Abstract
Background
The much-debated phylogenetic relationships of the five early branching metazoan lineages (Bilateria, Cnidaria, Ctenophora, Placozoa and Porifera) are of fundamental importance in piecing together events that occurred early in animal evolution. Comparisons of gene content between organismal lineages have been identified as a potentially useful methodology for phylogenetic reconstruction. However, these comparisons require complete genomes that, until now, did not exist for the ctenophore lineage. The homeobox superfamily of genes is particularly suited for these kinds of gene content comparisons, since it is large, diverse, and features a highly conserved domain.
Results
We have used a next-generation sequencing approach to generate a high-quality rough draft of the genome of the ctenophore Mnemiopsis leidyi and subsequently identified a set of 76 homeobox-containing genes from this draft. We phylogenetically categorized this set into established gene families and classes and then compared this set to the homeodomain repertoire of species from the other four early branching metazoan lineages. We have identified several important classes and subclasses of homeodomains that appear to be absent from Mnemiopsis and from the poriferan Amphimedon queenslandica. We have also determined that, based on lineage-specific paralog retention and average branch lengths, it is unlikely that these missing classes and subclasses are due to extensive gene loss or unusually high rates of evolution in Mnemiopsis.
Conclusions
This paper provides a first glimpse of the first sequenced ctenophore genome. We have characterized the full complement of Mnemiopsis homeodomains from this species and have compared them to species from other early branching lineages. Our results suggest that Porifera and Ctenophora were the first two extant lineages to diverge from the rest of animals. Based on this analysis, we also propose a new name - ParaHoxozoa - for the remaining group that includes Placozoa, Cnidaria and Bilateria.
Background
Ctenophores are a phylum of marine metazoans with uncertain phylogenetic affinity. Their signature morphological features include a set of eight ciliated comb rows that are used for swimming; these are controlled by an aborally located statocyst called the apical sense organ. Most ctenophores have a pair of feeding tentacles that contain specialized adhesive cells called colloblasts. Ctenophores are extremely fragile and difficult to culture and, as such, we know very little about their biology relative to other metazoans [1].
The unique ctenophore body plan has made it difficult to untangle its phylogenetic position in relation to other animal phyla. The earliest comparative classifications by Cuvier allied ctenophores with cnidarians and echinoderms in the Radiata [2]. Later, Leuckart grouped ctenophores with sponges and cnidarians in the Coelenterata [3]. Associations proposed between ctenophores and other taxa include groupings with Platyhelminthes, trochozoans, bilaterians and subsets of sponges and cnidarians (see [4] for a review). Many of the early molecular studies using 18 s ribosomal RNA (rRNA) sequences placed ctenophores sister to Placozoa, Cnidaria and Bilateria (for example, Wainright et al., 1993; Smothers et al. 1994; Bridge et al. 1995; Collins 1998; Kim et al. 1999 [5-9]).
The 18 S rRNA placement of ctenophores has recently been challenged using data generated in expressed sequence tag (EST)-based phylogenomic studies. One study re-allies ctenophores with the cnidarians [10], while another has ctenophores branching at the base of the animal tree [11,12]. Yet another study combined morphological, structural and sequence data, leading to the placement of ctenophores in a clade with all other non-bilaterians sister to the Bilateria [13]. These conflicting results could be due to the use of different methods, the inherent incomplete nature of transcript sequencing (in the case of the EST-based studies) or for other reasons. This series of studies have left most investigators waiting for further evidence to tilt the consensus convincingly in one direction or another.
Phylogenomic approaches hold the promise of reconstructing the true tree of life (reviewed in [14]). Thus far, most phylogenomic efforts that have included data from all four of the early branching phyla have been restricted to the aforementioned (and conflicting) EST-based analyses (for example [10-12,15]). Methods based on whole-genome content rather than large concatenated data matrices can provide an independent assessment of current phylogenetic hypotheses and, due to the rarity of events measured, may arguably be more appropriate in this context [14,16]. One such rare genomic change that has previously been employed involves the presence or absence of gene duplications of homeobox genes [17-19].
Homeobox genes encode transcription factor proteins characterized by the presence of a helix-loop-helix DNA-binding domain called the homeodomain [20]. Homeobox genes were present in the last common ancestor of plants, animals and fungi and underwent extensive independent diversification in each of these lineages [21,22]. In animals, the homeobox superfamily has been separated into 11 classes and more than 125 gene families [21,23,24].
Examination of the homeobox complement of species from early-branching metazoan phyla (such as Cnidaria [25,26], Placozoa [27,28] and Porifera [29,30]) has been an especially fertile area of research, one that has been fuelled by the recent availability of full genomic sequence data from several non-bilaterian genomes. The last remaining non-bilaterian phylum lacking a species with a sequenced genome (and, therefore, a completely examined homeobox repertoire) was Ctenophora.
We have used a next-generation sequencing approach to sequence and assemble the ~150 MB of the lobate ctenophore, Mnemiopsis leidyi. Here, we present the first whole-genome investigation of the ctenophore homeobox superfamily. Our results expand on previous studies that have utilized degenerate polymerase chain reaction (PCR) approaches to identify ctenophore homeoboxes from the PRD, ANTP and SINE classes [22,31-36]. In addition to these classes, we show that the POU, LIM and TALE classes were also present prior to the divergence of the ctenophores from the rest of Metazoa.
This is also the first study to compare the complete homeobox catalogue of species from all of the non-bilaterian phyla, along with that of the two major bilaterian lineages (Protostomia and Deuterostomia) where complete genomic sequence data is available. As such, this work provides a major missing piece of evidence that is critical to understanding the makeup of the homeodomain superfamily in early metazoan history. With these data in hand, we evaluate the congruency of the homeodomain data with the recently proposed phylogenetic relationships of the early branching phyla.
Results
Overview of homeobox genes in Mnemiopsis
We extracted 76 homeoboxes from the genome of Mnemiopsis leidyi. The corresponding homeodomains were aligned to the human and Drosophila dataset used in Holland et al. 2007 [23] and supplemented with eight amphioxus homeodomains known to be missing from humans. The sequence alignment is available as supplemental material (Additional File 1). We generated nine trees from this alignment using multiple methods (neighbor-joining, maximum likelihood (ML) and Bayesian inference), multiple starting trees and multiple implementations. For example, in the case of ML, we used RaxML [37] and PhyML [38]). In this case, we generated a likelihood value for each tree and then chose the one with the highest likelihood (Figure 1). We subsequently used this tree and secondary domain information, along with the classification scheme in the Homeo Database (HomeoDB) [24], to divide the 76 Mnemiopsis homeodomains into the following classes: ANTP (22 homeodomains); PRD (7); TALE (3); POU (4); LIM (4); and SINE (18). Eighteen homeodomains remained unclassified (Table 1).
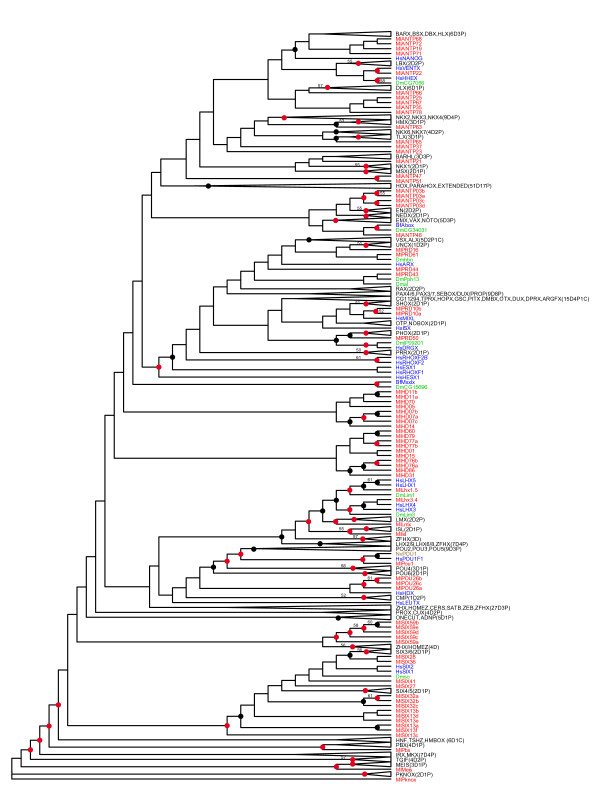
Figure 1.
Homeodomain superfamily tree. This tree is based on a RaxML tree that included homeodomains from human, Drosophila, Mnemiopsis and a few related species that serve as place-holders for homeodomains known to be missing from human and Drosophila (see Figure 2 legend for species codes). This tree is referred to as a 'superfamily tree' as it includes homeodomains from all classes of the homeodomain superfamily, in contrast to the trees in Figures 2, 3 and 4 that include only homeodomains from individual classes. Mnemiopsis sequences are shown in red; human and other deuterostome sequences are shown in blue; Drosophila and other protostome sequences are shown in green; and cnidarian sequences are shown in brown. This RaxML tree had a higher likelihood value compared to several other methods and variations of starting trees supplied to RaxML (see methods). Collapsed clades represent clades with no Mnemiopsis representative and include a code that indicates how many deuterostome, protostome and cnidarian homeodomains are in that particular clade (for example, 2D4P1C would signify 2 deuterostome, 4 protostome, and 1 cnidarian). ML bootstraps are included for clades with bootstrap values greater than 50. Black dots appear on clades with Bayesian posterior probability values greater than 50 and red dots on clades greater than 90. Rooting of this tree is for display purposes only; branch lengths are presented uniformly, also for display purposes. Actual branch lengths can be viewed by opening the Newick-formatted tree file (Additional File 4), which also includes bootstrap and Bayesian support values, in a tree viewing/editing program such as FigTree [53].
