of G243 (distance 2.7 Å) and an H-bonding network formed by the cluster of water molecules bound in the proximity. To provide an H-bond to the axial water, the middle portion of the I-helix accommodating G243 closely approaches the porphyrin plane (distance 4.2 Å between the G243 carbonyl group and the heme iron), imposing steric constraints on the binding of potential ligands in the active site and at the same time preventing the axial water from premature release. None of a dozen different compounds of varied structure examined here was able to expel the axial water from the CYP130 active site to generate a high-spin heme iron state.
A hydrogen-bonding network of water molecules that also stabilizes the distal water axial ligand is well defined in the crystal structure. This network leads from the distal water ligand along the N-terminal portion of the I-helix to the surface of the molecule (Fig. 3A, 4B). A chain of seven well-structured water molecules is interrupted only once between the third and fourth molecules, where the position of the missing water is taken by the hydroxyl group of I-helix residue T239, located one helical turn away from G243 toward the N-terminus.
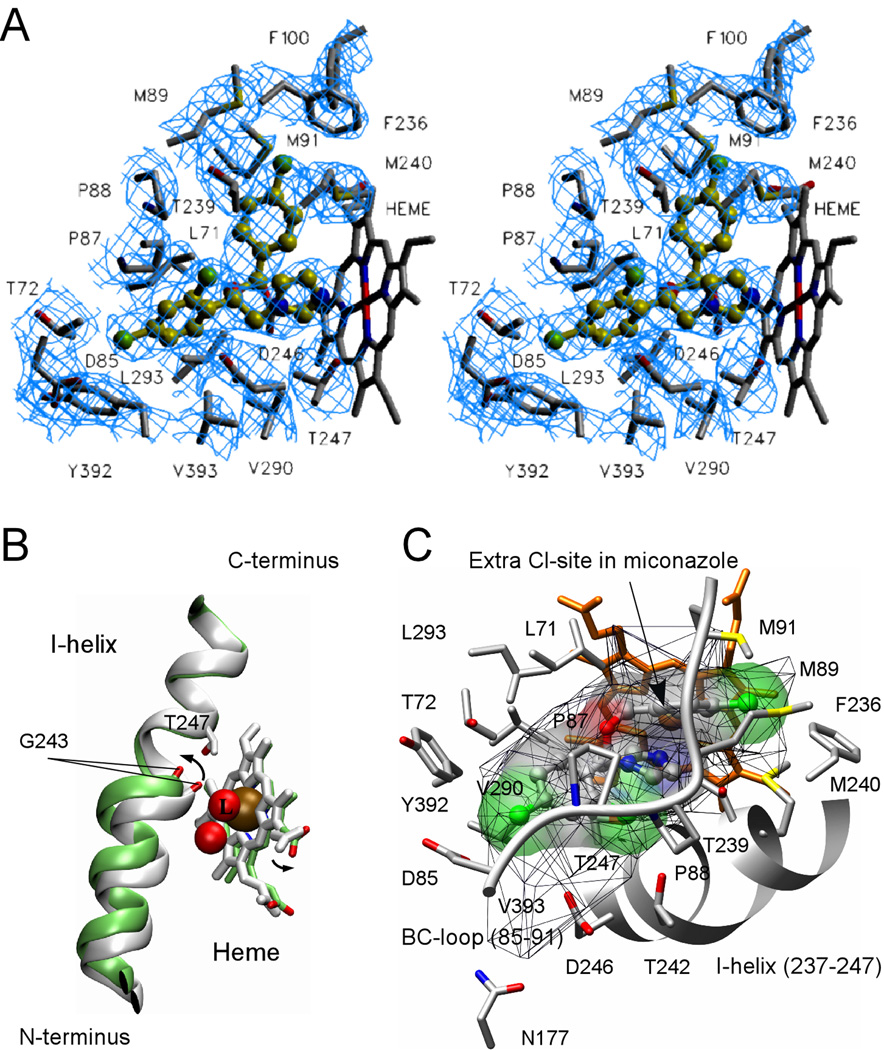
Econazole Binding Site
Despite the relatively low resolution (3.0 Å) of the econazole-bound crystal structure, the electron density for econazole is unambiguously defined in each of the two monomers in the asymmetric unit (Fig. 7A). Econazole binds to CYP130 through a set of predominantly hydrophobic interactions in addition to the coordination bond (length ~ 2.75 Å) formed between the heme iron and the lone pair of nitrogen electrons of the azole moiety. Econazole introduces a kink into the I-helix that displaces G243 by 2.3 Å from the hydrogen bonding position and releases the axial water stabilized by this H-bond (Fig. 7B). The econazole binding mode deviates from the expected geometry (47), including the length of the coordination bond (ideal ~2.1 Å) and the ~80° angle (ideal 90°) between the azole plane and the porphyrin macrocycle. These deviations are likely due to the steric constraints imposed by the I-helix, but are less pronounced than those observed elsewhere for the CYP121/fluconazole complex (19). Given a weakened coordination bond and a larger volume of the active site cavity (accessible volume 600 Å3, shown by mesh surface in Fig. 7C) than is required to accommodate econazole (330 Å3) (Fig. 7C), alternative coordination mode(s) may arise to account for the temperature dependent shift of the low-spin Soret band of the CYP130/econazole complex observed by spectroscopic analysis in solution (Fig. S1B). However, in contrast to the CYP121/fluconazole complex, no structured water molecules are observed in the vicinity (or in the active site in general) at this resolution to allow us unambiguously conclude that the formation of a low-spin heme iron complex involving indirect iron-nitrogen coordination through a water molecule (19) can occur in econazole-bound CYP130.
