Abstract
Background
Previous phylogenetics studies of Asparagales, although extensive and generally well supported, have left several sets of taxa unclearly placed and have not addressed all relationships within certain clades thoroughly (some clades were relatively sparsely sampled). One of the most important of these is sampling within and placement of Nolinoideae (Ruscaceae s.l.) of Asparagaceae sensu Angiosperm Phylogeny Group (APG) III, which subfamily includes taxa previously referred to Convallariaceae, Dracaenaaceae, Eriospermaceae, Nolinaceae and Ruscaceae.
Methods
A phylogenetic analysis of a combined data set for 126 taxa of Ruscaceae s.l. and related groups in Asparagales based on three nuclear and plastid DNA coding genes, 18S rDNA (1796 bp), rbcL (1338 bp) and matK (1668 bp), representing a total of approx. 4·8 kb is presented. Parsimony and Bayesian inference analyses were conducted to elucidate relationships of Ruscaceae s.l. and related groups, and parsimony bootstrap analysis was performed to assess support of clades.
Key Results
The combination of the three genes results in the most highly resolved and strongly supported topology yet obtained for Asparagales including Ruscaceae s.l. Asparagales relationships are nearly congruent with previous combined gene analyses, which were reflected in the APG III classification. Parsimony and Bayesian analyses yield identical relationships except for some slight variation among the core asparagoid families, which nevertheless form a strongly supported group in both types of analyses. In core asparagoids, five major clades are identified: (1) Alliaceae s.l. (sensu APG III, Amarylidaceae–Agapanthaceae–Alliaceae); (2) Asparagaceae–Laxmanniaceae–Ruscaceae s.l.; (3) Themidaceae; (4) Hyacinthaceae; (5) Anemarrhenaceae–Behniaceae–Herreriaceae–Agavaceae (clades 2–5 collectively Asparagaceae s.l. sensu APG III). The position of Aphyllanthes is labile, but it is sister to Themidaceae in the combined maximum-parsimony tree and sister to Anemarrhenaceae in the Bayesian analysis. The highly supported clade of Xanthorrhoeaceae s.l. (sensu APG III, including Asphodelaceae and Hemerocallidaceae) is sister to the core asparagoids. Ruscaceae s.l. are a well-supported group. Asparagaceae s.s. are sister to Ruscaceae s.l., even though the clade of the two families is weakly supported; Laxmanniaceae are strongly supported as sister to Ruscaceae s.l. and Asparagaceae. Ruscaceae s.l. include six principal clades that often reflect previously named groups: (1) tribe Polygonateae (excluding Disporopsis); (2) tribe Ophiopogoneae; (3) tribe Convallarieae (excluding Theropogon); (4) Ruscaceae s.s. + Dracaenaceae + Theropogon + Disporopsis + Comospermum; (5) Nolinaceae, (6) Eriospermum.
Conclusions
The analyses here were largely conducted with new data collected for the same loci as in previous studies, but in this case from different species/DNA accessions and greater sampling in many cases than in previously published analyses; nonetheless, the results largely mirror those of previously conducted studies. This demonstrates the robustness of these results and answers questions often raised about reproducibility of DNA results, given the often sparse sampling of taxa in some studies, particularly the earliest ones. The results also provide a clear set of patterns on which to base a new classification of the subfamilies of Asparagaceae s.l., particularly Ruscaceae s.l. (= Nolinoideae of Asparagaceae s.l.), and examine other putatively important characters of Asparagales.
Keywords: Aphyllanthes, Asparagaceae, Convallariaceae, Dracaenaceae, Eriospermum, monocot phylogenetics, Nolinaceae, Nolinoideae
INTRODUCTION
Asparagales are the largest order among the five orders of Lilianae (= Liliiflorae) sensu Dahlgren et al. (1985), who followed the concepts of Huber (1969). There are up to 29 families [APG (Angiosperm Phylogeny Group), 1998] in the order, which has been considered monophyletic on the basis of their phytomelan-containing seed coat and several other characteristics (Huber, 1969; Rudall et al., 2000; Chase et al., 2006). Chase et al. (1995a) performed the first extensively sampled phylogenetic analysis to examine their circumscription. This analysis led to the recircumscription of Asparagales to include Orchidaceae (including the former Apostasiaceae and Cypripediaceae) and Iridaceae (including the former Geosiridaceae), both families formerly Liliales/Orchidales, and to exclude Dasypogonaceae s.l., Hanguanaceae, Luzuriagaceae and Philesiaceae. The boundary between Asparagales and Liliales can be difficult to define with morphological data alone because several characters are shared by some lilioids and asparagoids, especially net-veined taxa (Conran, 1989; Rudall et al., 2000). The combined molecular–morphology analysis (Chase et al., 1995b) indicated that although the lilioid monocots were monophyletic, several asparagoid families were paraphyletic or polyphyletic (Chase et al., 1995a, 2006). Within Asparagales there was a paraphyletic grade (predominantly characterized by simultaneous microsporogenesis and inferior ovaries) and a ‘core asparagoid’ clade, uniformly characterized by successive microsporogenesis and mostly superior ovaries (Rudall et al., 1997; Furness and Rudall, 1999). The combined plastid DNA (including rbcL, atpB, trnL intron, and trnL-F intergenic spacer) analyses by Fay et al. (2000) and additional DNA sequences by Pires et al. (2006) further resolved phylogenetic relationships within Asparagales. To accord with the molecular and morphological studies (Chase et al., 1995b, b; Fay and Chase, 1996; Rudall et al., 1997, 2000; Fay et al., 2000), many families in Asparagales have been recircumscribed (APG, 1998; APG II, 2003), and several new families have been erected (Chase et al., 1996, 1997; Conran et al., 1997; Fay and Chase, 1996; Rudall and Chase, 1996).
Ruscaceae sensu lato are a recently recognized family in the broad sense (Chase et al., 1995a; Rudall et al., 2000; APG, 1998); they include Ruscaceae s.s., Convallariaceae, Nolinaceae, Dracaenaceae, Eriospermaceae and Comospermum (the last of highly speculative placement in Dahlgren et al., 1985). Ruscaceae s.l. can be distinguished from other higher asparagoid groups by usually possessing berries or other indehiscent fruit types and absence of phytomelan in the seed coat. One might suppose that indehiscent fruits and absence of phytomelan could be correlated characters, but in Asparagus (Asparagaceae s.s.) berries and phytomelan co-occur. The combined analysis of rbcL and morphology (Chase et al., 1995b; Rudall et al., 1997) indicated that several genera that had been included in Convallariaceae were members of other families or were embedded within a larger clade; this larger clade was recognized as the newly circumscribed broad-sense Convallariaceae (APG, 1998; Fay et al., 2000). Rudall et al. (2000) suggested Ruscaceae Sprengel (1826) had priority over Convallariaceae Horaninow (1834), and they are now generally referred to as Ruscaceae s.l. (Jang and Pfosser, 2002; APG II, 2003), which could also be included in a much-expanded circumscription of Asparagaceae. The latter was presented as an alternative classification in APG II. In APG III (2009), the broadly circumscribed families (including Asparagaceae s.l.) were accepted as the only circumscription in accord with APG, in which case this clade would be referred to as subfamily Nolinoideae.
Ruscaceae s.s., comprising three genera (Ruscus, Danae and Semele), are distributed in the Mediterranean–Macronesian area; they have woody stems, scale-like leaves, berries, and a basic chromosome number of x = 20. Dahlgren et al. (1985) and Takhtajan (1997) regarded Ruscaceae s.s. as the most closely related group to Asparagaceae s.s., but there has been no clear evidence on relationships of these families. The two families have several similarities including phylloclades (but even for this character there are questions about homology; Arber, 1924; Cooney-Sovetts and Sattler, 1986), baccate fruits and similar karyotypes (Sato, 1942; Tamura, 1995), and they have differences in the position of inflorescences and seed coat (Conran and Tamura, 1998). Rudall et al. (1998) recognized that the karyotype of Ruscaceae (x = 20) is more similar to Convallariaceae (usually x = 19, rarely 18, 20) than to that of Asparagaceae s.s. (mostly x = 10). Serological analyses and lack of phytomelan in the seed coat indicated a closer relationship between Ruscaceae s.s. and Convallariaceae than either to Asparagaceae s.s. (Chupov and Cutjavina, 1980).
Convallariaceae are rhizomatous perennial herbs distributed in the Northern Hemisphere; they are abundant in eastern and southeastern Asia and comprise four tribes: Polygonateae, Ophiopogoneae, Convallarieae and Aspidistreae (Dahlgren et al., 1985; Tamura, 1995). They share calcium oxalate crystals and two ovules (rarely or over) per locule, but it is not so easy to identify distinguishing morphological characters for the tribes in Convallariaceae, and Dahlgren et al. (1985) used plesiomorphic characters for the taxonomic key, including baccate fruits, non-phytomelaniferous seeds and nuclear endosperm formation. Polygonateae share a sympodial rhizome, an elongated aerial stem and berries, and the position and shape of inflorescences (axillary in Polygonatum and Disporopsis, terminal in Smilacina and Maianthemum, and axillary and terminal in Heteropolygonatum) are variable in the tribe. Ophiopogoneae have a sympodial rhizome, fruits that rupture at an early stage, seeds with sarcotesta, and basic chromosome number x = 18; this tribe comprises three genera (Liriope, Ophiopogon and Peliosanthes) distributed in eastern and southeastern Asia. Convallarieae and Aspidistreae have a monopodial rhizome and a short stem, usually berries (except drupes in Tricalistra), and basic chromosome number usually of x = 19, rarely 20 (Theropogon) or 18 (some Aspidistra). Conran and Tamura (1998) merged Aspidistreae with Convallarieae. The plastid trnK sequence analysis of Yamashita and Tamura (2000) supported the treatment of Conran and Tamura (1998).
Nolinaceae are arborescent, anomalously woody plants with terminal rosette leaves and indehiscent nutlets, and they comprise four genera, Nolina, Dasylirion, Calibanus and Beaucarnea, found in warm, dry regions of North America. Nolinaceae were often previously included in a broadly defined family Liliaceae near Dracaena, and they had been treated in the tribe Dracaeneae (Bentham and Hooker, 1883) or Nolineae (Krause, 1930). Hutchinson (1934) included Nolinaceae, Yuccoideae and Dracaenae in Agavaceae because of their anomalous woody growth (via a secondary thickening meristem) and fibrous leaves, but this treatment was not supported by other morphological characters (flowers, fruits and seeds) and karyology (Sharma and Chaudhuri, 1964). Nolinaceae were excluded from Agavaceae and arranged near Dracaenaceae in Dahlgren et al. (1985).