Table 1.
Mnemiopsis homeobox genes.
| Name | Domain | Signature | Intron codons | Accession |
|---|---|---|---|---|
| TALE | ||||
| MlPbx | Pbx | insert(NLA) 23 | 2,47/48 | HM444125 |
| MlMeis | MeisA, MeisD | insert(HLT) 23 | 25/26, 51 | HM444091 |
| MlPknox | Pbx | insert(HLG) 23 | 25/26, 51 | HM444122 |
| POU | ||||
| MlPou1 | Pou | HM444110 | ||
| MlPOU26a | Pou | HM444092 | ||
| MlPOU26b | HM444093 | |||
| MlPOU26c | HM444094 | |||
| LIM | ||||
| MlIsl | Lim | 39 | HM444123 | |
| MlLhx1.5 | Lim | 55/56 | HM444088 | |
| MlLhx3.4 | Lim | 27 | HM444089 | |
| MlLmx | Lim | 11,55/56 | HM444090 | |
| SIX | ||||
| MlSIX13a | Six | HM444111 | ||
| MlSIX13b | Six | HM444112 | ||
| MlSIX13c | Six | HM444113 | ||
| MlSIX13d | Six | 12,58 | HM444114 | |
| MlSIX13e | Six | HM444115 | ||
| MlSIX13f | Six | HM444130 | ||
| MlSIX27 | Six | 17 | HM444128 | |
| MlSIX28 | Six | HM444129 | ||
| MlSIX32a | HM444116 | |||
| MlSIX32b | Six | HM444144 | ||
| MlSIX32c | Six | 30 | HM444117 | |
| MlSIX36 | Six | NKL | 45 | HM444118 |
| MlSIX41 | Six | 9,45 | HM444127 | |
| MlSIX59a | Six | 57* | HM444131 | |
| MlSIX59b | Six | 57* | HM444119 | |
| MlSIX59c | Six | 57* | HM444120 | |
| MlSIX59d | Six | 57* | HM444121 | |
| MlSIX59e | Six | 57* | HM444126 | |
| ANTP | ||||
| MlANTP03a | HOXL2 | HM444145 | ||
| MlANTP03b | HOXL2 | HM444132 | ||
| MlANTP03c | HM444134 | |||
| MlANTP03d | HM444072 | |||
| MlANTP19 | NKL | 21/22 | HM444073 | |
| MlANTP21 | 10,52 | HM444074 | ||
| MlANTP22 | 13/14 | HM444075 | ||
| MlANTP23 | 18 | HM444140 | ||
| MlANTP25 | HM444076 | |||
| MlANTP35 | 44/45 | HM444077 | ||
| MlANTP37 | NKL, HOXL | 44-45 | HM444078 | |
| MlANTP47 | 46-47 | HM444079 | ||
| MlANTP48 | NKL, HOXL2 | 21-22 | HM444080 | |
| MlANTP51 | 36 | HM444136 | ||
| MlANTP63 | 12/13 | HM444137 | ||
| MlANTP65 | 12/13,45 | HM444081* | ||
| MlANTP66 | 46/47 | HM444082* | ||
| MlANTP67 | 17/18,47 | HM444083* | ||
| MlANTP68 | NKL | 14/15,44/45 | HM444084* | |
| MlANTP71 | NKL | 9,53 | HM444085 | |
| MlANTP72 | NKL | 44/45 | HM444086 | |
| MlANTP78 | 39 | HM444087 | ||
| PRD | ||||
| MlPRD10a | PRD | 37 | HM444097 | |
| MlPRD10b | PRD | 24,46/47 | HM444098* | |
| MlPRD16 | Octapeptide | PRD | 46/47 | HM444102* |
| MlPRD43 | Octapeptide | 46/47 | HM444104 | |
| MlPRD44 | PRD, HOXL2 | 46/47 | HM444105* | |
| MlPRD50 | Octapeptide | 14/15,46/47 | HM444141 | |
| MlPRD61 | Octapeptide | PRD | 12/13,46/47 | HM444147 |
| Unclassified | ||||
| MlHD01 | HM444143 | |||
| MlHD05 | HM444146 | |||
| MlHD07a | HM444139 | |||
| MlHD07b | HM444095 | |||
| MlHD07c | HM444096 | |||
| MlHD11a | HM444133 | |||
| MlHD11b | HM444099 | |||
| MlHD14 | HM444100 | |||
| MlHD15 | HM444101 | |||
| MlHD31 | HM444103 | |||
| MlHD60 | insert(LP) 33 | HM444135 | ||
| MlHD70 | HM444106 | |||
| MlHD76a | HM444107 | |||
| MlHD76b | HM444142 | |||
| MlHD77a | HM444108 | |||
| MlHD77b | HM444138 | |||
| MlHD79 | insert(N) 22 | HM444109 | ||
| MlHD86 | HM444124 |
The first column (Name) contains the names given to each gene. The second column (Domain) indicates any additional domains detected either in the predicted gene sequence or in close genomic proximity to the homeobox in the same orientation. If NKL, HOXL, HOXL2 or PRD sequence signatures or are present in the translated homeodomain (as defined in [40]), this is noted in the third column. The third column (Signature) also includes the amino acid sequences of atypical insertions if they are present. The format for insertions is the word 'insert' followed by the amino acids that make up the insertion in parentheses and the first codon of the homeobox occupied by the insertion. For those homeoboxes that are interrupted by one or more introns, the fourth column (Intron Codons) lists either the codon that is interrupted (noted by a single number) or the two codons that are separated by an intron (noted by two numbers separated by a forward slash). Commas separate codon positions for genes with multiple introns. The five MlSIX59 homeoboxes are truncated at the 57th codon and occur at the end of their corresponding GENSCAN gene prediction. The last column indicates the GenBank accession of the corresponding nucleotide sequences. An asterisk next to an accession indicates a previously described version of this homeodomain exists. The previously described homeodomains are as follows: MlANTP65 - ACD85820 (Tlx-like), MlANTP66 - ACD85819 (Dlx/NK-like), MlANTP67 - ACD85818 (BarH/BarX-like), MlANTP68 - ACD85817 (Bsh), MlPRD10b - ACD85823 (Prd3), MlPRD16 - ACD85821 (Prd1), MlPRD44 - ACD85822 (Prd2).
Most of these class-level assignments are confirmed by the presence of secondary domains, sequence signatures, and/or class-specific introns (Table 1). To all of these classes (with the exception of the 18 homeodomains that remained unclassified), we added corresponding homeodomain sequence data from the demosponge Amphimedon queenslandica [30], the placozoan Trichoplax adhaerens [27], the cnidarian Nematostella vectensis [26]and the choanoflagellate Monosiga brevis [39]: we then performed class-specific phylogenetic analyses. We named Mnemiopsis homeodomains that showed a strong affiliation for a particular family accordingly; otherwise, the name of the class is used in conjunction with a preliminary number that was originally assigned to the homeodomain.
ANTP class NKL subclass
Eighteen of the 22 ANTP homeodomains group are within the NKL subclass. There is only weak support for assigning any of the Mnemiopsis NKL homeodomains with particular families but, in some cases, there is consistency between our initial superfamily tree (Figure 1) and our ANTP-specific tree that included the additional Amphimedon, Nematostella, and Trichoplax sequences (Figure 2).
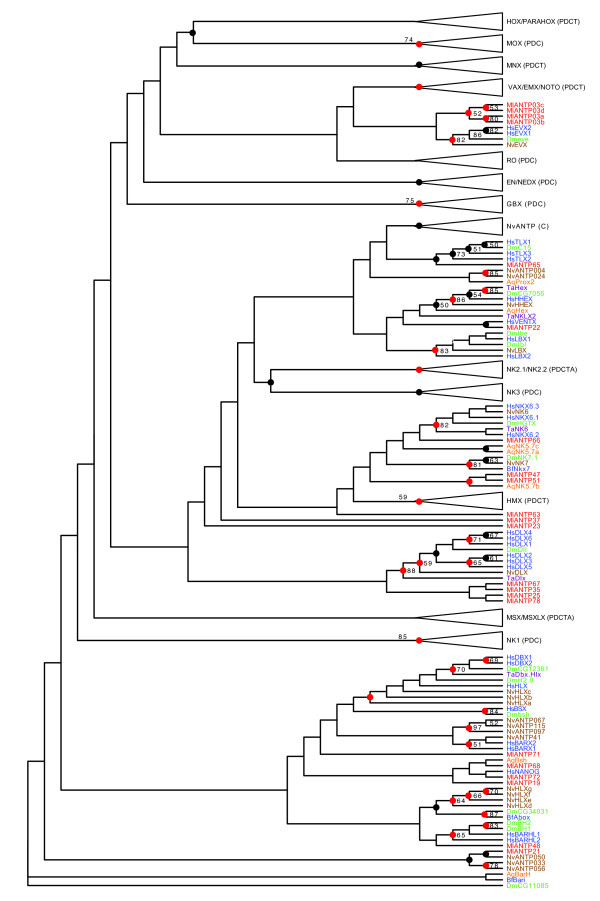
Figure 2.