Fig. 7. Econazole binding in the active site.
A, stereo view of econazole (yellow-green) bound in the active site of CYP130 is shown. Cl atoms are colored in green, N atoms in blue and O atoms in red. Amino acid residues within 4 Å of econazole are labeled in black. Fragments of the 2Fo−Fc electron density composite omit map contoured at 1.0 σ are in blue. To avoid excessive cluttering, heme was excluded from the map calculation and T242 was excluded from the view as projecting on top of econazole. The image was generated using the SETOR program (52). B, a kink of the I-helix introduced by the binding of econazole is shown. The ligand-free (gray) and econazole-bound (green) CYP130 structures were superimposed with an r.m.s.d. of 0.93 Å2 for all the protein residues. Iron (ochre) and oxygen (red) atoms are shown as spheres. The iron axial water ligand is labeled with a capital L. The arrows show the directions of movements upon transition from the ligand-free to the econazole-bound state. C, alignment of the BC-loop fragment (85–91) (grey) in the groove formed between the mono- and double-chlorinated phenyl rings of econazole is shown. The additional chlorination site in miconazole is indicated by an arrow. The solid surface represents the van der Waals surface of econazole (volume of 330 Å3) and is colored according to the underlying atoms: oxygen in red, nitrogen in blue, chlorine in green. The mesh surface shows the accessible space in the active site (600 Å3). A fragment of the I-helix (237–247) is represented by the grey ribbon. The heme is in orange. This image was generated using the program CHIMERA (53).
The rest of the econazole molecule forms mainly hydrophobic contacts with the amino acid side chains of L71, T72, D85, P87, P88, M89, M91, F100, F236, T239, M240, T242, G243, G244, D246, T247, V290, Y392, and V393 situated within interaction distance (6 Å) from econazole (Fig. 7C). Notably, D85 is the only charged amino acid side chain in the active site, with the exception of the catalytic negative charge of D246 that is highly conserved throughout the CYP family. These two charged residues and the neutral N177 surround an accessible but econazole-unoccupied extension of the otherwise hydrophobic active site cavity (Fig. 7C), suggesting that the enzyme may normally act upon a hydrophobic endogenous substrate, possibly larger than econazole that carries a positively charged functional group.