Dracaenaceae include perennial plants with a more or less woody trunk, but many do not have a trunk; they comprise two genera, Dracaena and Sansevieria (perhaps best combined into one genus), which occur in subtropical to tropical regions of the Old World. Dracaenaceae are distinguished from Nolinaceae in having berries, no oils in guard cells and mucilage-filled cells with crystal raphides in vegetative parts.
Eriospermaceae are perennial herbs with various types of tubers and free perianth parts. They comprise a single genus (Eriospermum) distributed in southern parts of Africa. This family shows seasonal developmental differences between leaves and inflorescences. Because they have extraordinary characters such as leaf appendages, epidermal hairs on the seeds and embryological attributes but have successive microsporogenesis and thin testa, Dahlgren et al. (1995) suggested that treatment as a family separate from related groups was probably best. The taxonomic position of Eriospermaceae has been controversial whether included (Rudall et al., 2000) or not (Jang and Pfosser, 2002) in Ruscaceae s.l.
Ruscaceae s.l. have no distinguishable synapomorphic characters from the other higher asparagoids except the absence of phytomelan in the seed coat, but analysis of the combined molecular and morphology matrix (Chase et al., 1995b) indicated that Ruscaceae s.l. was a well-supported clade that was largely unresolved relative to the related families and genera. This grouping of Ruscaceae s.l. (= Convallariaceae s.l.) was supported by plastid DNA restriction-site analyses of some taxa, although Bogler and Simpson (1995) lacked some of the core taxa such as Ruscaceae s.s. and Comospermum. Several molecular studies supported monophyly of Ruscaceae s.l. (Rudall et al., 1997, 2000). Yamashita and Tamura (2000) sequenced the plastid trnK region (including the matK exon) for 39 Convallariaceae species and related families, which indicated that there were six major clades; Convallariaceae s.s. were paraphyletic in this analysis. They compared the trnK tree with the rbcL tree and looked at basic chromosome numbers, but they occasionally had unresolved relationships due to a lack of informative characters and sampling of potential sister groups; they nonetheless found evidence to support the tribal limits in Convallariaceae of Conran and Tamura (1998). Jang and Pfosser (2002) performed a phylogenetic analysis based on rbcL and trnL-F intron/spacer sequences, but there were no improved assessments of relationships because of poor sampling of taxa in Ruscaceae s.l.
Asparagaceae s.s. have been usually considered sister to Ruscaceae s.l. due to their cytological and morphological similarities (Tamura, 1995). Aphyllanthes (Aphyllanthaceae) was also indicated as a possible sister group to Ruscaceae (Conran, 1998; Yamashita and Tamura, 2000), but Fay et al. (2000) made a cautious accessment of Aphyllanthes, a taxonomically isolated Mediterranean genus, because of its labile phylogenetic position. Laxmanniaceae were sister to Ruscaceae s.l. plus Asparagaceae (Rudall et al., 1997; Fay et al., 2000; Bogler et al., 2006; Givnish et al., 2006; Graham et al., 2006; Pires et al., 2006). APG II (2003) and APG III (2009) suggested a broader circumscription of Asparagaceae based largely on results of analysis for four plastid DNA regions (Fay et al., 2000); Ruscaceae s.l. was treated as an optional circumscription along with Agavaceae s.l. (including Anemarrhenaceae, Anthericaceae, Behniaceae, Herreriaceae and Hesperocallidaceae) and related families such as Aphyllanthaceae, Hyacinthaceae, Laxmanniaceae and Themidaceae.
A molecular phylogenetic study was conducted to re-evaluate delimitation of Ruscaceae s.l. of Rudall et al. (2000) and related families (APG, 1998; APG II, 2003; APG III, 2009; Chase et al., 2006), especially to assess their possible sister groups in Asparagales and evaluate phylogenetic relationships with the related families in the core asparagoids. The aim was to investigate relationships in Asparagales by sequencing three genes, 18S nuclear ribosomal DNA and plastid rbcL and matK, for 121 taxa of Asparagales. These genes were chosen because of their use in recent studies of familial and higher-level phylogenetics (Chase et al., 1995a, 2006; Soltis et al., 1997, 2000; Fay et al., 2000; Yamashita and Tamura, 2000; Hilu et al., 2003; Devey et al., 2006). The impact of these data on the classification of Ruscaceae s.l. and related families was also evaluated. New sequences from mostly new accessions of the sampled taxa were produced for this study; this was done to avoid possibile misidentification of taxa in the earlier published studies or sequences with errors due to the prelatively primitive techniques used to produce rbcL and 18S rDNA sequences in the early period of DNA sequencing.
MATERIALS AND METHODS
Plant materials
The taxa used for this study included all genera (except Heteropolygonatum) in Ruscaceae s.l. sensu Rudall et al. (2000) and representatives of all families of Asparagales (APG). The plant material used was either fresh, collected from the field and dried, taken from specimens in the herbarium, or was a DNA sample borrowed from the Royal Botanic Gardens, Kew, DNA Bank (http://data.kew.org/dnabank/DnaBankForm.html). Voucher specimens of the taxa were prepared; source, voucher information and database accession numbers are listed in the Appendix. Provenance and distributions were also prepared from voucher specimens and the World Checklist of Selected Plant Families (http://apps.kew.org/wcsp/home.do). For one taxon (Bulbine sp.), sequences from different species (B. succulenta and B. frutescens) in GenBank were used, and several sequences (six for 18S rDNA, nine for rbcL and ten for matK) were from GenBank and previous papers (Chase et al., 2006). Otherwise, new sequences were prepared.
DNA extraction, PCR, sequencing and alignment
Total genomic DNA was extracted from 0·5–1·0 g of fresh or silica gel-dried leaves using the 2× CTAB buffer method (Doyle and Doyle, 1987). Lipids were removed with SEVAG solution (24 : 1 chloroform : isoamyl alcohol), and DNA was precipitated with isopropanol at –20 °C. Total extracted DNA was dissolved in 1× TE buffer and stored at –70 °C, and the concentration of DNA was determined with GeneQuant pro (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA) before use.
The 18S rDNA gene was amplified using the primers and protocols of White et al. (1990), Nickrent and Soltis (1995), and Soltis and Soltis (1998); matK was amplified with primers and protocols of Johnson and Soltis (1995) and Hilu et al. (2003), and the rbcL gene was amplified with primers and protocols of Omstead et al. (1992), Shinwari et al. (1994) and Fay and Chase (1996). Amplifications were carried out in 50-μL reactions, containing 2 units Taq DNA polymerase, 5 µL 10× reaction buffer (100 mm Tris–HCl, 500 mm KCl, 15 mm MgCl2), 2·5 mm dNTPs, 5 pmol μL−1 forward and reverse primers, using Perkin-Elmer 9700 machine (Applied Biosystems, Inc., Beverly, MA, USA). DMSO (2 %) was added to reduce the secondary structure in PCR. PCR conditions were premelt of 94 °C for 2 min, followed by 30–35 cycles of denaturation at 94 °C for 1 min, annealing at 50–55 °C for 1 min, extension at 72 °C for 3 min, followed by a final extension of 7 min at 72 °C.
All PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH, USA) according to the manufacturer's protocols. Dideoxy cycle sequencing was performed using the chain-termination method and the ABI prism big dye reaction kit (ver. 3·1) following the manufacturer's protocols. Products were run on an ABI 3700 genetic analyser or MegaBace1000 (Amersham Pharmacia Biotech, Inc.) using the manufacturers' protocols. Sequence editing and assembly of contigs were carried out using Sequence Navigator and AutoAssembler software (ABI).
All sequences were aligned initially in ClustalX (ver. 1·83; Thompson et al., 1997) and MacClade (ver. 4·0; Maddison and Maddison, 2000) and then manually adjusted following the guidelines of Kelchner (2000). Alignment of sequences for these coding genes was easily performed because there were no insertions/deletions (indels) among the sequences of Ruscaceae s.l., but there were indels in the sequences of other Asparagales and outgroups: three in 18S rDNA and nine in matK; the aligned matrix is available from kimjh@dju.ac.kr or m.chase@kew.org. The three indels in 18S rDNA correspond to positions 496–501, 666–672 and 1363–1369 on the reference sequence of Glycine max (L.) Merr. (Soltis et al., 1997, 2000; Soltis and Soltis, 1998).
Parsimony analysis
Two separate sets of analyses were carried out. The first (analysis A) comprised the plastid sequences of 121 taxa representing all 29 families of Asparagales, and the second (analysis B) comprised the combined 18S rDNA and plastid DNA sequences for the same taxa. Orchidaceae were designated as the outgroup for both analyses based on previous results (Chase et al., 1995a, 2000b; Fay et al., 2000). PAUP* (ver. 4·10b; Swofford, 2007) was used for parsimony analysis and followed the widely used parsimony analysis with successive approximations weighting and bootstrapping (Fay et al., 2000; Clarkson et al., 2004; the bootstrap did not use the relative weights). In analyses A and B, tree searches were performed under the Fitch (equal weight, EW; Fitch, 1971) criterion with 1000 random sequence additions and tree–bisection–reconnection (TBR) branch swapping, permitting ten trees to be held at each step (Multrees on) to reduce time searching suboptimal ‘islands’ of trees (Chase et al., 2006). All shortest trees collected in the 1000 replicates were swapped on to completion without a tree limit. Successive approximation weighting (SW; Farris, 1989) was carried out to select the most stable trees (Carpenter, 1988) according to the rescaled consistency index, using the maximum value (best fit) criterion and a base weight of 1·0, followed by 100 replicates of heuristic search with random sequence additions and subtree pruning-regrafting (SPR) swapping. All shortest trees from these 100 replicates were then swapped to completion, after which another round of weighting was implemented. This process was repeated until the same tree length was obtained twice in succession. DELTRAN character optimization was used to illustrate branch length throughout. To evaluate internal support, 1000 bootstrap replicates were carried out with equal weights, TBR branch swapping with five trees held at each step and simple taxon addition (Felsenstein, 1985). The following descriptions for categories of bootstrap support were used: weak, 50–74; moderate; 75–84; well supported, 85–100 % (Chase et al., 2000a).
