Abstract
Background
Dwarfing genes have widely been used in barley breeding program. More than 30 types of dwarfs or semidwarfs have been reported, but a few has been exploited in barley breeding because pleiotropic effects of dwarfing genes cause some undesired traits. The plant architecture of newly discovered dwarfing germplasm "Huaai 11" consisted of desirable agronomic traits such as shortened stature and early maturity. Genetic factor controlling the plant height in dwarf line Huaai 11 was investigated.
Results
The Huaai 11 was crossed with tall varieties Monker, Mpyt, Zhenongda 3, Zaoshu 3, Advance, Huadamai 1, Huadamai 6, Hyproly and Ris01508. All the F1 plants displayed tall trait. Both tall and dwarf plants appeared in all the F2 populations with a 3:1 segregation ratio, suggesting that dwarfism of Huaai 11 is controlled by a single recessive gene, btwd1. Allelism test indicated that this dwarfing gene in the Huaai 11 is nonallelic with the gene br, uzu, sdw1 and denso. Using a double haploid population derived from a cross of Huadamai 6 and Huaai 11 and SSR markers the novel dwarfing gene was mapped onto the long arm of chromosome 7H, and closely linked to Bmac031 and Bmac167 with genetic distance of 2.2 cM.
Conclusion
Huaai 11 is a new source of dwarf for broadening the genetic base of dwarfism. This dwarf source was controlled by a recessive dwarfing gene btwd1, was mapped onto the long arm of chromosome 7H.
Background
Dwarfism is a valuable trait in crop breeding, because it increases lodging resistance and decreases damages due to wind and rain [1]. Successful use of a dwarfing gene is critical for developing dwarf cultivars [2]. In barley, more than 30 types of dwarfs or semidwarfs have been found including 'breviaristatum-ari', 'brachytic-br', 'curly dwarf-cud', 'denso dwarf-denso', 'erectoides-ert', 'lazy dwarf-lzd', 'many noded dwarf-mnd', 'narrow leaf dwarf-nld', 'semidwarf-sdw', 'single node dwarf-sid', 'slender dwarf-sld', 'uzu or semibrachytic-uzu' and 'vegetative dwarf-dwf' [2-5]. However, only a few of them have successfully been used in barley breeding program. The dwarfing gene uzu on the chromosome 3HL has widely been used in barley breeding in China, Japan and Korea peninsula [6-10]. The dwarfing gene sdw1 (named as sdw previously) and denso have been used in feed barley breeding in North America and Australia [11-13], and in barley breeding in European [2,13], respectively. The sdw1 and denso genes were allelic [14-17], and were mapped onto the same region of chromosome 3HL and tagged with a RFLP marker MWG 847 by 0.6 cM [12,18,19].
Some of the dwarfing genes have rarely or never been used in barley breeding programs, although have been genetically characterized. For example, the dwarfing gene GPert or ari-e was mapped on chromosome 5H [5,20,21]. The recessive GA3-insensitive dwarfing genes Rht-H1 and Dwf2 were mapped on the centromeric region of chromosome 2H [22], and on the short arm of 4H [16,23], respectively. Two recessive dwarfing mutants, gai and gal were mapped on the centromere region and on the long arm of the 2H, respectively [24].
In 1993, we found a dwarf individual in field of barley landrace 'Daofu Bai Qing Ke' in Daofu County, Sichuan. Plant height of 'Daofu Bai Qing Ke' is about 119 cm, while height of the dwarf individual is only about 40 cm. The dwarf individual is naked six-rowed barley, and its maturation date is about 4 days earlier than 'Daofu Bai Qing Ke'. Multi-year and -location experiments showed that the dwarfism is stable. The dwarf line was named as 'Huaai 11'.
Genetic factor controlling plant height of the barley dwarf line Huaai 11 was investigated in the present study. Here we reported the results of genetic analysis and molecular mapping of this novel dwarfism gene.
Methods
Plant materials and field experiments
Plant height of Huaai 11 and other four agronomic traits were evaluated for seven years at two locations (Yaan, Sichuan Province, and Wuhan, Hubei Province). The results showed that dwarfism is constantly expressed with height of 40.8 ± 1.84 cm (Table 1). Its maturity is about 7 days earlier than the control Zaoshu 3 (early maturity variety).
Table 1.