ANTP tree. Mnemiopsis homeodomains are in red. Arrows represent single genes or clades consisting entirely of Mnemiopsis sequences. Collapsed clades (triangles) represent clades with no Mnemiopsis representative. These clades have families represented in bold along with the phyla/subkingdom who have sequences represented in the collapsed clade (P = Protostomia; D = Deuterostomia; C = Cnidaria; T = Placozoa; A = Porifera). Support values shown are maximum likelihood (ML) bootstrap greater than 50%. Nodes with posterior probabilities generated by MrBayes greater than 50 are represented by a black dot and those clades with posterior probabilities greater than 90 are represented by a red dot. For visibility, trees were initially formatted in FigTree [53] as cladograms with decreasing ordered nodes and uniform branch lengths. They were later formatted by hand in Adobe Illustrator. Rooting of this tree is for display purposes only; branch lengths are presented uniformly, also for display purposes. Actual branch lengths can be viewed by opening the Newick-formatted tree file (Additional File 4), which also includes bootstrap and Bayesian support values, in a tree viewing/editing program such as FigTree [53]. Homeodomain names are prefixed with two letter species abbreviations as follows: Ml = Mnemiopsis leidyi; Aq = Amphimedon queenslandica (Porifera/demosponge); Ta = Trichoplax adhaerens (Placozoa); Nv = Nematostella vectensis (Cnidaria/starlet sea anemone); Dm = Drosophila melanogaster (Protostomia/fruitfly); Bf = Branchiostoma floridae (Deuterostomia/amphioxus); Hs = Homo sapiens (Deuterostomia/human). Other taxa codes either from collapsed clades of this tree or in other trees: Hv = Hydra vulgaris (Cnidaria/hydrozoan); Pd = Platynereis dumerilii (Protostomia/annelid worm); Am = Apis mellifera (Protostomia/honey bee); Ps = Phascolion strombus (Protostomia/sipunculan worm); Sm = Strigamia maritima (Protostomia/centipede); Mb = Monosiga brevicollis (choanoflagellate). Mnemiopsis sequences are shown in red; human and other deuterostome sequences are shown in blue; Drosophila and other protostome sequences are shown in green; cnidarian sequences are shown in brown; Trichoplax sequences are shown in purple; and Amphimedon sequences are in orange.
The following groupings are consistent in both trees and have support values over 50 in our best Bayesian tree: (1) MlANTP65 with the Tlx family and (2) MlANTP22 with the Human Ventx gene. MlANPT25, MlANTP35, MlANTP67 and MlANTP78 group with the Dlx family consistently in both trees but, in the full tree, the Dlx clade also includes MlANTP66. Similarly, MlANTP19, MlANTP68, MlANTP71 and MlANTP72 form clades positioned sister to the Barx, Bsx, Dbx and Hlx familes. However, the relationships between Mnemiopsis homeodomains is inconsistent between these trees. The other NKL homeodomains identified are MlANTP21, MlANTP23, MlANTP25, MlANTP35, MlANTP37, MlANTP47, MlANTP48, MlANTP51, MlANTP63, MlANTP66, MlANTP67 and MlANTP78.
Consistent with our analysis, a previous study classified MlANTP65 as a Tlx-like homeodomain [36]. The same study also associated MlANTP66 with the Dlx family, MlANTP67 with the Barh family and MlANTP68 with the Bsx family, an observation that was not consistently reproduced in our trees.
Evidence in the form of diagnostic residues can provide additional support to classifications [40]. The following homeodomains all contain the diagnostic residues associated with the NKL subclass ([AKST][DENPS][LAST][Q][V] at positions 41-45): MlANTP19, MlANTP37, MlANTP48, MlANTP68, MlANTP71 and MlANTP72. The only other Mnemiopsis homeodomain with the NKL signature is the SINE class homeodomain MlSIX36. (The position of MlSIX36 on the tree in Figure 1 and its upstream SIX domain led to its SINE class designation.) In addition to the NKL signature, the MlANTP37 homeodomain also contains the HOXL signature ([KT][IV]WFQNRR[AMV]K[DEHKLMQWY][KR][KR] at positions 46-58) and the MlANTP48 homeodomains contains the HOXL2 signature (LE[AGKNR]E at positions 16-19) (Table 1).
ANTP class HOXL-related
Four paralogous Mnemiopsis ANTP homeodomains (MlANTP03a, MlANTP03b, MlANTP03c and MlANTP03d) group with the engrailed family in our superfamily tree (Figure 1) and with the Evx family in the ANTP tree (Figure 2). Despite the engrailed family being assigned to the NKL subclass in HomeoDB [24], engrailed has been historically allied with the extended Hox subclass based on synteny [41,42] and phylogeny [43]. Evx is also considered a member of the extended Hox subclass. While it is difficult to pin down the exact relationship of the MlANTP03 homeodomains, it does appear that they are the most likely descendants of the homeodomain that gave rise to the HOXL genes in the lineage leading to Placozoa, Cnidaria and Bilateria. Consistent with this classification, the MlANTP03a and MlANTP03b genes both contain the HOXL2 diagnostic residue signature. There are no clear ParaHox or Hox genes in Mnemiopsis.
PRD class
We identified seven PRD class homeodomains in the Mnemiopsis genome. The PRD class is divided into three subclasses based on the amino acid residue at position 50: Q50, K50 and S50 [44]. As with most homeodomain studies, these subclasses are not monophyletic in our trees (Figure 3). However, given the extremely low support values at the subfamily level, this may not reflect their true relationship. All three subclasses are clearly present in the genomes of bilaterians, Nematostella and Trichoplax. Eight of the nine PRD class homeoboxes in Amphimedon possess the Q50 residue. The remaining PRD homeodomain is the Amphimedon PaxB homeodomain, which is has a degenerate homeodomain [45] and, as such, was not included in our phylogenetic analysis.
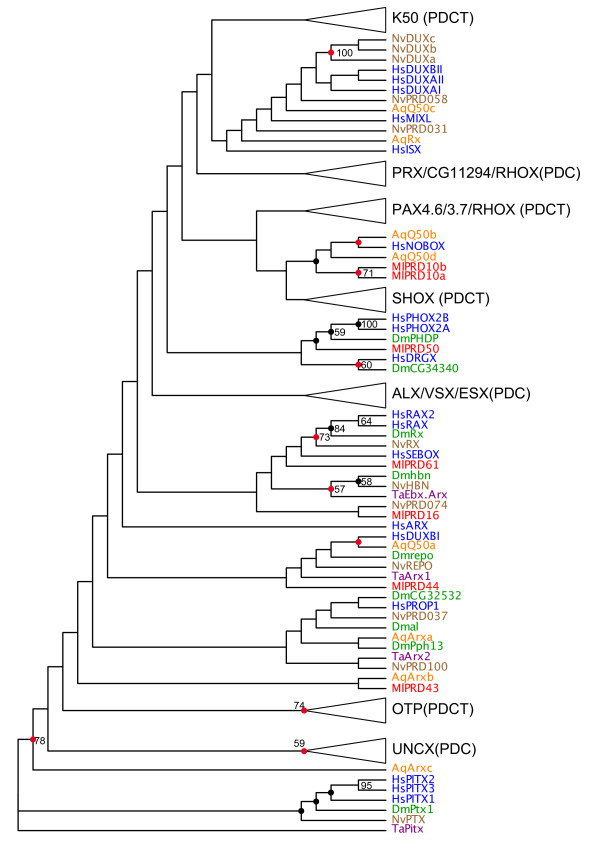
Figure 3.
PRD-HD tree. See Figure 2 for explanation of tree formatting, color codes and species codes. Rooting of this tree is for display purposes only; branch lengths are presented uniformly, also for display purposes. Actual branch lengths can be viewed by opening the Newick-formatted tree file (Additional File 4), which also includes bootstrap and Bayesian support values, in a tree viewing/editing program such as FigTree [53].
Of the seven Mnemiopsis PRD class homeodomains, six have a Q at position 50. The exception (MlPRD43) is missing sequence information at that position. (Note: just prior to the submission of this manuscript, a new assembly has revealed the likely 3' end of this homeodomain that includes a Q at position 50). We did not find any Mnemiopsis genes with an S at position 50. The only other Mnemiopsis genes with a K at position 50 are the 18 SINE class genes that, like the K50 PRD class genes, also characteristically have a K residue at position 50.
Consistent with the absence of lysine or serine residues at position 50 in Mnemiopsis and Amphimedon PRD homeodomains, we see no grouping of Mnemiopsis or Amphimedon homeodomains with S50 or K50 clades, with the following exceptions: (1) MlPRD16 groups with the Nematostella S50 homeodomain NvPRD074, albeit with virtually no support (ML bootstrap = 2, Bayesian posterior probability distribution = 2), within a larger clade of Q50 homedomains; and (2) the Amphimedon homeodomain AqQ50a groups with the highly divergent HsDUXBI (ML bootstrap = 27, Bayesian posterior probability distribution = 95), also within a larger clade of Q50 homedomains (Figure 1). The overwhelming evidence suggests that Mnemiopsis and Amphimedon are devoid of S50 and K50 PRD class homeodomains. Conversely, Nematostella and Trichoplax both have clear K50 and S50 homeodomains. The phylogenetic distribution of Q50, S50 and K50 PRD homeodomains in our study is consistent with the hypothesis that Q50 homeodomains were the founders of the PRD class [44].