Bayesian analysis
Further phylogenetic analyses were performed using Bayesian inference as implemented in MrBayes (ver. 3·12; Ronquist et al., 2005). MrModeltest (ver. 2·2; Nylander, 2005) was used to determine the best model of DNA substitution for each partition, evaluating all models against defaults of the program. The GTR + I + G model (a general time reversible model with a proportion of invariable sites and a gamma-shaped distribution of rates across sites) was chosen for the three genes as the best-fitting among the 24 models compared. Thus, all three genes were assigned a model of six substitution types (n = 6) with a proportion of invariable sites. Four simultaneous Markov Chain Monte Carlo (MCMC) chains were run for 5 × 106 generations and sampled every 100 generations, and the first 1 × 105 trees were excluded (‘burn-in’). Post-burn-in samples of trees drawn from the posterior probability distribution were summarized, and this tree is illustrated (see Fig. 3). Bayesian analysis was performed three times to ensure convergence of results.
Fig. 3.
Bayesian tree from combined DNA analysis (analysis C) for 121 taxa of Asparagales. The numbers above branches are posterior probabilities from 5 × 106 generations with the GTR + I + G model. A, Asparagaceae; B, Laxmanniaceae.
RESULTS
A summary of characteristics of the DNA data is presented in Table 1. The aligned number of characters was 4802, but 71 positions for 18S rDNA were excluded from phylogenetic analyses as in previous studies due to ambiguous alignments in these short sections of the matrix (Soltis et al., 1997, 2000; Soltis and Soltis, 1998). The total number of included bases was 4731 of which 1851 were variable (39·1 %) and 1301 (27·5 %) were potentially parsimony informative. The number of positions in the matrix included 1338 for rbcL, 1668 for matK and 1725 for 18S rDNA. The matK gene was the most variable among the three genes and gave the greatest number of parsimony informative sites; 18S rDNA showed the lowest variation. The number of parsimony-informative characters was 327 (25·1 %) for rbcL, 784 (60·3 %) for matK and 190 (14·6 %) for 18S rDNA.
Table 1.
Statistics for the three genes analysed in this study
| Characters | rbcL (I) | matK (II) | 18S rDNA (III) | Plastid data (I + II) | Combined (I + II + III) |
|---|---|---|---|---|---|
| Aligned | 1338 | 1668 | 1796 | 3006 | 4802 |
| Included | 1338 | 1668 | 1725 | 3006 | 4731 |
| Parsimony informative | 327 | 784 | 190 | 1111 | 1301 |
| Variable | 462 | 1072 | 317 | 1534 | 1851 |
| Constant | 876 | 596 | 1408 | 1472 | 2880 |
| Transition/transversion | 877/456 (1·87) | 1868/1189 (1·52) | 507/179 (2·52) | ||
| G + C (%) | 43·45 | 31·37 | 50·43 | ||
| Tree length (EW/SW) | (1572/407·660) | (3744/1156·147) | (902/290·889) | (5435/1537·590) | (6442/1811·619) |
| CI (EW/SW) | (0·40/0·71) | (0·44/0·70) | (0·46/0·80) | (0·42/0·70) | (0·42/0·71) |
| RI (EW/SW) | (0·69/0·84) | (0·74/0·86) | (0·65/0·82) | (0·72/0·85) | (0·70/0·84) |
EW, Equally weighted; SW, successive weighted; CI, consistence index; RI, retention index.
Parsimony analysis based on plastid DNA (analysis A)
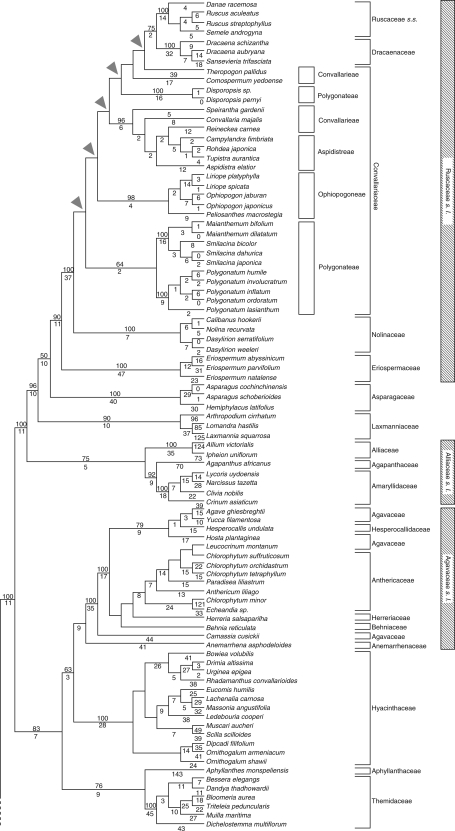
The final alignment of the combined (rbcL and matK) plastid DNA matrix comprised 3006 positions, of which 1534 were variable (51·0 %) and 1111 (37·0 %) were potentially parsimony informative. Fitch analysis (EW; Table 1) produced 5760 equally most-parsimonious trees [length = 5435 steps; CI (consistency index, including autapomorphies) = 0·42; RI (retention index) = 0·72]. Successive weighting (SW) identified one shortest tree as optimal with an SW score of 1537·59 (5435 Fitch length; CI = 0·70, RI = 0·85). The SW tree is therefore one of the trees found with equal weights; it is shown with its Fitch branch lengths (DELTRAN optimization) in Fig. 1. Groups (nodes) not found in the consensus tree of Fitch analysis are marked with triangles. Bootstrap percentages (BP) consistent with the strict consensus tree are shown below each branch; groups with BP < 50 are not indicated.
Fig. 1.
The single shortest tree from successive weighting of plastid rbcL and matK (analysis A) for Ruscaceae s.l. and related groups of Asparagales. Numbers of substitutions are indicated below each branch (DELTRAN optimization), and bootstrap percentages >50 % are given above each branch. Triangles indicate branches not present in the strict consensus tree of 5760 equally MP trees by Fitch analysis (equal weight). Tree length is 5435 steps with CI = 0·70 and RI = 0·85. The dashed line in the lower left-hand corner marks the point where the non-core asparagoids are attached to this part of the tree (non-core taxa are not shown; this part of the tree is identical to that show in Fig. 2).
In this study, only the core asparagoids are presented for the plastid DNA tree (Fig. 1) since it showed a topology similar to that of the combined DNA tree except for relationships among Ruscaceae s.l. and related families. The core asparagoids formed a strongly supported group (BP 100), and the other asparagoids were paraphyletic (not shown). The core asparagoids fell into two clades, one moderately (BP 84) and the other well supported (BP 90). The first consisted of four families including Agavaceae s.l. sensu APG I (BP 96), Hyacinthaceae (BP 100) and Themidaceae (BP 100), as well as Aphyllanthaceae. The second consisted of Ruscaceae s.l., Asparagaceae, Laxmanniaceae, Alliaceae, Agapanthaceae and Amaryllidaceae.
Within the second group, Ruscaceae s.l. were well supported (BP 90; Fig. 1). Asparagaceae s.s. were strongly supported (BP 100) and sister to Ruscaceae s.l., but the two families together were weakly supported (BP 50); Laxmanniaceae were strongly supported (BP 96) as a member of the clade with Ruscaceae s.l. and Asparagaceae s.s. Alliaceae s.l. sensu APG (1998) including Alliaceae s.s., Agapanthaceae and Amaryllidaceae form a moderately supported clade (BP 75) as the sister of the rest (Fig. 1).
The tree topology of Ruscaceae s.l. in this study did not accord or was only partly congruent with previous studies (Rudall et al., 1997; Yamashita and Tamura, 2000; Jang and Pfosser, 2002). Ruscaceae s.l. were strongly supported (BP 90), and within this clade fell Ruscaceae s.s., Dracaenaceae, Convallariaceae, Nolinaceae and Eriospermaceae (Fig. 1). The combined Ruscaceae s.s. and Dracaenaceae clade was moderately supported (BP 75), and they were interdigitated within clades of Convallariaceae. Within Convallariaceae, Aspidistreae (BP 96; including Campylandra, Rohdea, Tupistra and Aspidistra) and Ophiopogoneae (BP 98; including Liriope, Ophiopogon and Peliosanthes) were strongly supported. Convallarieae were not monophyletic, and Polygonateae were only weakly supported as monophyletic (BP 64) and excluded Disporopsis (BP 100). Eriospermaceae (BP 100) were sister to highly supported Nolinaceae (BP 100).
Parsimony analysis based on combined DNA (analysis B)
The number of positions included in the combined analysis (18S rDNA, rbcL and matK) was 4731. The number of bases contributed by each individual gene was 1338 for rbcL, 1668 for matK and 1725 for 18S rDNA. The number of variable sites was 1851 (39·1 %), and 1301 (27·5 %) were potentially parsimony informative. Fitch analysis (EW), including 121 asparagoid monocots (Table 1), produced 5721 equally most-parsimonious trees of 6442 steps with CI (including autapomorphies) = 0·42 and RI = 0·70. Successive weighting (SW) identified one shortest tree as optimal with an SW score of 1811·62 (6442 Fitch length; CI = 0·71, RI = 0·85). The SW tree was one of the Fitch trees, and it is shown with its Fitch branch lengths (DELTRAN optimization) in Fig. 2. Groups not found in the strict consensus tree of the Fitch analysis are marked with triangles. Bootstrap percentages (BP; equal weights) consistent with the strict consensus tree are shown below each branch, but groups with BP < 50 are not indicated (Fig. 2).
Fig. 2.
One MP tree from combined DNA data (analysis B) for 121 taxa of Asparagales. Numbers of substitutions are indicated below each branch, and bootstrap percentages >50 % are given above each branch. Triangles indicate groups not present in the strict consensus tree of the Fitch analysis. Bars show points at which changes in microsporogenesis have taken place. Tree length is 6442 steps with CI = 0·71 and RI = 0·85. A, Asparagaceae; B, Laxmanniaceae; C, Alliaceae s.l.; D, Agavaceae s.l.; E, Hyacinthaceae; F, Aphyllanthaceae; G, Themidaceae; H, Xanthorrhoeaceae s.l.