A stretch of the hydrophobic BC-loop residues (80–91) (highlighted in pink in Fig. 3B) is relocated by up to 18 Å when econazole binds. One of the two consecutive proline residues, P87, residing in this region binds in the groove formed between the mono- and double-chlorinated econazole phenyl moieties (Fig. 7A, C). This interaction appears to be critical for positioning of this portion of the BC-loop, which is directly involved in formation of the CYP130/econazole dimerization interface. The additional chlorine atom in miconazole is expected to protrude toward P87, altering the local configuration of the BC-loop and, hence, the dimerization interface. Should such alterations occur, they may account for the lack of binding cooperativity observed with miconazole (Fig. 1C) and failure of the CYP130/miconazole complex to crystallize from >400 different crystallization conditions, including those which reproducibly generate ligand-free or econazole-bound crystals.
Thermodynamic Parameters of CYP130-Inhibitor Interactions by ITC
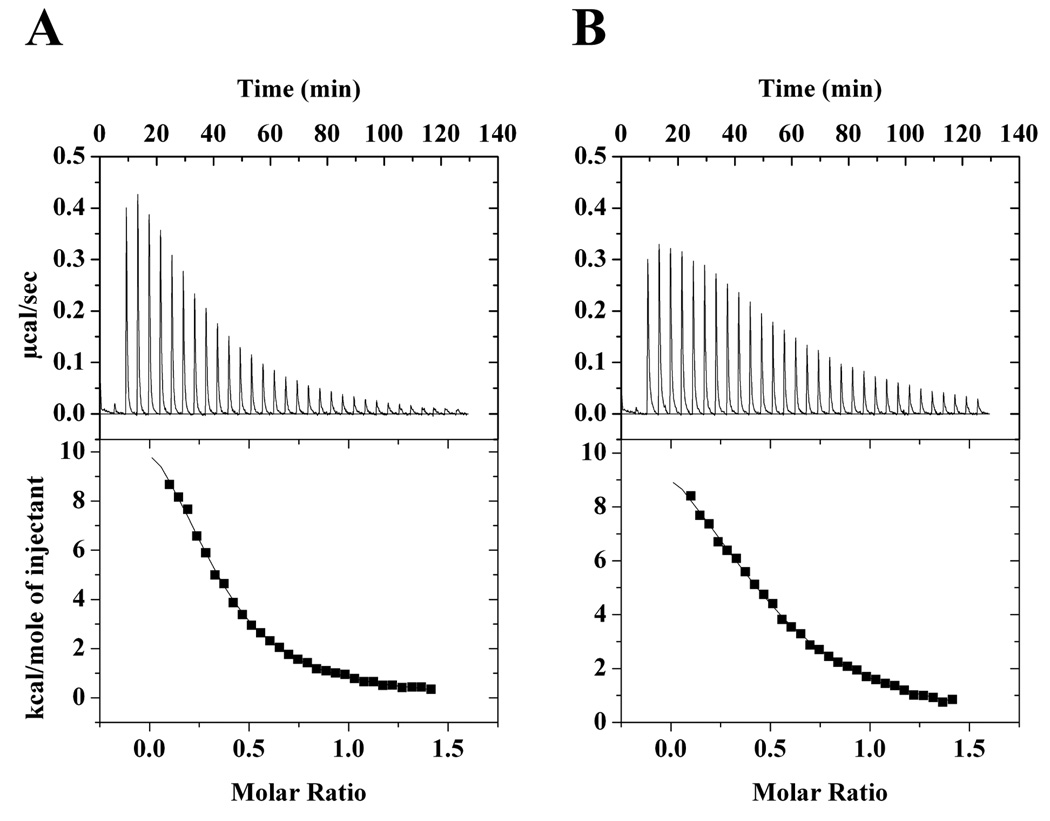
Binding of econazole and miconazole to CYP130 was addressed by ITC to examine the thermodynamics of protein-inhibitor interactions independently of the accompanying spectroscopic changes. Clotrimazole was excluded from the analysis because its low solubility in aqueous solutions obviated data acquisition. The binding isotherms were obtained in the reverse titration mode (Fig. 8). A control titration of the protein into the buffer alone was also conducted and did not reveal any significant heat of dilution, confirming the monomeric state of the protein in solution at the concentration employed (not shown). The data were best fitted to a sequential two-step binding model. The thermodynamic parameters derived from the analysis are summarized in Table 4. Binding of both inhibitors is an endothermic and entropy-driven process, as evidenced by the large and positive ΔS values. Two sequential binding steps were deduced from the ITC data for both econazole and miconazole with one set of association constants close to those obtained from the optical titrations.