Five of the seven Mnemiopsis PRD class homeodomains contain the diagnostic residues (L[EINQRV][^DGHMPTVWY][^CDGKMNPQR][FL][^CFILPTWY][AEFHKQRV][ADEGKNSTW][CHKMPQR][FHY]P at positions 16-26) associated with paired homeodomains in bilaterians: MlPRD10a, MlPRD10b, MlPRD16, MlPRD44, MlPRD61 (Table 1). No other Mnemiopsis homeodomains display this pattern. MlPRD10b, MlPRD16 and MlPRD44 had been identified as Paired class genes in a previous study and were named Prd3, Prd1 and Prd2 respectively [36]. MlPRD44 also contains the HOXL2 diagnostic residues (Table 1).
MlPRD16 and MlPRD61 have clear octapeptide sequences upstream of the homeodomain (SSISSLLS and HSIDDILG, respectively), a hallmark characteristic of a subset of the PRD class homeodomains. MlPRD43 and MlPRD50 have less-conserved but possible octapeptides as well (QRILGILS and YNIEGLLG, respectively). There are no paired domains associated with any Mnemiopsis homeodomains, but there are two independent paired domain sequences that appear to be direct orthologs of the two identified in the ctenophore Coeloplana willeyi [33].
Like most PRD class homedomains [46], all but one of the Mnemiopsis PRD homeodomains have an intron that occurs in the vicinity of the 46th and 47th codons. The one exception, MlPRD10a, has a single intron that interrupts the 37th codon. This might be the result of a retrotransposition event involving a transcript from its paralog (MlPRD10b) followed by an intron gain event. There are additional introns in the N-termini of the homeodomains of MlPRD10b, MlPRD50, and MlPRD61.
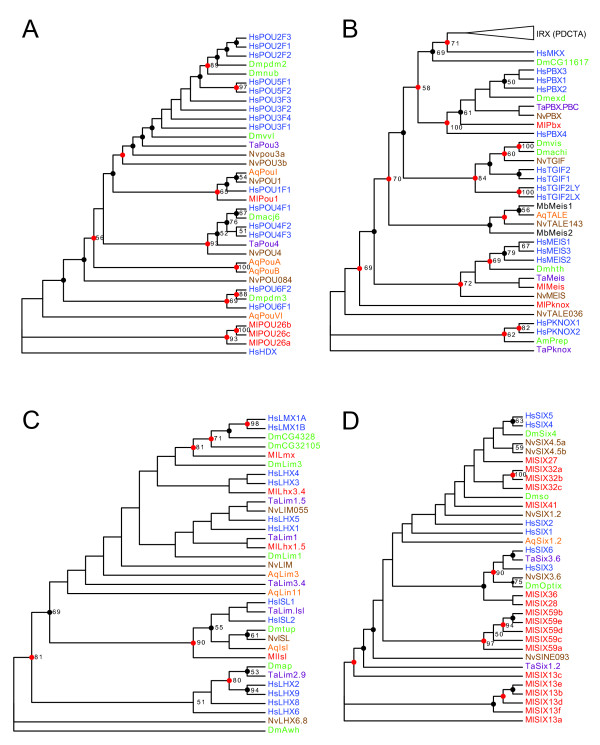
POU class
MlPOU1, MlPOU26a, MlPOU26b, and MlPOU26c make up the four Mnemiopsis POU class homeodomains. MlPOU1 has relatively strong support values, placing it in the POU1 family (ML bootstrap = 65; Bayesian posterior probability distribution = 98; Figure 4A). In addition, it has a POU-specific domain upstream of the homeodomain, a defining factor of the POU class [47]. There is weak support uniting MlPOU26a, MlPOU26b and MlPOU26c with the human HDX (highly divergent homeobox) homeodomain of POU class genes (ML bootstrap = 19; Bayesian posterior probability distribution = 45). Only one of the three MlPOU26 homeodomains (MlPOU26a) contains an upstream POU-specific domain.
Figure 4.
Trees for other classes. The remaining class trees are as follows: (A) POU class; (B) TALE class; (C) LIM class; and (D) SIX class. See Figure 2 for an explanation of tree formatting, colour codes and species codes. Branch lengths are presented uniformly for display purposes. Actual branch lengths can be viewed by opening the Newick-formatted tree file (Additional File 4), which also includes bootstrap and Bayesian support values, in a tree viewing/editing program such as FigTree [53].
TALE class
MlPbx, MlMeis and MlPknox, like other TALE class homeodomains, have a three amino acid insertion in the loop between the first and second alpha-helices (Table 1). MlPbx, MlMeis and MlPknox consistently group with the Pbx, Meis and Pknox families, respectively, in both trees with moderate support (Figures 1 and 4B). In all three cases, the phylogenetic assignment of these homeodomains is reinforced by the identification of several conserved motifs outside of the homeodomain, as well as by conserved intron positions (Table 1).
Like other Pbx genes (and unlike other TALE genes), MlPbx has a glycine residue at position 50 of the homeodomain. In addition, a Basic Local Alignment Search Tool (BLAST) search to the contig containing MlPBX shows significant similarity to the PBC domain [48] located ~1.5 KB upstream of the homeodomain, as assessed by BLAST [percent identity (ID)= 25/67, expectation (E)-value = 2 × 10-7). Like the cnidarian and human PBX genes, MlPbx has an intron that interrupts the second codon and one that falls between the 47th and 48th codon of the 63-codon TALE homeobox.
Meis homeodomain proteins have several conserved motifs in addition to the homeodomain [49]. A GENSCAN prediction containing the MlMeis homeodomain shows similarity to the upstream MEIS A domain (ID = 19/69, E-value = 0.005), as well as weaker similarity to the MEIS D domain downstream of the homeodomain (ID = 16/48, E-value = 0.014). Similar to bilaterians and cnidarians, MlMeis has two introns. One falls between the 25th and 26th codons, while another interrupts the 51st codon.
The GENSCAN-predicted peptide that contains the MlPknox homeodomain also includes the abbreviated MEIS A domain that is characteristic of the Pknox family, as well as the MEIS B motif (ID = 33/139, E-value = 8 × 10-5). MlPknox, like the human PKNOX1 and PKNOX2 genes, has an intron that separates the 25th and 26th codons and one that interrupts the 51st codon of the homeobox.
We were unable to identify an Irx homeodomain in Mnemiopsis, despite there being Irx family members from Amphimedon, Trichoplax and Nematostella. Also absent was the Tgif homeodomain, found only in cnidarians and bilaterians.
LIM class
MlIsl, MlLhx1.5, MlLhx3.4 and MlLmx make up the four LIM class homeodomains of Mnemiopsis (Figure 4C). We assigned these four Mnemiopsis homeodomains to the Isl, Lhx1/5, Lhx3/4 and Lmx families, respectively, based on the consistency between tree runs (Figures 1 and 4C) and moderate support in the full homeodomain tree (Figure 1). BLAST searches of the genomic scaffolds containing Mnemiopsis LIM homeodomains reveal LIM-type zinc finger domains immediately upstream of these four homeodomains. Additional BLAST searches also reveal traces of LIM domains independent of homeodomains in the Mnemiopsis genome (data not shown), suggesting the existence of LIM domain transcription regulator genes.
SINE class
Eighteen SINE class homeodomains representing seven distinct SINE lineages were recovered from the Mnemiopsis genome (Figure 4D, Table 1). Of these, all but one have the characteristic lysine at position 50 (as described in [21]). The exception is MlSIX41, for which we are missing the sequence information from the C-terminus of the homeodomain (including position 50). Additionally, 17 of the 18 SINE class homeodomains have a SIX domain upstream of the homeodomain. The exception, MlSIX32a, is situated on the N-terminal end of a small scaffold in our current assembly, so its absence may be due to the resolution of our assembly.
The SINE class is monophyletic in our superfamily tree except for a clade of five Mnemiopsis homeodomains (MlSIX59a, MlSIX59b, MlSIX59c, MlSIX59 d and MlSIX59e), which group with Zhx/Homez (Figure 1). This exception is perhaps not completely unexpected given that, like the Zhx/Homez genes, MlSIX59 homeodomains are quite divergent; they are five of only six homeodomains in our entire Mnemiopsis set that do not include a tryptophan at position 48, which is characteristic of the typical homeodomain. The other homeodomain, MlSIX45, is also a member of the SINE class.
The Mnemiopsis SINE class homeodomains do not clearly separate into the three families recognized in bilaterians. Only two of the 18 maintain the four family-defining diagnostic residues (positions 3-6) in the homeodomain [50]. MlSIX41 and MlSIX27 have the SIX1/2 family 'ETSY' pattern in positions 3-6 of the homeodomain. However, neither MlSIX41 nor MlSIX27 group convincingly with the Six1/2 group. The Mnmemiopsis SIX class is the result of extensive ctenophore-specific diversification. A more in-depth phylogenetic analysis that includes SIX domains may provide additional insight into these relationships.
Unclassified Mnemiopsis homeodomains
Two clades consisting of 18 Mnemiopsis homeodomains appear as separate offshoots in our superfamily tree (Figure 1, Table 1). None of these 18 homeodomains have introns, or any of the known class signatures, that would hint that they might belong to an existing class. MlHD60 and MlHD79 have insertions that interrupt the homeodomain but these insertions are unlike the known insertions seen in the TALE, HNF and PROS classes. The MlHD60 insertion consists of two amino acids that occur in the third alpha-helix. The other insertion occurs in the loop region between the first and second alpha-helices but, unlike the TALE insertions, it consists only of a single amino acid. The average branch length of the homeodomains in these clades is 5% shorter than for the other Mnemiopsis homeodomains, confirming that these unclassified Mnemiopsis homeodomains do not simply comprise a clade of unusually long branches.