The topology of the combined DNA tree for Asparagales largely followed the previous analyses in the broad sense of the core asparagoids concept (Chase et al., 1995a; Fay et al., 2000; Pires et al., 2006). The core asparagoids formed a strongly supported group (BP 100) with the rest of the families of Asparagales forming a grade relative to the core group (Fig. 2). The core asparagoids fell into two big clades, one with strong support (BP 86; group B) and the other with weak support (BP 56; group A). The former consisted of four families including Agavaceae s.l. sensu APG II (BP 94), Hyacinthaceae (BP 100), Themidaceae (BP 100) and Aphyllanthaceae. The other consisted of Ruscaceae s.l., Asparagaceae s.s., Laxmanniaceae and Alliaceae s.l.
Within group A in the core asparagoids, Ruscaceae s.l. were well-supported (BP 88; Fig. 2). Asparagaceae s.s. were strongly supported (BP 100) as sister to Ruscaceae s.l., even though the clade of the two was weakly supported (BP 50), and Laxmanniaceae appeared as sister (BP 96) to Ruscaceae s.l. and Asparagaceae. Alliaceae s.l. sensu APG (1998) were weakly supported (BP 73) as sister to group A (Fig. 2).
The tree topology of Ruscaceae s.l. from the combined analysis did not accord with or was only partly congruent with previous plastid analyses (Rudall et al., 1997; Yamashita and Tamura, 2000; Jang and Pfosser, 2002). Ruscaceae s.l. were grouped together in one strongly supported clade (BP 88; Fig. 2), and Ruscaceae s.s., Dracaenaceae, Nolinaceae and Eriospermaceae received strong bootstrap support (BP > 99 %) even though Convallariaceae were polyphyletic (Fig. 2). For the tribes of non-monophyletic Convallariaceae, Aspidistreae (BP 64) and Ophiopogoneae (BP 99) were monophyletic, but Convallarieae were not monophyletic. Polygonateae were monophyletic but weakly supported (BP 60). Nolinaceae were sister to the rest of Ruscaceae s.l. minus Eriospermum (BP 99). Strongly supported Eriospermaceae (BP 100) were sister to the rest of Ruscaceae s.l.
Outside the core asparagoids, the tree topology from analysis of the combined DNA data was congruent with those from previous analyses (Fay et al., 2000; Pires et al., 2006). The major differences between combined and plastid results were mostly not in topology but rather in levels of support. The core asparagoids were sister to Xanthorrhoeaceae s.l. sensu APG III (2009) with strong support (BP 100). Xeronemataceae were sister to the large clade (core asparagoids and Xanthorrhoeaceae s.l.; BP 99), which was strongly supported. Iridaceae were strongly supported (BP 100) as sister to the above clade. The relationships among the next asparagoid families [(Ixioliriaceae + Tecophilaceae) Doryanthaceae] were strongly supported (BP < 94), and the sister to the above clade (BP 100) consisted of <Branfordiaceae {Boryaceae [Asteliaceae (Hypoxidaceae + Lanariaceae)]}>. The final clade in Asparagales was Orchidaceae, which was designated in this study as outgroup to the rest of the order (BP 100), following results of broader monocot analyses that demonstrated Orchidaceae to be sister to the rest of Asparagales (Chase et al., 2006).
Bayesian analysis of combined matrix (Analysis C)
The Bayesian tree (Fig. 3) shows the posterior probabilities summarized from the set of recovered post-burn-in trees; parameters of the GTR + I + G model used in this analysis are listed in Table 2. Although one node in the core asparagoids had low posterior probability (PP), 0·62, the majority of nodes in the tree are supported by PPs >0·95. Bayesian analysis produced a similar overall topology to that of the maximum parsimony analysis (Fig. 3), but it showed a few differences in the core asparagoids. The core asparagoids were strongly supported (1·00 PP; Fig. 3). Within the core asparagoids, a big clade consisting of Ruscaceae s.l. (PP 0·99), Asparagaceae (PP 1·00) and Laxmanniaceae (PP 1·00) was highly supported (PP 1·00). Among the taxa of Ruscaceae s.l., Dracaenaceae (PP 1·00), Ruscaceae s.s. (PP 1·00) and Eriospermaceae (PP 1·00) were strongly supported, but the former four tribes of Convallariaceae were not monophyletic except for Ophiopogoneae (PP 1·00). Agavaceae s.l. (sensu APG III) including Anemarrhenaceae, Anthericaceae, Behniaceae and Herreriaceae were weakly supported (PP 0·86). A combined clade with Agavaceae s.l. and Aphyllanthaceae showed low PP (PP 0·37), and the node with Themidaceae (PP 1·00) and Hyacinthaceae (PP 1·00) was highly supported (PP 0·92). Amaryllidaceae s.l. (sensu APG III) consisting of Alliaceae (PP 1·00), Amaryllidaceae s.s. (PP 1·00) and Agapanthaceae were strongly supported (PP 1·00).
Table 2.
Parameters of models for each gene as estimated by MrModeltest 2·1
| Parameters* | rbcL | matK | 18S rDNA |
|---|---|---|---|
| r(G ↔ T) | 1 | 1 | 1 |
| r(C ↔ T) | 4·0874 | 2·9017 | 11·9400 |
| r(C ↔ G) | 1·1394 | 0·9554 | 0·4848 |
| r(A ↔ T) | 0·4645 | 0·2741 | 2·1859 |
| r(A ↔ G) | 2·6483 | 3·2593 | 1·9161 |
| r(A ↔ C) | 0·8335 | 1·7435 | 0·9482 |
| freqA | 0·2854 | 0·3060 | 0·2573 |
| freqC | 0·1900 | 0·1488 | 0·2134 |
| freqG | 0·2278 | 0·1425 | 0·2724 |
| freqT | 0·2968 | 0·4062 | 0·2569 |
| Shape | 0·7818 | 1·1016 | 0·5748 |
| Pinvar | 0·5254 | 0·0879 | 0·6916 |
* r(N ↔ N), Substitution rates for each nucleotide pair; freqA, freqC, freqG, freqT, empirical base frequency; Shape, gamma distribution shape parameter; Pinvar, proportion of invariable sites.
The spine of the tree among the non-core asparagoids was nearly congruent to that of the maximum-parsimony (MP) tree with high PP (1·00; Fig. 3). All nodes were strongly supported (PP > 0·89) with only one exceptional branch (PP 0·30), that linking Doryanthaceae (PP 1·00), Ixiolirionaceae and Tecophilaceae (PP 1·00). Also Xanthorrhoaceae s.l. (PP 1·00) were sister to the core asparagoids (Fig. 3); Xeronemataceae were sister to Xanthorrhoaceae s.l. plus core asparagoids.
DISCUSSION
The tree topology in Asparagales from analysis of three genes is nearly congruent with those of previous analyses, although this study used Orchidaceae as the only outgroup (Chase et al., 1995a, b; Rudall et al., 2000; Fay et al., 2000; Pires et al., 2006). The overall results produced here, with different accesissions of species and a different set of taxa, indicate that the tree topologies from the previous studies are robust with respect to the samples used to represent genera and the taxa sampled. The core asparagoid clade was strongly supported, and the tree topology of the asparagoids characterized by simultaneous microsporogenesis and inferior ovaries, is congruent with the previous analyses and has strong support (Figs 2 and 3; Chase et al., 1995a; Fay et al, 2000; Pires et al., 2006). The family composition of the core asparagoids is the same as that in APG (1998) and characterized by a reversal to successive microsporogenesis, although there are a few parallel occurrences in Xanthorrhoeaceae and Hypoxidaceae (Rudall et al., 1997). In this study, the core asparagoids was split into two subclades: (1) Ruscaceae s.l. + Asparagaceae s.s. + Laxmanniaceae + Alliaceae s.l. sensu APG II; and (2) Agavaceae s.l. sensu APG II + Hesperocallidaceae + Hyacinthaceae + Themidaceae with Aphyllanthaceae. These two major clades differ from the two identified in the study by Pires et al. (2006), upon which the APG III set of families was based (see below for more discussion). The present study also supports Xanthorrhoeaceae s.l. sensu APG III as sister to all core asparagoids.
The most variable gene was matK, and 18S rDNA exhibited the lowest level of variation. The variable positions in the two plastid DNA genes changed twice as fast as those in 18S rDNA. The topologies exhibited similar patterns in the asparagoids for each analysis from three genes separately (not shown) as in the previous combined analyses (Rudall et al., 2000; Fay et al., 2000).
Phylogenetics of Ruscaceae s.l. and related families
Ruscaceae s.l. are a recently recognized family (APG, 1998; Rudall et al, 2000), which can be distinguished by the absence of phytomelan in the seed coat and indehiscent or berry-like fruits (Rudall et al., 2000). Ruscaceae s.l. represent a well-supported clade in DNA alone (Fay et al., 2000; Pires et al., 2006) and combined DNA–morphological analyses (Rudall et al., 2000). This study strongly supports monophyly of Ruscaceae s.l. (BP 90 from plastid DNA alone, and BP 88 from the combined data). Asparagaceae s.s. were monophyletic (BP 100, plastid; BP 100, combined data), and the sister group to Ruscaceae s.l. (BP 90, plastid; BP 93, combined) was Laxmanniaceae, as in previous analyses (Fay et al., 2000; Pires et al., 2006). The clade with Ruscaceae s.l., Asparagaceae and Laxmanniaceae was sister to Amaryllidaceae s.l. (APG III, 2009), including Alliaceae, Amaryllidaceae s.s. and Agapanthaceae. This set of relationships, particularly with respect to the position of Amaryllidaceae s.l., was a little different from previous results. However, the relationships identified here were only moderately supported and contradicted by Pires et al. (2006), who found Asparagaceae s.l. (sensu APG II) to be sister to Amaryllidaceae s.l.; the core asparagoids were thus composed of three clades in the strict consensus tree (not shown) of the equally weighted analysis of the plastid DNA data. Yamashita and Tamura (2000) suggested that the outgroups for Convallariaceae were Eriospermum, Aphyllanthes and former Anthericaceae genera in their trnK region analyses, but the present study shows that Aphyllanthes and Anthericaceae have a more remote relationship to that family than Asparagaceae s.s.