Fig. 8. Calorimetric binding studies of azole drugs.
The isothermal calorimetric enthalpy changes (upper panel) and the resulting binding isotherms (lower panel) are shown for reverse titrations of CYP130 with econazole (A) and miconazole (B). The data were best fitted to a two-step sequential binding model. The binding parameters obtained are listed in Table 4.
TABLE 4.
Thermodynamic parameters of CYP130-inhibitor interactions derived from ITC and optical titration.
| Ka | ΔGa | ΔH | ΔSb | Kdcalorimetry | Kdoptical | |
|---|---|---|---|---|---|---|
| Inhibitor | (1/Ka) | |||||
| × 10−4 (M−1) | (kcal/mol) | (kcal/mol) | (cal/mol/K) | (µM) | (µM) | |
| Econazole | 1.93 ± 0.03 | |||||
| Step-1 | 2.78 ± 0.25 | −6.07 | 2.87 ± 0.34 | 30.0 | 35.9 | |
| Step-2 | 32.7 ± 3.5 | −7.51 | 9.48 ± 0.61 | 57.0 | 3.0 | |
| Miconazole | 1.70 ± 0.21 | |||||
| Step-1 | 18.7 ± 1.2 | −7.18 | 6.05 ± 0.09 | 44.4 | 5.3 | |
| Step-2 | 3.47 ± 0.37 | −6.19 | 8.47 ± 0.71 | 49.2 | 28.8 |
ΔG = −RT ln Ka.
ΔS = (ΔH − ΔG)/T.
While the first step in econazole binding detected by ITC has a Kd (35.9 µM) much higher than that obtained from the optical titration (1.9 µM), the second step occurs with a Kd of 3.0 µM that closely matches the spectroscopic dissociation constant, suggesting that full ‘Soret binding’ is achieved during the second step. Apparently, the second step represents the energies associated with the conformational changes accompanying the primary ligand recognition in the active site, presumably including protein dimerization. Collectively, the ITC and spectroscopic data indicate that these conformational changes/dimerization are beneficial to the Soret status of econazole and concur with the assumption of multimer binding cooperativity. Unlike econazole, the first step of miconazole binding occurs with a Kd (5.3 µM) close to that obtained from the optical titration (1.7 µM), suggesting that the majority of the Soret response is achieved during the first step. Although the second step (Kd =28.8 µM) significantly impacts the binding energetics (Table 4), it is virtually silent spectrally and, hence, undetectable by the spectroscopic techniques.
DISCUSSION
Econazole is an antifungal antibiotic with a potent activity against the latent and multidrug-resistant forms of tuberculosis (3–7). Mtb P450s, including CYP130, are therefore plausible therapeutic targets for the azole class of antifungal agents. CYP130 binds a number of antifungal drugs, including econazole, miconazole, clotrimazole, and ketoconazole with poorer affinities than those observed for the other two characterized Mtb P450 enzymes, CYP121 (17,48) and CYP51 (Table 2) (11, 49). Nevertheless, the potential use of P450 enzymes as therapeutic targets in Mtb depends not only on the binding affinity toward currently available azole drugs, but also on their biological roles.
The binding of azole inhibitors to CYP130 is an endothermic entropy-driven two-step process apparently complicated by protein-protein interactions manifested in the ligand-specific binding cooperativity observed for econazole and clotrimazole (Fig. 1). While virtually full Soret binding response for miconazole (the inhibitor lacking positive binding cooperativity) is achieved during the first binding step, econazole requires the second step to be completed before full spectral shift of the Soret band occurs (Table 4). We attribute the second binding step to the conformational changes associated with CYP130 dimerization. An apparent ability of CYP130 to dimerize in solution is supported by covalent cross-linking of the protein in the presence or absence of azole ligands (Fig. 6). The crystal structure indicates that dimerization is likely to involve the closed form of the protein favored by econazole binding (Fig. 5A). However, in the absence of a ligand, the equilibrium distribution of accessible protein conformers can be shifted towards the closed form as the cross-linked dimer is formed and is thus removed from the equilibrium.