Missing classes
There are no Mnemiopsis homeodomains that grouped with HNF, CUT, PROS, or CERS classes in our analyses. Consistent with this result, no Mnemiopsis homeodomains exhibit insertions between the second and third helices, like those seen in the bilaterian HNF and PROS class homeoboxes. Besides the five apparent SINE class homeodomains, no other Mnemiopsis homeodomains group with zinc finger (ZF) homeodomains.
Homeobox linkage
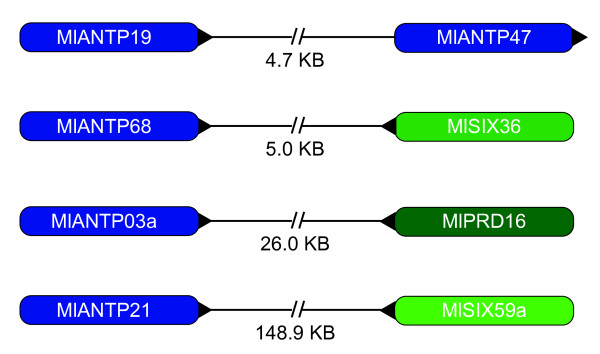
There are four pairs of linked homeoboxes in our current Mnemiopsis genome assembly (Figure 5). The tightest linkage is between two ANTP class homeoboxes (MlANTP19 and MlANTP47), which are 4.7 KB apart. A different ANTP class homeobox (MlANTP68) is situated 5.0 KB downstream from the SINE class homeobox MLSIX36. The HOXL-related ANTP class homeobox MlANTP03a is separated by 26.0 KB from the PRD class homeobox MLPRD16. The ANTP class MlANTP21 and the SINE class homeobox MLSIX59 are on the same contig, 148.9 KB apart. None of the linked homeoboxes are obvious paralogs, suggesting that these pairs are not the result of recent duplication events.
Figure 5.
Homeobox linkage in Mnemiopsis. These four pairs of homeoboxes are linked on four pairs of genomic scaffold. The distance between homeoboxes is noted in between each pair. Arrowheads applied to one side or the other represent the orientation of the linkage.
Evolutionary dynamics of the Mnemiopsis homeodomain superfamily
In order to better-understand the nature of the homeodomain superfamily in Mnemiopsis, we compared average branch lengths and the number of species-specific homeodomain clades in the Mnemiopsis, Amphimedon, Trichoplax, Nematostella, Drosophila, Caenorhabditis elegan, and human genomes (Table 2). We performed ML analyses with homeodomain sequences from this set of species. Degenerate homeodomains (for example, Amphimedon PaxB) and homeodomains from pseudogenes were not included. The resulting tree and alignments are included as supplemental material (Additional file 2).
Table 2.
Paralog count and estimated branch lengths of seven species.
| Species | No. of HDs in this test | No. of HDs in species-specific clades | No. of species-specific clades | Average branch length |
|---|---|---|---|---|
| Nematostella vectensis | 127 | 66 | 22 | 1.010 |
| Drosophila melanogaster | 102 | 25 | 12 | 1.052 |
| Trichoplax adhaerens | 35 | 0 | 0 | 1.162 |
| Amphimedon queenslandica | 31 | 7 | 2 | 1.286 |
| Homo sapiens | 256 | 197 | 74 | 1.293 |
| Mnemiopsis leidyi | 76 | 45 | 14 | 1.344 |
| Caenorhabditis elegans | 113 | 34 | 11 | 1.480 |
| Average | 105.71 | 53.43 | 19.29 | 1.232 |
The second column specifies the number of homeodomains (HDs) used in the neighbor-joining analyses that produced the tree from which this table is based. The third column indicates the number of homeodomains that were more closely related to a homeodomain from the same species. The number of species-specific clades is denoted in the fourth column. The final column shows the average length of all branches from a particular species based on a midpoint rooted tree. Files used in this analysis are (Additional File 2).
For each species, we recorded the number of species-specific clades that included more than one homeodomain, as well as the total number of homeodomains in those species-specific clades (Table 2). These numbers give us an approximation of the number of lineage-specific homeodomain duplications that have been preserved in a specific lineage since it split from its closest relative in the analysis [51]. A species that has recently undergone extensive genome reduction would be expected to harbour less species-specific clades than a genome that has experienced a recent genomic expansion. Our data shows that very few paralogous homeodomains exist in the Amphimedon (7) and Trichoplax (0) genomes, whereas the human genome has a remarkably high level of paralogs (197). Mnemiopsis (45) and Nematostella (66) are both very close to the mean (Table 2).
Branch lengths provide a means of measuring the level of divergence for a particular homeodomain. Longer branches correspond to higher levels of divergence. We rooted the same neighbor-joining tree described above at its midpoint and determined the average branch lengths for each species' set of homeodomains (Table 2). In our tree, the Mnemiopsis branches tend to be longer than for all the other species except for C. elegans, which is known to have very long branches [52]. The Mnemiopsis average branch length is slightly closer to the mean than it is to the C. elegans average, suggesting that the Mnemiopsis homeodomains are moderately divergent. The trees used in this analysis are included as supplemental material (Additional Files 2 and 3) and branch lengths can be visualized directly using a tree-viewing program such as Figtree [53].
Discussion
There is strong evidence suggesting that Mnemiopsis has homeodomains belonging to six of the 11 defined homeodomain classes (ANTP, PRD, LIM, POU, SINE and TALE). Mnemiopsis appears to be missing the other five homeodomain classes (HNF, CUT, PROS, ZF and CERS). It is also missing ¬the Hox/ParaHox and extended Hox subclasses of the ANTP class, as well as the S50 and K50 subclass of the PRD class. Given that Trichoplax, Nematostella and the bilaterians examined in our study clearly possess all of these classes and subclasses, the most parsimonious animal tree would involve Ctenophora and Porifera branching off the main animal trunk prior to the Placozoa, Cnidaria and Bilateria (Figure 6). This configuration is congruent with most of the previously published 18 S phylogenies and results from several of the EST-based phylogenomic studies [12,15].
Figure 6.
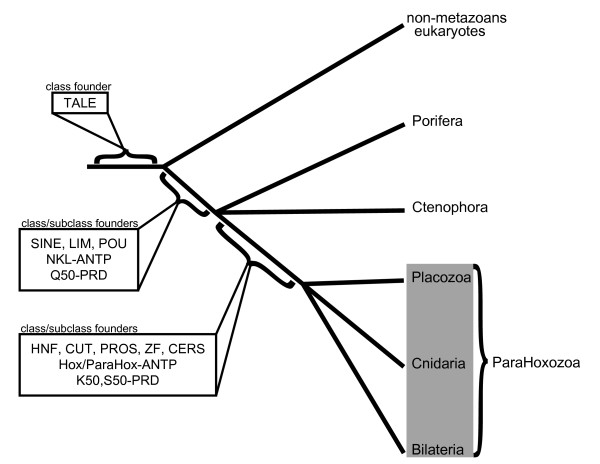
Phylogenetic relationship of early-branching animal lineages. Given the conflicting results of several recent studies and the current existing phylogenomic evidence presented in this study, we have constructed an animal phylogeny that includes two polytomies. It is uncertain whether Porifera or Ctenophora branched at the base. The evidence from this study (and others) unites Placozoa, Cnidaria and Bilateria into the subkingdom ParaHoxozoa. The origins of class and subclass founders are indicated along major branches of the tree.
ParaHoxozoa
The apparent absence of Hox/ParaHox, S50 and K50 PRD and HNF class homeodomains in Amphimedon and Mnemiopiopsis supports an organismal clade that unites cnidarians, bilaterians and placozoans. Although previous analyses have strongly suggested a monophyletic relationship of these three phyla (particularly Collins [6]), we are not aware of any studies to date that have given this subkingdom a name. We propose the name ParaHoxozoa for the clade containing Bilateria, Placozoa and Cnidaria.
Studies have shown that the ancestor of the ParaHoxozoa had at least one Hox/ParaHox gene [26,54] and that these genes are missing from both ctenophores and sponges ([55] and this study). While cnidarians and bilaterians have homeodomains that are clearly descended from both ancestral Hox and ParaHox genes, Trichoplax, has only a single Hox/ParaHox gene, Trox2 [54]. This homeodomain consistently forms a moderately supported clade with the ParaHox GSX family ([28,56] and this study) and may be a true ParaHox gene. However, it has been postulated that the Trox2 gene may be a direct descendant of an ancestral 'ProtoHox' gene rather than a proper ParaHox gene [28,54,56]. The name ParaHoxozoa was chosen since, based strictly on the trees themselves, a ParaHox clearly unites this group.
Evidence distinguishing Mnemiopsis from the ParaHoxozoa
The presence of ample paralogs in the Mnemiopsis genome suggests that it has not undergone extensive genome reduction. Currently, we cannot rule out the possibility that recent duplication events have masked more ancient gene losses or that the paralog count in the homeobox superfamily is not typical of most superfamilies. Regardless, the level of paralog retention does give some initial insight into the evolutionary dynamics of the Mnemiopsis genome, while also lowering the probability that the Mnemiopsis lineage at one time had (and subsequently lost) these missing homeodomains. The availability of other ctenophore genomes for future studies and analysis of other gene superfamilies will help to better-resolve the evolutionary dynamics of the Mnemiopsis genome.