Aphyllanthes has been a problem taxon in core asparagoid phylogenetics. In this study Aphyllanthes was found to be sister to Themidaceae (BP 100) in both MP analyses, and this combined clade (77/81 BP) of Aphyllanthes and Themidaceae was the sister to Hyacinthaceae (100/100 BP) and Agavaceae s.l. (94/94 BP). Also, in the present MP analyses that excluded Aphyllanthes (results not shown) there was no change in tree topology and a small increase in internal support, but in the Bayesian tree (Fig. 3) Aphyllanthes was sister to Anemarrhenaceae in Agavaceae s.l., although this result was weakly supported (37 PP). Further detailed studies are required to establish the phylogenetic relationships of Aphyllanthes. If Asparagaceae s.l. is recognized as in APG III (2009), then at least the problem becomes one of within-family phylogenetics.
Phylogenetics within Ruscaceae s.l.
Although Asparagales were established with phytomelaneous seeds as the synapomorphic character by Huber (1969), Ruscaceae s.l., which have non-phytomelaneous seeds, were controversially included within the core asparagoids that exhibit successive microsporogenesis. Most taxa in Ruscaceae s.l. have several additional synapomorphies, such as articulate pedicels, septal nectaries and berries. Chase et al. (1995a) first mentioned the expanded range of taxa in Ruscaceae s.l. including Convallariaceae s.s., Ruscaceae s.s., Nolinaceae, Dracaenaceae, Eriospermaceae and Comospermum, and this group of taxa was treated as Convallariaceae s.l. in some papers (Rudall et al., 1997; APG, 1998; Fay et al., 2000), but Ruscaceae has priority (Rudall et al., 2000).
This study confirmed the monophyly of Ruscaceae s.l. with strong support (BP 90, Fig. 1), including Eriospermaceae (BP 100, Figs 1 and 2; Rudall et al., 2000). Based on the combined three-gene analyses, Ruscaceae s.l. consist of six subclades: (1) Polygonateae (excluding Disporopsis), (2) Ophiopogoneae, (3) Convallarieae (excluding Theropogon), (4) Ruscaceae s.s. + Dracaenaceae + Theropogon + Disporopsis + Comospermum, (5) Nolinaceae and (6) Eriospermum. This result corresponds with that of Rudall et al. (2000): (1) Eriospermum, (2) Comospermum, (3) nolinoids (Nolinaceae, Ophiopogoneae except Peliosanthes), (4) dracaenoids (Dracaenaceae), (5) Polygonateae, (6) Convallarieae with ruscoids (Ruscaceae s.s.) and Peliosanthes. Yamashita et al. (2000) also found six groups: (1) Polygonateae, (2) Ophiopogoneae, (3) Convallarieae, (4) Nolinaceae, (5) Ruscaceae (with Dracaenaceae) and (6) Comospermum. Only Eriospermum and Polygonateae were consistent in the results from all three sets of analyses.
Within the Ruscaceae s.l. clade, Eriospermaceae (BP 100/PP 1·00), Nolinaceae (BP 100/PP 1·00), Ruscaceae s.s. (BP 100/PP 1·00) and Dracaenaceae (BP 100/PP 1·00) were well supported. However, Convallariaceae are paraphyletic (Figs 1 and 2) as in previous studies (Rudall et al., 2000; Yamashita and Tamura, 2000; Jang and Pfosser, 2002). If Convallariaceae is to be recognized, it should be recircumscribed; this result has been well supported by the results of molecular and combined molecular and morphological data (Chase et al., 1995b; Rudall et al., 1997; Fay et al., 2000; Rudall et al., 2000; Yamashita et al., 2000; Tamura and Yamashita, 2004).
Relationships of Ophiopogoneae
In this study, Ophiopogoneae (BP 98/PP 100) were the only monophyletic tribe among the four previously recognized in Convallariaceae. Ophiopogoneae share hypodermal fibres and well-developed fruits with a thin, papery pericarp and fleshy seeds (Conran and Tamura, 1998). The leaf epidemal cells are ridged and sculptured with the subsidary cells surrounding the guard cells in Liliope and Ophiopogon, and flowers are perigynous in Ophiopogon and Peliosanthes (Cutler, 1992).
Polygonateae
Monophyly of Polygonateae has been supported in previous studies (Rudall et al., 2000; Yamashita et al., 2000; Tamura and Yamashita, 2004). Polygonateae share sympodial rhizomes, elongate stems and broad leaves relative to those of Ophiopogoneae. Their chromosome numbers and karyotypes are diverse: Polygonatum, x = 9–15; Heteropolygonatum, x = 16; Maianthemum (including Smilacina, x = 18); Disporopsis, x = 20. It was reported recently that variation in chromosome numbers of Polygonateae was derived from an ancestral basic one (x = 19) in Ruscaceae s.l. (Yamashita and Tamura, 2004). Polygonateae including Disporopsis was strongly supported as monophyletic in Bayesian tree (PP 0·95; Fig. 3).
Smilacina was treated within Maianthemum by LaFrankie (1985a, b; 1986), and many studies have agreed with combining these two genera (Conran and Tamura, 1998; Yamashita et al, 2000; Rudall et al., 1997; 2000; Shinwari, 2000). The two genera exhibit several distinguishing characters. For example, Smilacina has trimerous flowers, multiple (>6) leaves, and adventitious roots from both nodes and internodes of the rhizome, whereas Maianthemum has dimerous flowers, 2–5 leaves and adventitious roots only from the internodes of the rhizome. Kim and Lee (2007) also proposed to merge the two genera based on analyses of the trnK data (including matK). We also agree with the previous studies that proposed their merger (LaFrankie, 1985a; Conran and Tamura, 1998; Yamashita et al, 2000; Rudall et al., 1997, 2000; Shinwari, 2000), but more intensive studies including distributional diversity and more samples are needed to elucidate this relationship more clearly.
Convallarieae clade
Dahlgren et al. (1985) divided Convallariaceae into four tribes, Polygonateae, Ophiopogoneae, Convallarieae and Aspidistreae, but they did not suggest any obvious characteristics to delimit Convallarieae relative to Aspidistreae. After Dahlgren et al. (1985), most of studies treated Convallariaceae as composed of three tribes and merged Aspidistreae with Convallarieae (Conran and Tamura, 1998; Yamashita et al. 2000; Rudall et al, 2000), which was supported here. Theropogon differs from Convallarieae in anatomical features (Utech, 1979), basic chromosome number and floral morphology. Rudall et al. (2000) mentioned close relationships of Convallarieae, Ruscaceae s.s. and Peliosanthes, but Ruscaceae s.s. and Peliosanthes are different in their basic chromosome numbers (x = 20 and x = 18, respectively) and septal nectaries from Convallarieae. Also, Peliosanthes is included in Ophiopogoneae, which have some special fruit features and perigynous flowers. Convallarieae/Aspidistreae have several synapomorphies such as basic chromosome numbers (x = 19), monopodial rhizomes and shoots and non-septal nectaries (Dahlgren et al., 1985; Tamura, 1995). In the Convallariae/Aspidistreae clade, Campylandra, Rohdea, Tupistra, Aspidistra, Convallaria and Speirantha formed a group (BP 96/PP 0·96), but the genera are not monophyletic.
Ruscaceae s.s. + Dracaenaceae + Theropogon + Comospermum clade
The close relationships of Ruscaceae s.s., Dracaenaceae and Comospermum have been found in previous studies (Tamura, 1995; Rudall et al., 1997), which all have tenuinucellate parietal cells and the same basic chromosome number (x = 20). The basic chromosome numbers of Theropogon (x = 19) differs from Convallarieae and Polygonateae, and it has septal nectaries, otherwise found only in Convallarieae. Additional molecular and morphological studies should be pursued to resolve the phylogenetic problems and controversies concerning relationships of Theropogon.
Nolinaceae clade
It has been previously reported that Nolinaceae have a close relationship with Dracaenaceae. They were often treated in tribe Dracaeneae (Bentham and Hooker, 1883) or Nolineae (Krause, 1930). Recently several studies suggested that they are close to Dracaenaceae and Convallariaceae (Bogler and Simpson, 1995, 1996), particularly Ophiopogoneae, even though there are no obvious morphological characters to support this (Rudall et al., 2000). Nolinaceae are sister to Convallariaceae–Ruscaceae s.s.–Dracaenaceae (BP 100) in the MP tree but sister to Convallariae/Aspidistreae alone with BA (PP 0·96).
Eriospermum clade
Eriospermum, endemic to southern Africa, is strongly supported (BP 100/PP 1·00) as sister to Ruscaceae s.l. (BP 100/PP 0·99; Figs 2 and 3). In previous studies, Eriospermum with Aphyllanthes were close to Ruscaceae s.l. (Rudall et al., 1997; Fay et al., 2000) or proposed to be included in Convallariaceae (Yamashita and Tamura, 2000). However, Jang and Pfosser (2002) suggested Aphyllanthes should go with Anthericaceae and Eriospermum should be included in Ruscaceae s.l.; Eriospermum and Ruscaceae s.l. share many characters such as seeds without phytomelan, articulate peduncles and septal nectaries, but Eriospermum differs in its seed trichomes, special leaf appendages, large ovules and oily perisperm (Dahlgren et al., 1985; Lu, 1985). The phylogenetic position of Eriospermum seems secure; it shares many of the traits of Ruscaceae s.l. Little is gained by recognizing it as a family on its own.
Conclusions
This study with different taxon sampling and different species representing genera than in previous phylogenetic studies documents the stability of relationships within Asparagales. Moreover, a better-supported topology for relationships within Ruscaceae (Nolinoideae of Asparagaceae sensu APG III, 2009) than in any previous study is provided here, and it is documented that there are still subjects for more detailed future studies of genera and tribes in this clade. The higher-level relationships (interfamilial) found in this study are not totally in agreement with other broad studies (e.g. Pires et al., 2006), in particular the parsimony analysis in this study does not find support for the broader circumscription of Asparagaceae sensu APG III. Amaryllidaceae s.l. are supported, but in this study Asparagaceae s.l. are paraphyletic to Amaryllidaceae s.l. However, this set of relationships is not strongly supported. In contrast, the Bayesian analysis found that Asparagaceae s.l. were sister to Amaryllidaceae s.l. but with PP < 95. All other aspects of the higher-level relationships within Asparagales are similar to those found previously. We intend to collect more data to evaluate this disagreement in greater detail and also to investigate relationships in Ruscaceae further by increasing both taxa and numbers of loci.