It is impossible at this stage to predict whether protein-protein interactions play any physiological role in modulating the functional activities of CYP130 or other known bacterial P450 enzymes, e.g., via alteration of dimer affinity for an electron donor partner or other mechanisms. Both the native substrate(s) and an electron donor for CYP130, as for the majority of bacterial P450 enzymes, remain unknown. A similarity of dimerization patterns for two unrelated bacterial P450 proteins, CYP130 and CYP154C1 (Fig. 5), suggests that the association between two monomers may not be random. In this regard, P450-P450 interactions have been reported to modulate the catalytic activities of drug metabolizing mammalian microsomal P450 enzymes, although the occurrence and physiological significance of such interactions in intact cellular membranes remains to be confirmed. Nevertheless, a precedent for an inherent dimerization propensity among P450 enzymes is relevant to our understanding of P450-drug- and drug-drug interactions.
The binding of econazole was addressed in more detail by crystallographic studies. The position of econazole in the active site of CYP130 exhibits notable deviations from the ideal geometry that result from steric constraints imposed by the I-helix analogous to those observed for the CYP121/fluconazole complex (19). In addition, the volume of the active site cavity is larger than is required to accommodate econazole and provides room for possible alternative ligation mode(s) to the heme iron, such as that in which a water molecule is placed between the iron and the azole nitrogen. The less than perfect protein/inhibitor fit presumably contributes to the attenuated binding affinity of the complex. For instance, the affinity of the CYP121/fluconazole complex (10 µM) with the strongest observed perturbations of the triazole-heme iron coordination geometry (19) is 50- and 5-fold reduced compared to that of the CYP121/econazole (17) and CYP130/econazole complexes, respectively. Therefore, a better fit between the compound and the spatial and chemical features of the P450 active site would yield stronger inhibitors. In this regard, a portion of the active site cavity surrounded by the charged and/or hydrophilic residues D85, D246, and N177 (Fig. 7C), contrasts with the almost exclusively hydrophobic environment of the rest of the CYP130 active site. This pocket could serve as a landmark for substrate (or inhibitor) recognition, similar to that observed in the macrolide monooxygenase PikC, where a salt-bridge formed between the positively charged tertiary amino group of the macrolide substrate and a negatively charged carboxylic amino acid residue is essential to achieve catalytically competent binding (50).
The high resolution (1.46 Å) of the ligand-free structure has allowed us to define a hydrogen-bonded network that includes seven water molecules and the hydroxyl group of I-helix residue T239. These water molecules are evenly distributed and spaced by H-bond distances along the N-terminal portion of the I-helix leading from the distal water ligand to the molecular surface (Fig. 3A, 4B). The involvement of the I-helix in this hydrogen bonded network stabilizes the distal water ligand and suggests that the movement of the I-helix N-terminus that transiently exposes the active site for substrate access may, at the same time, facilitate displacement of the distal water, providing yet another level of regulation of CYP130 catalysis. Therefore, the N-terminal portion of the I-helix may mediate coupling of (i) the binding of substrate possibly assisted by protein dimerization, and (ii) the release of the axial water ligand. This coupling may be part of a regulatory mechanism aimed at preventing unproductive oxygen binding under limited access to nutrients such as oxygen (e.g., in granulomas, avascular environments where dormant infectious tubercle bacilli adapt for long-term asymptomatic survival). The reluctance of CYP130 to release the water axial ligand and thus to be reduced and bind molecular oxygen in response to a small range of potential substrates examined in this study is consistent with this assumption.