The unusually high level of conservation of the homeobox genes and their vital roles in early development further lowers the possibility that the loss of entire classes and subclasses of these genes could be tolerated. There are numerous examples of homeobox families missing from a wide array of phylogenetically disparate lineages. For example, Ciona intestinalis and multiple species of parasitic platyhelminthes appear to have lost several Hox genes [57,58]. In addition, the genomes of Drosophila and human are each missing several homeobox families [26]. However, it is important to note that no species examined thus far has been shown to be missing entire classes or subclasses of homeobox genes.
Long branches do have the ability to distort phylogenies and the presence of longer branches in our Mnemiopsis data set was initially a concern. However, if ctenophores are among the earliest metazoan branches and substantial expansion of the homeodomain superfamily occurred along the lineage leading to Mnemiopsis, these long branches are not entirely unexpected. Conversely, the somewhat shorter branches in Amphimedon homeodomains, despite its early-branching phylogenetic position, are likely due to it having far fewer paralogs.
Another possible concern is that our paralog results are an artifact caused by the attraction of long branches and that these long branches represent class members that appear to be missing but are perhaps unrecognizable in Mnemiopsis. Two pieces of evidence undermine this assertion. First, C. elegans, which has 9% longer branches than Mnemiopsis and 67% more homeodomains, would be expected to have more species-specific homeodomain clades, but in fact has 23% fewer (Table 2). Second, when we remove the C. elegans homeodomains from the dataset (which vastly reduces the number of long branches in our tree) and redo the analysis, the number of Mnemiopsis-specific clades does not change (Additional File 3).
Incongruence with the 'Coelenterate' hypothesis
A scenario grouping cnidarians and ctenophores into a 'coelenterate' clade consistent with the phylogenomic study by Philippe et al. [10] seems unlikely based on the number of clades that include a Nematostella homeodomain to the exclusion of a Mnemiopsis representative. For example, Nematostella has representatives of the ANTP homeodomain families Mox, Emx, Gbx, Ro, Mnx, Vax, Not, Nk1, NK2 and NK3, as well as two ParaHox and three Hox-related homeoboxes (Figure 2). If Ctenophora and Cnidaria were, in fact, sister taxa, an extraordinary number of gene losses would have been required in the Mnemiopsis lineage, given the observation that Mnemiopsis is lacking these families. This amount of loss seems very unlikely given the pattern of paralog retention in Mnemiopsis. A more recent study by this group also failed to recover the 'coelenterate' clade [15].
Mnemiopsis extended Hox?
The origin of the four paralogous Mnemiopsis ANTP homeodomains (MlANTP03a, MlANTP03b, MlANTP03c and MlANTP03d) that group with the engrailed family in the superfamily tree (Figure 1) and with the Evx family in the ANTP tree is difficult to interpret. The presence of the HOXL signature in two of these four homeodomains, combined with their tendency to form a clade with hox-related genes, suggests that these four homeodomains may have descended from an ancestral homeodomain; this ancestral homeodomain may then, in turn, have led to the formation of the extended Hox (Evx, Gbx, Meox, Mnx, Rough), Hox and ParaHox classes through a series of duplications in the ParaHoxozoa stem.
Functional analysis of the MlANTP03 genes may give further insight into the role of this ancestral gene, which would be particularly interesting given the roles that extended Hox genes play in critical biological processes such as neurogenesis, myogenesis, axial patterning, segmentation, gastrulation and photoreception.
Identifying the basal branch
Despite strong evidence uniting the ParaHoxozoa to the exclusion of Porifera and Ctenophora, it does not appear that homeodomain sequences alone (at least based on the current set of available sequences) will be powerful enough to incontrovertibly determine the basal metazoan branch. There are several unambiguous clades containing Mnemiopsis homeodomains grouping with parahoxozoan clades to the exclusion of Amphimedon representatives (for example, Lmx, Hlx and Pbx). The many occurrences of this pattern insinuate that sponges branched off at the base of the Metazoa. This pattern is consistent with many of the 18 S phylogenies (see Table 1 in [59] and references therein), as well as a recent EST-based phylogeny [15]. However, there is substantial evidence suggesting the alternative relationship.
A number of parahoxozoan clades include Amphimedon homeodomains but lack a corresponding Mnemiopsis homeodomain (for example, NK2 and Irx). The scarcity of paralogous homeodomains could be a sign that the Amphimedon genome is reduced. This, in turn, increases the chance that multiple homeodomain families were lost in Amphimedon. In addition, the lack of a homeobox associated with a paired domain in Mnemiopsis opens the possibility that the fusion of the homeobox and paired domain postdates the divergence of ctenophores from the rest of Metazoa. This would be consistent with other phylogenomic studies [11,12].
It is possible that similar analyses of additional gene families with the same set of taxa might be able to resolve the basal branch. Alternatively, it could be that sequence data from additional sponge and ctenophore genomes will be required to satisfactorily settle the debate. The current study does make a strong case that this uncertainty might eventually be resolved using a gene family-type approach.
Implications for the evolution of homeobox clusters
Extensive genomic clustering of ANTP homeoboxes in multiple metazoan genomes suggests that the ANTP class homeobox genes were formed by a series of tandem duplications (reviewed in [60]). A recent study showed that six of the eight Amphimedon NKL homeoboxes are clustered [30] and are likely descendants of the ancestral ANTP megacluster. One of the four Mnemiopsis homeobox clusters - specifically, the cluster containing MlANTP19 and MlANTP47 - is potentially a remnant of this ANTP megacluster as well (Figure 5). In our phylogenetic analyses, MlANTP47 consistently groups with the Amphimedon NK5.7b homeodomain with moderate support (ML bootstrap = 24, Bayesian posterior probability distribution = 95; Figure 2). The family-level identity of MlANTP19 is uncertain but it does not appear to be paralogous with MlANTP47.
It is very difficult to draw any conclusions as to the implications of the other three linked homeoboxes, particularly given the difficulty of assigning family-level orthology to the Mnemiopsis homeodomains. The process for detecting ancestral/functional linkages involves identifying orthologs in multiple evolutionarily disparate genomes. Therefore, it may be possible that these linkages are significant and, perhaps, representative of some ancestral cluster, but the current phylogenetic resolution of the Mnemiopsis homeoboxes may make this relationship difficult to detect.
Conclusions
We have identified, named and classified 76 homeoboxes in the Mnemiopsis leidyi genome. In many cases, we have provided additional evidence for our classifications through the detection of diagnostic residues, presence of surrounding domains and identification of conserved intron positions. We have shown that several classes and subclasses are present in placozoan, cnidarian and bilaterian species but are missing from both Mnemiopsis and the sponge Amphimedon (Table 3). Using a phylogenomic approach, we have determined that it is very likely, based on the presence and absence of homeodomains, that the phyla Placozoa, Cnidaria and Bilateria are more closely related to each other than they are to Ctenophora or Porifera. Based on this evidence, we have proposed the name ParaHoxozoa for the clade that includes Placozoa, Cnidaria and Bilateria.
Table 3.
Distribution of classes and subclasses of homeobox genes among early branching taxa.
| Ancestral eukaryote | Amphimedon | Mnemiopsis | Nematostella | Trichoplax | Human/Drosophila | |
|---|---|---|---|---|---|---|
| ANTP | ||||||
| Hox | -- | -- | -- | + | + | + |
| ParaHox | -- | -- | -- | + | + | + |
| Extended Hox | -- | -- | + | + | + | + |
| NKL | -- | + | + | + | + | + |
| PRD | ||||||
| Q50 | -- | + | + | + | + | + |
| N50 | -- | -- | -- | + | + | + |
| K50 | -- | -- | -- | + | + | + |
| LIM | -- | + | + | + | + | + |
| POU | -- | + | + | + | + | + |
| SINE | -- | + | + | + | + | + |
| TALE | + | + | + | + | + | + |
| HNF | -- | -- | -- | + | + | + |
| CUT | -- | -- | -- | -- | -- | + |
| PROS | -- | -- | -- | -- | -- | + |
| ZF | -- | -- | -- | -- | -- | + |
| CERS | -- | -- | -- | -- | -- | + |
The first column contains the class (bold) or subclass (indented). A plus indicates the presence of a clear member of the class or subclass. Alternatively, a minus indicates the absence of a clear member of the class or subclass. Inference of the homeodomain complement of the ancestral eukaryote is based on Derelle et al. [22].
The expansion of the homeobox superfamily has played a major role in the evolution of animal phyla [61]. An understanding of this expansion, along with an accurate animal phylogeny, is critical to understanding metazoan evolution. With this new dataset and phylogeny, as well as the help of functional genomic techniques, we can start piecing together the evolutionary steps that led to such astounding evolutionary feats such as the development of nervous systems, muscular systems, and complex symmetry.
Methods
Sequencing and assembly
Genomic DNA was isolated from the larvae of two separate self-fertilizing hermaphroditic individuals. A library from one source was sequenced to 10× coverage using 454 sequencing: 8.1 million reads totaling 2.7 gigabases were assembled into 29,877 contigs (contig-N50 = 11KB) using the Phusion assembler [62]. Subsequently, we constructed and sequenced a paired-end DNA library with insert sizes of ~4kb from the second genomic source using Illumina sequencing. These 2.8 million paired end reads were used to compile the contigs into 10,106 scaffolds (scaffold-N50 = 123 KB) bringing the physical coverage to ~50×.