ACKNOWLEDGEMENTS
This research was supported by the Mid-career Reseacrher Program through an NRF grant funded by MEST to J.-H. Kim (No. R01-2008-000-11910-0) and by the Korean Research Foundation Grant Fund (MOEHRD, Basic Research Promotion Fund; KRF-2007-C00036).
APPENDIX
Voucher data and GenBank accession numbers for Ruscaceae s.l. and related groups in the order Asparagales. Order and family circumscriptions are as in APG (1988) with slight modification (Chase et al., 2000). Names with asterisks are the family circumscriptions of Dahlgren et al. (1985).
| Family/Tribe | Taxon | Source/voucher | Origin/distribution | 18S | matK | rbcL |
|---|---|---|---|---|---|---|
| Agapanthaceae | Agapanthus africanus | Chase 627 (K) | SW Cape, S Africa | HM640715 | HM640599 | HM640485 |
| Agavaceae | Agave ghiesbreghtii | Chase 3467 (K) | Mexico, N and C America | HM640709 | HM640592 | HM640478 |
| Yucca filamentosa | DK Kim 06-077 (TUT) | E USA, N America | HM640713 | HM640596 | HM640482 | |
| Leucocrinum montanum | Chase 795 (K) | S USA. N America | HM640712 | HM640595 | HM640481 | |
| Hosta plantaginea | JX Feng s.n. (HZU) | Hangzhou, China, E Asia | HM640711 | HM640594 | HM640480 | |
| Alliaceae | Allium victorialis var. platyphyllum | DK Kim 04-142 (TUT) | Korea, E Asia | HM640714 | HM640597 | HM640483 |
| Ipheion uniflorum | Murakami 631 (KYO) | N Argentina, S America | – | HM640598 | HM640484 | |
| Amaryllidaceae | Lycoris uydoensis | DK Kim 05-102 (TUT) | Korea, E Asia | HM640716 | HM640600 | HM640486 |
| Narcissus tazetta var. chinensis | DK Kim 06–167 (TUT) | W Mediterranean | HM640717 | HM640601 | HM640487 | |
| Crinum asiaticum var. japonicum | GH Tae s.n. (TUT) | Korea, E Asia | HM640718 | HM640602 | HM640488 | |
| Clivia nobilis | Chase 3080 (K) | E Cape, S Africa | AF206889 | HM640603 | Chase et al., 2006 | |
| Anemarrhenaceae* | Anemarrhena asphodeloides | TCMK 312 (K) | Korea, NE Asia | HM640719 | HM640604 | HM640489 |
| Anthericaceae* | Anthericum liliago | Chase 515 (K) | N, C and S Europe | HM640720 | HM640605 | HM640490 |
| Chlorophytum minor | BY Ding s.n. (KUN) | Zambia, Africa | HM640721 | HM640606 | HM640491 | |
| Chlorophytum suffruticosum | Chase 1043 (K) | E Africa | HM640723 | HM640608 | HM640493 | |
| Chlorophytum orchidastrum | Chase 2155 (K) | W and C Africa | HM640722 | HM640607 | HM640492 | |
| Chlorophytum tetraphyllum | Chase 1044 (K) | Ethiopia, N Africa | HM640724 | HM640609 | L05031 | |
| Comospermum yedoense | Chase 833 (K) | Japan, E Asia | HM640725 | HM640610 | HM640494 | |
| Echeandia sp. | Chase 602 (K) | S and C America | HM640727 | HM640612 | HM640495 | |
| Paradisea liliastrum | Chase 826 (K) | Pyrenees, Alps, S Europe | HM640728 | HM640613 | HM640496 | |
| Aphyllanthaceae | Aphyllanthes monspeliensis | Chase 614 (K) | W and C Mediterranean | HM640729 | HM640614 | Z77259 |
| Asparagaceae | Asparagus cochinchinensis | DK Kim 04–122 (TUT) | Korea, E and SE Asia | HM640730 | HM640615 | HM640497 |
| Asparagus schoberioides | DK Kim 04–165 (TUT) | Korea, NE Asia | HM640731 | HM640616 | HM640498 | |
| Hemiphylacus latifolius | Chase 668 (K) | Mexico, N America | HM640732 | HM640617 | HM640499 | |
| Behniaceae | Behnia reticulata | Goldbladtt 9273 (MO) | S and E Africa | HM640733 | HM640618 | HM640500 |
| Convallariaceae* | ||||||
| Convallarieae | Convallaria majalis | DK Kim 04–082 (TUT) | Korea, NE Asia, Europe | HM640672 | HM640557 | HM640443 |
| Reineckea carnea | DK Kim 05–008 (TUT) | Korea, E Asia | HM640673 | HM640558 | HM640444 | |
| Speirantha gardenii | Chase 495 (K) | SE China | HM640674 | HM640559 | HM640445 | |
| Theropogon pallidus | Chase 2933 (K) | SW China, Himalaya | HM640675 | HM640560 | HM640446 | |
| Aspidistreae | Aspidistra elatior | DK Kim 05-013 (TUT) | Korea, E Asia | HM640676 | HM640561 | HM640447 |
| Campylandra fimbriata | Liu Yang 484 (KUN) | Himalaya, NW China | HM640677 | HM640562 | HM640448 | |
| Rohdea japonica | DK Kim 05-005 (TUT) | Korea, E Asia | HM640678 | HM640563 | HM640449 | |
| Tupistra aurantiaca | Chase 1100 (K) | Yunnan, SW China, E Asia | HM640679 | HM640564 | HM640450 | |
| Polygonateae | Disporopsis pernyi | Chase 493 (K) | S China, E Asia | HM640681 | HM640566 | HM640452 |
| Disporopsis sp. | DK Kim 05-136 (TUT) | Sichuan, China, E Asia | HM640680 | HM640565 | HM640451 | |
| Maianthemum bifolium | DK Kim 04-182 (TUT) | Korea, temperate Eurasia | HM640682 | HM640567 | HM640453 | |
| Maianthemum dilatatum | DK Kim 04-165 (TUT) | Korea, NE Asia | HM640683 | HM640568 | HM640454 | |
| Polygonatum humile | DK Kim 04-029 (TUT) | Korea, C and E Asia | HM640684 | HM640569 | HM640455 | |
| Polygonatum inflatum | DK Kim 04-043 (TUT) | Korea, NE Asia | HM640685 | HM640570 | HM640456 | |
| Polygonatum involucratum | DK Kim 04-059 (TUT) | Korea, NE Asia | HM640686 | HM640571 | HM640457 | |
| Polygonatum lasianthum var. coreanum | DK Kim 04-046 (TUT) | Korea, NE Asia | HM640687 | HM640572 | HM640458 | |
| Polygonatum odoratum var. pluriflorum | DK Kim 04-067 (TUT) | Korea, NE Asia | HM640688 | HM640573 | HM640459 | |
| Smilacina bicolor | DK Kim 04-077 (TUT) | Korea, E Asia | HM640689 | HM640574 | HM640460 | |
| Smilacina dahurica | DK Kim 04-082 (TUT) | Korea, NE Asia | HM640690 | HM640575 | HM640461 | |
| Smilacina japonica | DK Kim 04-039 (TUT) | Korea, NE Asia | HM640691 | HM640576 | HM640462 | |
| Ophiopogoneae | Liriope platyphylla | DK Kim 07-001 (TUT) | Korea, E Asia | HM640692 | HM640577 | HM640463 |
| Liriope spicata | DK Kim 07-002 (TUT) | Japan, E Asia | HM640693 | HM640578 | HM640464 | |
| Ophiopogon jaburan | DK Kim 07-004 (TUT) | Korea, E Asia | HM640694 | HM640579 | HM640465 | |
| Ophiopogon japonicus | DK Kim 07-003 (TUT) | Korea, E Asia | HM640695 | HM640580 | HM640466 | |
| Peliosanthes macrostegia | G Murata 44832 (KYO) | S China, E Asia | HM640696 | HM640581 | HM640467 | |
| Dracaenaceae* | Dracaena schizantha | Chase 21514 (K) | Ethiopia, NE Africa | HM640698 | HM640582 | HM640469 |
| Dracaena aubryana | Chase 1102 (K) | Uganda, WC Africa | HM640699 | HM640583 | HM640470 | |
| Sansevieria trifasciata | DK Kim 07-05 (TUT) | Nigeria, WC Africa | HM640700 | HM640584 | HM640471 | |
| Eriospermaceae* | Eriospermum abyssinicum | Chase 2051 (K) | S Africa | HM640706 | HM640589 | HM640475 |
| Eriospermum natalense | Chase 2052 (K) | S Africa | HM640707 | HM640590 | HM640476 | |
| Eriospermum parvifolium | Chase 2053 (K) | W Cape, S Africa | HM640708 | HM640591 | HM640477 | |
| Herreriaceae* | Herreria salsaparilha | Chase 2154 (K) | Brazil, S America | HM640734 | HM640619 | HM640501 |
| Hesperocallidaceae | Hesperocallis undulata | Cranfill & Schmid s.n. (JEPS) | SW USA, N America | HM640735 | HM640620 | HM640502 |
| Hyacinthaceae* | Bowiea volubilis | Chase 176 (K) | Uganda, C and S Africa | HM640736 | HM640621 | HM640503 |
| Camassia cusickii | Cronquist 6549 (RSA) | C USA, N America | HM640710 | HM640593 | HM640479 | |
| Dipcardi filifolium | Chase 1783 (K) | C Asia, Africa, India | HM640737 | HM640622 | HM640504 | |
| Drimia altissima | Chase 1870 (K) | C and S Africa | HM640738 | HM640623 | HM640505 | |
| Eucomis humilis | Chase 1847 (K) | Lesotho, S Africa | HM640739 | HM640624 | HM640506 | |
| Lachenalia carnosa | Chase 2261 (K) | W Cape, S Africa | HM640740 | HM640625 | HM640507 | |
| Ledebouria cooperi | Chase 1786 (K) | S Africa | HM640741 | HM640626 | HM640508 | |
| Massonia angustifolia | Chase 5666 (K) | Cape, S Africa | HM640742 | HM640627 | HM640509 | |
| Muscari aucheri | Chase 21845 (K) | Turkey, Med. to Caucasus | HM640743 | HM640628 | HM640510 | |
| Ornithogalum armeniacum | Chase 1682 (K) | Turkey to Macedonia | HM640744 | HM640629 | HM640511 | |
| Ornithogalum shawii | Chase 1012 (K) | S Africa | HM640745 | HM640630 | HM640512 | |
| Rhadamanthus convallarioides | Goldblatt 10852 (A) | Cape, S Africa | HM640746 | HM640631 | HM640513 | |
| Scilla scilloides | DK Kim 05-039 (TUT) | Korea, E Asia | HM640747 | HM640632 | HM640514 | |
| Urginea epigea | Chase 2055 (K) | S Africa | HM640748 | HM640633 | HM640515 | |
| Laxmanniaceae | Arthropodium cirrhatum | Chase 651 (NCU) | New Zealand, Australia | HM640749 | HM640634 | HM640516 |
| Laxmannia squarrosa | Chase 2214 (K) | W and S Australia | HM640751 | HM640636 | HM640518 | |
| Lomandra hastilis | Brummitt et al. 21239 (K) | W and SW Australia | HM640750 | HM640635 | HM640517 | |
| Nolinaceae* | Calibanus hookeri | Chase 1006 (K) | Mexico, N America | HM640702 | HM640585 | HM640472 |