In summary, we report expression, purification, biophysical characterization, and crystallization of CYP130 in its ligand-free and econazole-bound forms. The crystal structure of the econazole-bound CYP130 is the first of a P450/econazole complex. Econazole binding in the active site involves conformational selection mediated by direct coordination to the heme iron and largely hydrophobic contacts with the active site amino acid residues. The interactions between CYP130, econazole, and other potent azole antifungal drugs were characterized in some detail by UV-visible spectroscopy, ITC, and chemical cross-linking. Overall, binding of azole inhibitors is a complex entropy-driven two-step process that appears to be assisted for econazole and clotrimazole by protein-protein interactions resulting from a propensity of the closed form of CYP130 to dimerize both in solution and in the crystal, providing evidence in support of a possible role for P450-P450 interactions in biology.
Supplementary Material
Acknowledgments
We acknowledge Dr. Chris Waddling for his assistance with the software and instrumentation in the UCSF X-Ray Facility, Dr. Vladimir N. Podust for mass spectrometric analysis of the Se-methionine CYP130 derivative, Dr. Youngchang Kim for fruitful discussions and valuable contributions, and Marco Moschini for excellent technical assistance.
Footnotes
This work was supported by NIH RO1 grants GM25515 and AI74824 (to P.O.M.), and GM078553 (to L.M.P.). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Protein Data Bank accession numbers. Atomic coordinates and structure factors determined in this study (Protein Data Bank IDs 2UUQ and 2UVN) have been deposited in the Protein Data Bank, Macrolomecular Structure Database Group, European Bioinformatics Institute.
The abbreviations are: Mtb, Mycobacterium tuberculosis; TAP, tetracycline/aminoglycoside-resistance protein; P450 or CYP, cytochrome P450; ITC, isothermal titration calorimetry; TB, terrific broth; LB, Luria-Bertani; FPLC, fast protein liquid chromatography; KPi, potassium phosphate; MAD, multiwavelength anomalous dispersion.
REFERENCES
- 1.Zhang Y. Annu. Rev. Pharmacol. Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 2.Sheehan DJ, Hitchcoch CA, Sibley CM. Clin. Microbiol. Rev. 1999;12:40–79. doi: 10.1128/cmr.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad Z, Sharma S, Khuller GK. FEMS Microbiology Letters. 2005;251:19–22. doi: 10.1016/j.femsle.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad Z, Sharma S, Khuller GK, Singh P, Faujdar J, Katoch VM. Int. J. Antimicrob. Agents. 2006;28:543–544. doi: 10.1016/j.ijantimicag.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad Z, Sharma S, Khuller GK. FEMS Microbiology Letters. 2006;261:181–186. doi: 10.1111/j.1574-6968.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad Z, Sharma S, Khuller GK. FEMS Microbiology Letters. 2006;258:200–203. doi: 10.1111/j.1574-6968.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 7.Banfi E, Scialino G, Zampieri D, Mamolo MG, Vio L, Ferrone M, Fermeglia M, Paneni MS, Pricl S. J. Antimicrob. Chemother. 2006;58:76–84. doi: 10.1093/jac/dkl182. [DOI] [PubMed] [Google Scholar]
- 8.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. Proc. Natl. Acad. Sci. U S A. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recchi C, Sclavi B, Rauzier J, Gicquel B, Reyrat JM. J. Biol. Chem. 2003;278:33763–33773. doi: 10.1074/jbc.M305963200. [DOI] [PubMed] [Google Scholar]
- 11.Bellamine A, Mangla AT, Nes WD, Waterman MR. Proc. Natl. Acad. Sci. U S A. 1999;96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatfield J, Pieters J. Science. 2000;288:1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 13.McLean KJ, Dunford AJ, Neeli R, Driscoll MD, Munro AW. Arch. Biochem. Biophys. 2007;464:228–240. doi: 10.1016/j.abb.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Ainsa JA, Blokpoel MC, Otal I, Young DB, De Smet KA, Martin C. J. Bacteriol. 1998;180:5836–5843. doi: 10.1128/jb.180.22.5836-5843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rossi E, Ainsa JA, Riccardi G. FEMS Microbiol. Rev. 2006;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- 16.Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. Infection. 2004;32:109–111. doi: 10.1007/s15010-004-3097-x. [DOI] [PubMed] [Google Scholar]