Retrieval of Mnemiopsis homeodomains
A TBLASTN search of the Mnemiopsis assembly was conducted using a set of bilaterian homeodomains downloaded from the Homeodomain Resource [63]. This set was compared to and supplemented by previous Mnemiopsis homeodomains that were generated from degenerate PCR and RACE [36].
Gene isolation via RACE PCR
MlPRD86, MlPbx, MlSIX59b, MlSIX41, MlSIX13f, MlPRD10b, MlANTP65, MlANTP67, MlPRD76b, MlPRD60, MlPRD77b and MlPRD86 homeoboxes had been isolated but not published prior to the sequencing of the genome and are included in this study. These were isolated as previously described [36].
Superclass alignment
The Mnemiopsis homeodomains were aligned by eye to the dataset used by Holland et al. 2007, which consisted of all human homeodomains and a representative set of protostome set consisting mostly of homeodomains from Drosophila melanogaster [23]. We supplemented these sequences with eight Branchiostoma floridae homeodomains that are known to be missing from humans. Insertions in the loop-region of the Mnemiopsis homeodomains were removed as done in [23] and other studies. In determining which amino acids to remove from atypical homeodomains, we realized that there was inconsistency in the Holland set as to which three amino acids were removed between the Drosophila and the human sequences. As with the human sequences, we removed the 23rd, 24th and 25th amino acids from the Mnemiopsis atypical TALE sequences and adjusted the Drosophila sequences so they conformed to this rule. Alignment is available as supplementary material (Additional File 1).
Class alignment
The superclass alignment was divided into six separate alignments, (1) ANTP, (2) PRD, (3) SIX, (4) LIM, (5) POU and (6) TALE based on the best superclass tree and the class membership of the bilaterian homeodomains determined from HomeoDB [24]. The three amino acid insertions were reinserted into the TALE alignment. To each of these datasets we added homeodomains from other non-bilaterian species based on published classifications. We added 32 homeodomains from the demosponge Amphimedon queenslandica [30], 37 homeodomains from the placozoan Trichoplax adhaerens [27], 127 homedomains from the cnidarian Nematostella vectensis [26] and two homeodomains from the choanoflagellate Monosiga brevis [39]. These alignments are available as supplemental material (Additional File 1).
Phylogenetic analyses
The Perl script proteinModelSelection.pl (available from the RaxML [37] web site) was used to determine the best scoring amino acid substitution model for our supertree alignment (RTREV +GAMMA). All subsequent analyses used this model.
Three independent runs of RaxML version 7.0.4 [37] were conducted. Two runs used random starting trees with the following command line (raxmlHPC-MPI -m PROTGAMMARTREV -s ALN.phy -#10 -n NAME -k). One run used a neighbor joining starting tree that was generated with default parameters in Phylip version 3.6a3 [64]. The command line for this run was (raxmlHPC -m PROTGAMMARTREV -s ALN.phy -t NJ.tre -n NAME -k).
Three independent runs of PhyML version 3.0 [38] were conducted. Two runs used random starting with the following command line (phyml -i ALN.phy -d aa -m RtRev -a e -q --rand_start -s SPR --r_seed 'cat FILE_W_RANDSEED'). One run used a neighbor joining starting tree with the following command line (beorun phyml -i ALN.phy -d aa -m RtRev -a e -q -s SPR).
Two independent runs with the MPI version of Mr. Bayes version 3.1.2 [65] were conducted with the following execution block (prset aamodelpr = fixed(RTREV); lset rates = gamma; mcmcp mcmcdiagn = no nruns = 1 ngen = 5000000 printfreq = 5000 samplefreq = 500 nchains = 5 savebrlens = yes; mcmc; sumt filename = tale_w_insert.nex nRuns = 1 Relburnin = YES BurninFrac = .25 Contype = Allcompat;). Log likelihood values were plotted and their progression was visually examined over time. All runs were found to be asymptotic before the .25 burnin fraction.
Likelihood values for all runs (3 RaxML, 3 PhyML, 2 Bayes and 1 neighbor joining) were generated using PhyMl version 3.0 [38] with the following command line (phyml -i ALN.phy -c 4 -m RtREV -a e -o lr -f d -u TREE.tre -d aa -b 0 -s NNI). The tree with the highest likelihood value was chosen for all downstream analysis. The best trees were as follows: Supertree = RaxML-randomstart, ANTP = RaxML-randomstart, PRD = RaxML-randomstart, SIX = PhyML-randomstart, LIM = RaxML-randomstart, POU = RaxML-NJstart, and TALE = RaxML-randomstart. The best trees are available as supplemental material (Additional File 4).
Support indices
Support was assessed by 100 replicates of the bootstrap using the method that provided the highest likelihood value. Bootstrap values greater than 50 were applied to the best tree. To these three trees, Bayesian posterior probabilities were also added from the Bayesian tree with the highest likelihood score. The best trees with support values are available as supplemental material (Additional File 4).
Paralog retention analysis
From our initial superfamily alignment, we removed all sequences that were not Mnemiopsis, Drosophila, or human. To this, we added all Trichoplax, Nematostella and Amphimedon sequences that were used in our class-level phylogenies. Finally we added all the C. elegans homeodomains that were used in [66]. We ran a neighbor-joining analysis using default parameters in Phylip [64]. This tree was then parsed with a Perl script (count_species_specific_clades.pl) that identified occurrences of homogeneous clades of taxa with identical two-letter prefixes. This Perl script, tree and alignment are included as supplemental material (Additional File 2). A version of this analysis was run without the C. elegans data (see Additional File 3).
Branch length analysis
The tree used in the paralog analysis was subsequently used to estimate average branch lengths (Table 2). This tree was opened in FigTree v.1.2.3 [53], rooted at the midpoint, and saved. This rooted tree was then opened in TreeStat v.1.2 [67] and the Root-Tip Lengths were calculated. A Perl script (calculate_average_root_tip.pl) was used to parse the output of TreeStat and calculate the average branch lengths. This Perl script, tree, and alignment are included as supplemental material (Additional File 2). In order to compare lengths of subsets of Mnemiopsis data, we ran a separate instance of this analysis, substituting the two-letter 'Ml' prefix of the subset to be analysed with 'Zz'. In order to assess the feasibility of using this technique to assess the relative branch lengths from trees based on 60 amino acid matrices, we performed a simulation study, and the details of this simulation study are available as supplemental material (Additional File 5).
Abbreviations
BLAST: Basic Local Alignment Search Tool; E-value: expectation value; EST: expressed sequence tag; HomeoDB: Homeo Database; ID: percent identity; ML: maximum likelihood; PCR: polymerase chain reaction; PHYML: phylogenetic inferences using ML; rRNA: ribosomal RNA; RAxML: randomized accelerated ML; ZF: zinc finger.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JFR designed and conceived the study, isolated Mnemiopsis homeodomains from the genomic assembly, performed alignment, performed phylogenetic analyses and drafted the manuscript. KP isolated Mnemiopsis DNA and RNA for sequencing and performed RACE PCR. NISC performed sequencing. JCM assembled Mnemiopsis genome. MQM participated in the design of the study and helped to draft the manuscript. ADB participated in the design and conception of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Zip file of alignments. Includes all alignments used for the supertree and class trees in Phylip format.
Zip file with data from paralog and branch length analysis. Includes files necessary to generate Table 2. See 00-README in the zip file for additional information.
Paralog count and estimated branch lengths of all species in Table 2 plus C. elegans. Includes a table similar to Table 2 and the necessary files to create this table. See 00-README in the zip file for additional information.
Zip file of trees. Includes the trees with the highest likelihood generated for the supertree and the class trees in Newick format. Trees include support values from MrBayes and maximum likelihood bootstraps. See 00-README in the zip file for additional information.
Simulation analysis to assess the feasibility of estimating relative branch lengths from 60 amino acids. Includes data used to determine the feasibility of the technique used to assess relative branch lengths from trees based on 60-amino acid matrices (as described in the main text).
Contributor Information
Joseph F Ryan, Email: jfryan@mail.nih.gov.
Kevin Pang, Email: kpang@hawaii.edu.
James C Mullikin, Email: mullikin@mail.nih.gov.
Mark Q Martindale, Email: mqmartin@hawaii.edu.
Andreas D Baxevanis, Email: andy@nhgri.nih.gov.
Acknowledgements
This work was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. Additional support was provided by the NSF Graduate Research Fellowship Program to KP and grants from NASA and NSF to MQM. We would like to thank Alice Young, Brian Schmidt, Natalie Gurson, Richelle Legaspi and Betsy Novotny, who were largely responsible for the sequencing work performed at NISC. JR would like to thank Arjun Prasad, Adam Reitzel, Bhavesh Borate and Tyra Wolfsberg for insightful conversations regarding this work. We are also thankful to the three anonymous reviewers whose thoughtful suggestions, corrections and insightful comments greatly improved this finished manuscript.