| Dasylirion serratifolium | Abisai et al., s.n. (RSA) | Mexico, N America | HM640704 | HM640587 | AB029847 | |
| Dasylirion wheeleri | Chase 3469 (K) | Texas, S USA, N America | HM640705 | HM640588 | HM640474 | |
| Nolina recurvata | Chase 3466 (K) | Mexico, N America | HM640703 | HM640586 | HM640473 | |
| Ruscaceae* | Danae racemosa | Chase 121 (K) | Turkey, Syria, Iran, Caucasus | HM640668 | HM640553 | HM640439 |
| Ruscus aculeatus | Bohuslavek 1348 (RSA) | W and C Europe, Medit. | HM640669 | HM640554 | HM640440 | |
| Ruscus streptophyllus | Chase 21990 (K) | Madeira | HM640670 | HM640555 | HM640441 | |
| Semele androgyna | Chase 997 (K) | Canary Is., Madeira | HM640671 | HM640556 | HM640442 | |
| Themidaceae | Bessera elegans | Chase 626 (K) | Mexico, N America | HM640752 | HM640637 | HM640519 |
| Bloomeria aurea | Chase 1010 (K) | SW USA, N America | HM640753 | HM640638 | HM640520 | |
| Dandya thadhowardii | Chase s.n. (K) | Mexico, N America | HM640754 | HM640639 | HM640521 | |
| Dichelostemma multiflorum | Chase 1830 (K) | SW USA, N America | HM640755 | HM640640 | HM640522 | |
| Muilla maritime | Chase 779 (K) | SW USA to Mexico, N America | HM640757 | HM640642 | HM640524 | |
| Triteleia peduncularis | Chase 1860 (K) | California, W USA, N America | HM640758 | HM640643 | HM640525 | |
| Asphodelaceae | Eremurus chinensis | Qing 00317 (KUN) | Tibet to S Gansu, W China | HM640759 | HM640644 | HM640526 |
| Asphodelus aestivus | Chase 482 (K) | Portugal, Spain, SW Europe | HM640760 | HM640645 | HM640527 | |
| Bulbine semibarbata | K Dixon, s.n. (KPBG) | S and E Australia | HM640761 | HM640646 | HM640528 | |
| Bulbine succulenta | Chase 5518 (K) | Cape, S Africa | AF206876 | Z73684 | ||
| Bulbine frutescens | Chase 9215 (K) | S Africa | AJ511414 | |||
| Asteliaceae | Astelia alpina | Chase 1103 (K) | NSW to Tasmania, S Australia | HM640762 | HM640648 | HM640530 |
| Milligania stylosa | Chase 511 (K) | Tasmania, S Australia | HM640763 | HM640649 | HM640531 | |
| Brandfordiaceae | Brandfordia punicea | MRK Rambert 787 (K) | Tasmania, S Australia | HM640764 | HM640650 | HM640532 |
| Boryaceae | Borya septentrionalis | Chase 2205 (K) | Perth, W Australia | HM640765 | HM640651 | HM640533 |
| Doryanthaceae | Doryanthes excelsa | Chase 188 (K) | NSW, SE Australia | HM640766 | HM640652 | HM640534 |
| Doryanthes palmeri | Chase 19153 (K) | Queensland, SE Australia | HM640767 | HM640653 | HM640535 | |
| Hemerocallidaceae | Dianella ensifolia | Nakai 5510 (KYO) | Taiwan, SE and Tropical Asia | HM640768 | HM640654 | HM640536 |
| Hemerocallis minor | DK Kim 05-091 (TUT) | Korea, NE Asia | HM640769 | HM640655 | HM640537 | |
| Hemerocallis littorea | Chase 3833 (K) | Korea, Japan, E Asia | Chase et al., 2006 | AJ581422 | AY149364 | |
| Hypoxidaceae | Curculigo capitulata | SW Lee 05-001 (TUT) | Yunnan, S Asia to N Australia | HM640770 | HM640656 | HM640538 |
| Rhodohypoxis milloides | Chase 479 (K) | E Cape, S Africa | AF207008 | AY368377 | Z77280 | |
| Rhodohypoxis baurii | Chase 16460 (K) | Cape, S Africa | HM640772 | HM640658 | HM640540 | |
| Hypoxis leptocarpa | Chase 108 (NCU) | Duke, SE USA, N America | AF135209 | AY368375 | Z73702 | |
| Hypoxis hemerocallidea | Chase 1045 (K) | Tropical and S Africa | HM640771 | HM640657 | HM640539 | |
| Iridaceae | Iris rossii | DK Kim 05-048 (TUT) | Korea, NE Asia | HM640773 | HM640659 | HM640541 |
| Gladiolus illyricus | Chase 9907 (K) | Portugal, SW Europe | HM640774 | HM640542 | ||
| Gladiolus papilio | Goldblatt & Manning 9841 (MO) | S Africa | AJ579956 | |||
| Ixioliriaceae | Ixiolirion tataricum | Chase 489B (K) | E Turkey to Kashmir, W Asia | HM640775 | HM640660 | HM640543 |
| Lanariaceae | Lanaria lanata | Goldblatt 9410 (MO) | Cape, S Africa | Chase et al., 2006 | AY368376 | Z77313 |
| Orchidaceae | Calanthe discolor | DK Kim 05-035 (TUT) | Korea, E Asia | HM640776 | HM640665 | HM640548 |
| Cephalanthera longibracteata | DK Kim 05-016 (TUT) | Korea, NE Asia | HM640777 | HM640666 | HM640549 | |
| Cypripedium calceolus | Chase 9484 (K) | Estonia, Europe to Asia | HM640778 | HM640667 | HM640550 | |
| Oncidium ensatum | Chase 9671 (K) | Tropical C and S America | HM640779 | AY368423 | HM640551 | |
| Apostasia wallichii | Chase 15744 (K) | Sri Lanka, S Asia to N Australia | HM640780 | AY557212 | HM640552 | |
| Tecophilaceae | Tecophilaea cyanocrocus | Chase 447 (K) | Chile, S America | HM640781 | HM640661 | HM640544 |
| Cyanella orchidiformis | Chase 5896 (K) | Cape, S Africa | HM640782 | HM640662 | HM640545 | |
| Xanthorrhoeaceae | Xanthorrhoea resinosa | Chase 192 (NCU) | NSW, Australia | HM640783 | HM640663 | HM640546 |
| Xanthorrhoea quadrangulata | Hahn 6978 (WIS) | S Australia | U42064 | |||
| Qiu 97039 (NID) | DQ401345 | |||||
| Unvouchered | Z73710 | |||||
| Xeronemataceae | Xeronema callistemon | Chase 653 (K) | Poor Night Is., New Zealand | HM640784 | HM640664 | HM640547 |
LITERATURE CITED
- APG (Angiosperm Phylogeny Group) An ordinal classification for the families of flowering plants. Annals of the Missouri Botanical Garden. 1998;18:531–553. [Google Scholar]
- APG II. An update of the Angiosperm Phylogeny Group Classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- APG III. An update of the Angiosperm Phylogeny Group Classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Arber A. Danae, Ruscus and Semele: a morphological study. Annals of Botany. 1924;38:229–260. [Google Scholar]
- Bentham G, Hooker JD. Genera Plantarum. London: Reeve; 1883. Liliaceae. [Google Scholar]
- Bogler DJ, Simpson BB. A chloroplast DNA study of the Agavaceae. Systematic Botany. 1995;20:191–205. [Google Scholar]
- Bogler DJ, Simpson BB. Phylogeny of Agavaceae based on ITS rDNA sequence variation. American Journal of Botany. 1996;83:1225–1235. [Google Scholar]
- Bogler DJ, Pires JC, Francisco-Ortega J. Phylogeny of Agavaceae based on ndhF, rbcL and ITS sequences: implications of molecular data for classification. In: Columbus JT, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 313–328. [Google Scholar]
- Carpenter JM. Choosing among multiple equally parsimonious cladograms. Cladistics. 1988;4:291–296. doi: 10.1111/j.1096-0031.1988.tb00476.x. [DOI] [PubMed] [Google Scholar]
- Chase MW, Duvall MR, Hills HG, et al. Molecular phylogenetic of Lilianae. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. Richmond: Royal Botanic Gardens, Kew; 1995a. pp. 109–137. [Google Scholar]
- Chase MW, Stevenson DW, Wilkin P, Rudall PJ. Monocots systematics: a combined analysis. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. Richmond: Royal Botanic Gardens, Kew; 1995b. pp. 685–730. [Google Scholar]
- Chase MW, Rudall PJ, Conran JG. New circumscription and a new family of asparagoid lilies: genera formerly included in Anthericaceae. Kew Bulletin. 1996;51:667–680. [Google Scholar]
- Chase MW, Rudall PJ, Conran JG. Validation of the family name Boryaceae. Kew Bulletin. 1997;52:416. [Google Scholar]
- Chase MW, de Bruijn AY, Cox AV, et al. Phylogenetics of Asphodelaceae (Asparagales): an analysis of plastid rbcL and trnL-F DNA sequences. Annals of Botany. 2000a;86:935–951. [Google Scholar]
- Chase MW, Solitis DS, Soltis PS, et al. Higher-level systematics of the monocotyledons: an assessment of current knowledge and a new classification. In: Wilson KL, Morrison DA, editors. Monocots, systematics and evolution. Melbourne: CSIRO; 2000b. pp. 3–16. [Google Scholar]
- Chase MW, Fay MF, Devey DS, et al. Multigene analyses of monocot relationships: a summary. In: JT Columbus, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG., editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 63–75. [Google Scholar]
- Chupov VS, Cutjavina NG. Phylogeny of some groups of Liliales based on the data of serological analysis. In: Zhilin SG, editor. Systematics and evolution of higher plants. Leningrad: Nauka; 1980. pp. 101–110. [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Molecular Phylogenetics and Evolution. 2004;33:75–90. doi: 10.1016/j.ympev.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Conran JG. Cladisitic analyses of some net-veined Liliiflorae. Plant Systematics and Evolution. 1989;168:123–141. [Google Scholar]
- Conran JG. Aphyllanthaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Vol. 3. Berlin: Springer-Verlag; 1998. [Google Scholar]
- Conran JG, Tamura MN. Convallariaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Vol. 3. Berlin: Springer-Verlag; 1998. [Google Scholar]
- Conran JG, Chase MW, Rudall PJ. Kew Bulletin. Vol. 52. Anemarrhenaceae and Behniaceae (Lilianae: Asparagales); 1997. Two new monocot families; pp. 995–999. [Google Scholar]