- 17.McLean KJ, Marshall KR, Richmond A, Hunter IS, Fowler K, Kieser T, Gurcha SS, Besra GS, Munro AW. Microbiology. 2002;148:2937–2949. doi: 10.1099/00221287-148-10-2937. [DOI] [PubMed] [Google Scholar]
- 18.Podust LM, Poulos TL, Waterman MR. Proc. Natl. Acad. Sci. USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seward HE, Roujeinikova A, McLean KJ, Munro AW, Leys D. J. Biol. Chem. 2006;281:39437–39443. doi: 10.1074/jbc.M607665200. [DOI] [PubMed] [Google Scholar]
- 20.Barnes HJ, Arlotto MP, Waterman MR. Proc. Natl. Acad. Sci. U S A. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omura T, Sato R. J. Biol. Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 22.Job P. Ann. Chim. 1928;9:113–203. [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 26.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crysallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.Xiang H, Tschirret-Guth RA, Ortiz De Montellano PR. J. Biol. Chem. 2000;275:35999–36006. doi: 10.1074/jbc.M005811200. [DOI] [PubMed] [Google Scholar]
- 33.Yoon MY, Campbell AP, Atkins WM. Drug Metab. Rev. 2004;36:219–230. doi: 10.1081/dmr-120033998. [DOI] [PubMed] [Google Scholar]
- 34.Cupp-Vickery J, Anderson R, Hatziris Z. Proc. Natl. Acad. Sci. U S A. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekroos M, Sjogren T. Proc. Natl. Acad. Sci. U S A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. J. Biol. Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 37.Kaminsky LS, Guengerich FP. Eur. J. Biochem. 1985;149:479–489. doi: 10.1111/j.1432-1033.1985.tb08950.x. [DOI] [PubMed] [Google Scholar]
- 38.Backes WL, Eyer CS. J. Biol. Chem. 1989;264:6252–5259. [PubMed] [Google Scholar]
- 39.Cawley GF, Batie CJ, Backes WL. Biochemistry. 1995;34:1244–1247. doi: 10.1021/bi00004a018. [DOI] [PubMed] [Google Scholar]
- 40.Cawley GF, Zhang S, Kelley RW, Backes WL. Drug. Metab. Dispos. 2001;29:1529–1534. [PubMed] [Google Scholar]
- 41.Backes WL, Batie CJ, Cawley GF. Biochemistry. 1998;37:12852–12859. doi: 10.1021/bi980674a. [DOI] [PubMed] [Google Scholar]
- 42.Hazai E, Kupfer D. Drug. Metab. Dispos. 2005;33:157–164. doi: 10.1124/dmd.104.001578. [DOI] [PubMed] [Google Scholar]
- 43.Jamakhandi AP, Kuzmic P, Sanders DE, Miller GP. Biochemistry. 2007;46:10192–10201. doi: 10.1021/bi7003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hazai E, Bikadi Z, Simonyi M, Kupfer D. J. Comput. Aided Mol. Des. 2005;19:271–285. doi: 10.1007/s10822-005-4995-4. [DOI] [PubMed] [Google Scholar]
- 45.Podust LM, Kim Y, Arase M, Neely BA, Beck BJ, Bach H, Sherman DH, Lamb DC, Kelly SL, Waterman MR. J. Biol. Chem. 2003;278:12214–12221. doi: 10.1074/jbc.M212210200. [DOI] [PubMed] [Google Scholar]
- 46.Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. Biotechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 47.Patchkovskii S, Ziegler T. Inorg. Chem. 2000;39:5354–5364. doi: 10.1021/ic0005691. [DOI] [PubMed] [Google Scholar]
- 48.McLean KJ, Cheesman MR, Rivers SL, Richmond A, Leys D, Chapman SK, Reid GA, Price NC, Kelly SM, Clarkson J, Smith WE, Munro AW. J. Inorg. Biochem. 2002;91:527–541. doi: 10.1016/s0162-0134(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 49.Guardiola-Diaz HM, Foster LA, Mushrush D, Vaz AD. Biochem. Pharmacol. 2001;61:1463–1470. doi: 10.1016/s0006-2952(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 50.Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. J. Biol. Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Evans SV. J. Mol. Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.