References
- Pang K, Martindale MQ. Ctenophores. Curr Biol. 2008;18:R1119–1120. doi: 10.1016/j.cub.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Cuvier G. Le Règne Animal Distribué Selon son Organisation, pour Servir de Base à l'Histoire Naturelle des Animaux et d'Introduction à l'Anatomie Comparée. Paris: Deterville; 1817. [Google Scholar]
- Leuckart R. Ueber die Morphologie und die Verwandtschaftsverhältnisse der wirbellosen thiere. Ein Beitrag zur Charakteristik und Classification der thierischen Formen. Braunschweig,: F. Vieweg und Sohn;; 1848. [Google Scholar]
- Harbison GR. In: The Origins and Relationships of Lower Iinvertebrates. Conway Morris S, George JD, Gibson R, Platt HM, editor. Vol. 28. London, UK: The Systematics Assocation; 1985. On the classification and evolution of the Ctenophora; pp. 78–100. [Google Scholar]
- Bridge D, Cunningham CW, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol. 1995;12:679–689. doi: 10.1093/oxfordjournals.molbev.a040246. [DOI] [PubMed] [Google Scholar]
- Collins AG. Evaluating multiple alternative hypotheses for the origin of Bilateria: an analysis of 18 S rRNA molecular evidence. Proc Natl Acad Sci USA. 1998;95:15458–15463. doi: 10.1073/pnas.95.26.15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim W, Cunningham CW. A new perspective on lower metazoan relationships from 18 S rDNA sequences [2] Molec Biol Evolution. 1999;16:423–427. doi: 10.1093/oxfordjournals.molbev.a026124. [DOI] [PubMed] [Google Scholar]
- Smothers JF, von Dohlen CD, Smith LH, Spall RD. Molecular evidence that the myxozoan protists are metazoans. Science. 1994;265:1719–1721. doi: 10.1126/science.8085160. [DOI] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E. et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD. et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U. et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, Kolokotronis SO, Eitel M, Desalle R. The Diploblast-Bilateria Sister hypothesis: Parallel revolution of a nervous systems may have been a simple step. Commun Integr Biol. 2009;2:403–405. doi: 10.4161/cib.2.5.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Pick KS, Philippe H, Schreiber F, Erpenbeck D, Jackson DJ, Wrede P, Wiens M, Alie A, Morgenstern B, Manuel M, Worheide G. Improved phylogenomic taxon sampling noticeably affects non-bilaterian relationships. Mol Biol Evol. 2010;27:(9): 1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/S0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- Balavoine G. Are Platyhelminthes Coelomates without a Coelom? An argument based on the evolution of hox genes. Amer Zool. 1998;38:843–858. [Google Scholar]
- Cook CE, Smith ML, Telford MJ, Bastianello A, Akam M. Hox genes and the phylogeny of the arthropods. Curr Biol. 2001;11:759–763. doi: 10.1016/S0960-9822(01)00222-6. [DOI] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308:428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Bürglin TR Meyers RA. Homeodomain Proteins Encyclopedia or Molecular Cell Biology and Molecular Medicine 20052Weinheim: Wiley-VCH Verlag; 16307171 [Google Scholar]
- Derelle R, Lopez P, Le Guyader H, Manuel M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol Dev. 2007;9:212–219. doi: 10.1111/j.1525-142X.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong YF, Butts T, Holland PW. HomeoDB: a database of homeobox gene diversity. Evol Dev. 2008;10:516–518. doi: 10.1111/j.1525-142X.2008.00266.x. [DOI] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, de Jong P, Steele RE, Technau U. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442:684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7:R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML. et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Schierwater B, Kamm K, Srivastava M, Rokhsar D, Rosengarten RD, Dellaporta SL. The early ANTP gene repertoire: insights from the placozoan genome. PLoS One. 2008;3:e2457. doi: 10.1371/journal.pone.0002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- Bebenek IG, Gates RD, Morris J, Hartenstein V, Jacobs DK. Sine oculis in basal Metazoa. Devel Genes Evolution. 2004;214:342–351. doi: 10.1007/s00427-004-0407-3. [DOI] [PubMed] [Google Scholar]
- Finnerty JR. Homeobox genes in the Ctenophora: identification of paired-type and Hox homologues in the atentaculate ctenophore, Beroe ovata. Molec Marine Biol Biotech. 1996;5:249–258. [PubMed] [Google Scholar]
- Hoshiyama D, Iwabe N, Miyata T. Evolution of the gene families forming the Pax/Six regulatory network: isolation of genes from primitive animals and molecular phylogenetic analyses. FEBS Lett. 2007;581:1639–1643. doi: 10.1016/j.febslet.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Lee SE, Gates RD, Jacobs DK. Gene fishing: the use of a simple protocol to isolate multiple homeodomain classes from diverse invertebrate taxa. J Mol Evol. 2003;56:509–516. doi: 10.1007/s00239-002-2421-2. [DOI] [PubMed] [Google Scholar]
- Martinelli C, Spring J. T-box and homeobox genes from the ctenophore Pleurobrachia pileus: comparison of Brachyury, Tbx2/3 and Tlx in basal metazoans and bilaterians. FEBS Lett. 2005;579:5024–5028. doi: 10.1016/j.febslet.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Pang K, Martindale MQ. Developmental expression of homeobox genes in the ctenophore Mnemiopsis leidyi. Dev Genes Evol. 2008;218:307–319. doi: 10.1007/s00427-008-0222-3. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I. et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca NA, Vieira CP, Holland PW, Vieira J. Protein evolution of ANTP and PRD homeobox genes. BMC Evol Biol. 2008;8:200. doi: 10.1186/1471-2148-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LF, Holland PW. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev. 2003;5:459–465. doi: 10.1046/j.1525-142X.2003.03052.x. [DOI] [PubMed] [Google Scholar]
- Pollard SL, Holland PW. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr Biol. 2000;10:1059–1062. doi: 10.1016/S0960-9822(00)00676-X. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, Baxevanis AD. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29:3258–3269. doi: 10.1093/nar/29.15.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Liubicich D, Hinman VF, Gauthier M, Gongora M, Green K, Worheide G, Leys SP, Degnan BM. Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol Dev. 2006;8:150–173. doi: 10.1111/j.1525-142X.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- Groger H, Callaerts P, Gehring WJ, Schmid V. Characterization and expression analysis of an ancestor-type Pax gene in the hydrozoan jellyfish Podocoryne carnea. Mech Dev. 2000;94:157–169. doi: 10.1016/S0925-4773(00)00286-0. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Bürglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Seo HC, Curtiss J, Mlodzik M, Fjose A. Six class homeobox genes in drosophila belong to three distinct families and are involved in head development. Mech Dev. 1999;83:127–139. doi: 10.1016/S0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- Fitch DHA. Introduction to nematode evolution and ecology (August 31 2005) The C. elegans Reseach Community, WormBook; http://www.wormbook.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, ed. Book FigTree. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh; 2006. FigTree. [Google Scholar]
- Jakob W, Sagasser S, Dellaporta S, Holland P, Kuhn K, Schierwater B. The Trox-2 Hox/ParaHox gene of Trichoplax (Placozoa) marks an epithelial boundary. Dev Genes Evol. 2004;214:170–175. doi: 10.1007/s00427-004-0390-8. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17:706–710. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Monteiro AS, Schierwater B, Dellaporta SL, Holland PW. A low diversity of ANTP class homeobox genes in Placozoa. Evol Dev. 2006;8:174–182. doi: 10.1111/j.1525-142X.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Wada S, Tokuoka M, Shoguchi E, Kobayashi K, Di Gregorio A, Spagnuolo A, Branno M, Kohara Y, Rokhsar D, Levine M. et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev Genes Evol. 2003;213:222–234. doi: 10.1007/s00427-003-0321-0. [DOI] [PubMed] [Google Scholar]
- Koziol U, Lalanne AI, Castillo E. Hox genes in the parasitic platyhelminthes Mesocestoides corti, Echinococcus multilocularis and Schistosoma mansoni: evidence for a reduced Hox complement. Biochem Genet. 2009;47:100–116. doi: 10.1007/s10528-008-9210-6. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Baxevanis AD. Hox, Wnt and the evolution of the primary body axis: insights from the early-divergent phyla. Biol Direct. 2007;2:37. doi: 10.1186/1745-6150-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Valentine JW. Why no new phyla after the cambrian? Genome and ecospace hypotheses revisited. PALAIOS. 1995;10:190–194. doi: 10.2307/3515182. [DOI] [Google Scholar]
- Mullikin JC, Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland RT, Ryan JF, Pan C, Baxevanis AD. The homeodomain resource: a comprehensive collection of sequence, structure, interaction, genomic and functional information on the homeodomain protein family. Database (Oxford) 2009;2009:bap004. doi: 10.1093/database/bap004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package). Version 3.63. 2004. [Distributed by the author: Department of Genetics, University of Washington, Seattle, USA]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, eds. TreeStat. v1.1:Tree Statstic Calculaton Tool. http://tree.bio.ed.ac.uk/software/treestat/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Zip file of alignments. Includes all alignments used for the supertree and class trees in Phylip format.
Zip file with data from paralog and branch length analysis. Includes files necessary to generate Table 2. See 00-README in the zip file for additional information.
Paralog count and estimated branch lengths of all species in Table 2 plus C. elegans. Includes a table similar to Table 2 and the necessary files to create this table. See 00-README in the zip file for additional information.
Zip file of trees. Includes the trees with the highest likelihood generated for the supertree and the class trees in Newick format. Trees include support values from MrBayes and maximum likelihood bootstraps. See 00-README in the zip file for additional information.
Simulation analysis to assess the feasibility of estimating relative branch lengths from 60 amino acids. Includes data used to determine the feasibility of the technique used to assess relative branch lengths from trees based on 60-amino acid matrices (as described in the main text).