- Cooney-Sovetts C, Sattler R. Phylloclade development in Asparagaceae: an example of homoeosis. Botanical Journal of the Linnean Society. 1986;94:327–371. [Google Scholar]
- Cutler DF. Vegetative anatomy of Ophiopogoneae (Convallariaceae) Botanical Journal of the Linnean Society. 1992;110:385–419. [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. The families of the monocotyledons. Berlin: Springer-Verlag; 1985. [Google Scholar]
- Devey DS, Leitch I, Rudall PJ, Pires JC, Pillon Y, Chase MW. Systematics of Xanthorrhoeaceae sensu lato, with an emphasis on Bulbine. In: JT Columbus, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG., editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 345–351. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Farris JS. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Fay MF, Chase MW. Resurrection of Themidaceae for Brodiaea alliance and recircumscription of Alliaceae, Amaryllidaceae and Agapanthoideae. Taxon. 1996;45:441–451. [Google Scholar]
- Fay MF, Rudall PJ, Sullivan S, et al. Phylogenetic studies of Asparagales based on four plastid DNA regions. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 360–371. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Furness CA, Rudall PJ. Microsporogenesis in monocotyledons. Annals of Botany. 1999;84:475–499. [Google Scholar]
- Givnish TJ, Pires JC, Graham SW, et al. Phylogenetic relationships of monocots based on the highly informative plastid gene ndhF: evidence for widespread concerted convergence. In: Columbus JT, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 28–51. [Google Scholar]
- Graham SW, Zgurski JM, McPherson MC, et al. Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. In: Columbus JT, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 3–21. [Google Scholar]
- Hilu K, Borsch DT, Muller K, et al. Angiosperm phylogeny based on matK sequence information. American Journal of Botany. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- Huber H. Die Samenmerkale und Verwandtschaftsverhältnisse der Liliifloren. Mitteilungen der Botanischen Staatssammlung München. 1969;8:219–538. [Google Scholar]
- Hutchinson J. Monocotyledons. Vol. 2. Oxford: Clarendon Press; 1934. The families of flowering plants. [Google Scholar]
- Jang CG, Pfosser M. Phylogenetics of Ruscaceae sensu lato based on plastid rbcL and trnL-F DNA sequences. Stapfia. 2002;80:333–348. [Google Scholar]
- Johnson LA, Soltis DE. Phylogenetic inference in Saxifragaceae sensu lato and Gilla (Polemoniaceae) using matK sequences. Annals of the Missouri Botanical Garden. 1995;82:149–175. [Google Scholar]
- Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden. 2000;87:482–498. [Google Scholar]
- Kim S-C, Lee NS. Generic delimitation and biogeography of Maianthemum and Smilacina (Ruscaceae sensu lato): preliminary results based on partial 3′ matK gene and trnK 3' intron sequences of cpDNA. Plant Systematics and Evolution. 2007;265:1–12. [Google Scholar]
- Krause K. Liliaceae. In: Engler A, Prantl K, editors. Die Natürlichen Pflanzanfamilien. 15a. Leipzig: Engelmann; 1930. pp. 227–386. [Google Scholar]
- LaFrankie JV. Morphological and systematic studies in the genus Smilacina (Liliaceae) Harvard University, Cambridge: MA; 1985a. Ph.D. Dissertation. [Google Scholar]
- LaFrankie JV. A note on seedling morphology and establishment growth in the genus Smilacina (Liliaceae) Bulletin of the Torrey Botanical Club. 1985b;112:313–317. [Google Scholar]
- LaFrankie JV. Transfer of species of Smilacina to Maianthemum (Liliaceae) Taxon. 1986;35:584–589. [Google Scholar]
- Lu AM. Embryology and possible relationships of Eriospermum. Nordic Journal of Botany. 1985;5:229–240. [Google Scholar]
- Maddison WP, Maddison DR. MacClade: analysis of phylogeny and character evolution. ver. 4·0. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Nickrent DL, Soltis DE. A comparison of angiosperm phylogenies from nuclear 18S rDNA and rbcL sequences. Annals of the Missouri Botanical Garden. 1995;82:208–234. [Google Scholar]
- Nylander JAA. Evolutionary Biology Centre, Uppsala University: Uppsala; 2005. MrModeltest ver. 2·2. Program distributed by the author. [Google Scholar]
- Olmstead RG, Michaels HJ, Scott KM, Palmer JD. Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Annals of the Missouri Botanical Garden. 1992;79:249–265. [Google Scholar]
- Pires JC, Maureira IJ, Givnish TJ, et al. Phylogeny, genome size, and chromosome evolution of Asparagales. In: Columbus JT, Friar EA, Hamilton CW, Porter JM, Prince LM, Simpson MG, editors. Monocots: comparative biology and evolution. Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 287–304. [Google Scholar]
- Ronquist F, Huelsenbeck JP, van der Mark P. MrBayes 3·1 Manual. 2005 http://mrbayes.csit.fsu.edu/manual.php . [Google Scholar]
- Rudall PJ, Campbell G. Flower and pollen structure of Ruscaceae in relation to Aspidistreae and other Convallariaceae. Flora. 1999;194:201–214. [Google Scholar]
- Rudall PJ, Chase MW. Systematics of Xanthorrhoeaceae sensu lato: evidence for polyphyly. Telopea. 1996;6:629–647. [Google Scholar]
- Rudall PJ, Furness CA, Chase MW, Fay MF. Microsporogenesis and pollen sulcus type in Asparagales (Lilianae) Canadian Journal of Botany. 1997;75:408–430. [Google Scholar]
- Rudall PJ, Engelman EM, Hanson L, Chase MW. Systematics of Hemiphylacus, Anemarrhena and Asparagus. Plant Systematics and Evolution. 1998;211:181–199. [Google Scholar]
- Rudall PJ, Conran JG, Chase MW. Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Botanical Journal of the Linnean Society. 2000;134:73–92. [Google Scholar]
- Sato D. Karyotype alteration and phylogeny in Liliaceae and allied families. Japanese Journal of Botany. 1942;12:57–161. [Google Scholar]
- Sharma AK, Chaudhuri U. Cytological studies as an aid in assessing the status of Sansevieria, Ophiopogon and Curculigo. Nucleus. 1964;7:43–58. [Google Scholar]
- Shinwari ZK. Chloroplast DNA variation in Polygonatae sensu lato (Liliaceae) Pakistan Journal of Botany. 2000;32:7–14. [Google Scholar]
- Shinwari ZK, Kato H, Terauchi R, Kawano S. Phylogenetic relationships among genera in the Liliaceae-Asparagoideae-Polygonatae s.l. inferred from rbcL gene sequence data. Plant Systematics and Evolution. 1994;192:263–277. [Google Scholar]
- Soltis DE, Soltis PS, Nickrent DL, et al. Angiosperm phylogeny inferred from 18S ribosomal DNA sequences. Annals of the Missouri Botanical Garden. 1997;84:1–49. [Google Scholar]
- Soltis DE, Soltis PS, Chase MW, et al. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Botanical Journal of the Linnean Society. 2000;133:381–461. [Google Scholar]
- Soltis PS, Soltis DE. Molecular evolution of 18S rDNA in angiosperms: implications for character weighting in phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants. II. DNA sequencing. Boston, MA: Kluwer; 1998. pp. 188–210. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony, ver. 4·10b. Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- Takhtajan AL. Diversity and classification of flowering plants. New York, NY: Columbia University Press; 1997. [Google Scholar]
- Tamura MN. A karyological review of the orders Asparagales and Liliales (Monocotyledonae) Feddes Report. 1995;106:83–111. [Google Scholar]
- Tamura MN, Yamashita J. Molecular phylogeny of monocotyledons inferred from combined analysis of plastid matK and rbcL gene sequences. Journal of Plant Research. 2004;117:109–120. doi: 10.1007/s10265-003-0133-3. [DOI] [PubMed] [Google Scholar]
- Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utech FH. Floral vascular anatomy of the Himalayan Theropogon pallidus Maxim. (Liliaceae-Convallarieae) Annals of Carnegie Museum. 1979;48:25–41. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. A guide to methods and applications. New York, NY: Academic Press; 1990. pp. 315–322. [Google Scholar]
- World Checklist of Selected Plant Families. 2008 Published by the Board of Trustees of the Royal Botanic Gardens, Kew, on the Internet (http://www.kew.org/wcsp/ ), accessed in January 2008. [Google Scholar]
- Yamashita J, Tamura MN. Molecular phylogeny of the Convallariaceae (Asparagales) In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 387–400. [Google Scholar]
- Yamashita J, Tamura MN. Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato), with some taxonomic treatments. Journal of Plant Research. 2004;117:363–370. doi: 10.1007/s10265-004-0169-z. [DOI] [PubMed] [Google Scholar]