Evaluation of plant height and other agronomic traits of Huaai 11 in different years and locations (Yaan, Sichuan Province and Wuhan, Hubei Province)
| Year | Location | Height (cm) | Days from Seedling emerging to heading | No. of Spikelets | No. of spikes per plant | Weight per1000 grains |
|---|---|---|---|---|---|---|
| 1993-1994 | Yaan | 39.9 | 138 | 56 | 4.8 | 26.5 |
| 1994-1995 | Wuhan | 41.4 | 141 | 54 | 5.1 | 25.7 |
| 1996-1997 | Wuhan | 41.8 | 142 | 61 | 5.5 | 23.6 |
| 1997-1998 | Wuhan | 43.2 | 140 | 56 | 6.1 | 24.7 |
| 2007-2008 | Wuhan | 37.8 | 132 | 48 | 5.0 | 24.1 |
| 2008-2009 | Wuhan | 39.5 | 127 | 54 | 8.6 | 24.9 |
| 2009-2010 | Wuhan | 42.1 | 138 | 63 | 10.8 | 25.4 |
| Mean | 40.8 | 136.9 | 56 | 6.6 | 25.0 | |
| SD | 1.84 | 5.43 | 4.933 | 2.28 | 0.98 |
For genetic analysis, Huaai 11 was crossed with tall varieties Monker, Mpyt, Zhenongda 3, Zaoshu 3, and Advance. The 5 F2 populations, their F1 and parents were planted in field in 1998. Additional 4 tall varieties, Huadamai 1, Huadamai 6, Hyproly and Ris01508 were crossed with Huaai 11 in year 2000. The 9 F2 populations, their F1 and parents were planted in 2002 (Table 2).
Table 2.
Plant height in the F2 populations derived from the crosses between Huaai 11 and several tall cultivars
| Cross | year | No. of F1 plants examined | Plant height of F1 (cm) | No. of F2 plants | No. of F2 dwarf plants (< 60 cm) | No. of F2 tall plants (≥60 cm) | F2 χ2 (3:1) |
|---|---|---|---|---|---|---|---|
| Huaai 11/Monker | 1998 | 15 | 97 ± 1.13 | 215 | 52 | 163 | 0.59 |
| Huaai 11/Mpyt | 1998 | 17 | 95 ± 1.16 | 183 | 45 | 138 | 0.02 |
| Huaai 11/Zhenongda 3 | 1998 | 13 | 99 ± 1.21 | 257 | 75 | 182 | 2.40 |
| Huaai 11/Zaoshu 3 | 1998 | 15 | 102 ± 1.05 | 217 | 65 | 152 | 2.84 |
| Huaai 11/Advance | 1998 | 11 | 99 ± 1.09 | 211 | 57 | 154 | 0.46 |
| Huaai 11/Monker | 2002 | 13 | 98 ± 1.20 | 223 | 48 | 175 | 1.44 |
| Huaai 11/Mpyt | 2002 | 11 | 96 ± 1.09 | 161 | 45 | 116 | 0.75 |
| Huaai 11/Zhenongda 3 | 2002 | 12 | 99 ± 1.12 | 259 | 57 | 202 | 1.24 |
| Huaai 11/Zaoshu 3 | 2002 | 15 | 105 ± 1.23 | 215 | 50 | 175 | 0.35 |
| Huaai 11/Advance | 2002 | 15 | 101 ± 1.28 | 195 | 37 | 158 | 3.78 |
| Huaai 11/Huadamai 1 | 2002 | 12 | 98 ± 1.05 | 242 | 56 | 186 | 0.45 |
| Huaai 11/Huadamai 6 | 2002 | 12 | 87 ± 1.16 | 192 | 45 | 147 | 0.25 |
| Huaai 11/Hyproly | 2002 | 13 | 111 ± 1.10 | 196 | 41 | 155 | 1.74 |
| Huaai 11/Ris01508 | 2002 | 15 | 110 ± 1.30 | 205 | 55 | 150 | 0.36 |
χ21, 0.05 = 3.841
To test the allelic relationship of Huaai 11 with other dwarfing genes in barley, The F1 of Huaai 11 × Monker that is a popular commercial cultivar developed in USA was crossed with four dwarf varieties, Himalaya (br, 65.7 cm), Aiganqi (uzu,71.7 cm), India dwarf (sdw1, 61.3 cm) and Maris Mink (denso, 69.2 cm).
All abovementioned generations and their parents were grown on the Experimental Farm of Huazhong Agricultural University, Wuhan. A randomized complete block design with three replications was adopted. The materials were planted in five-row plots with a row length of 1.1 m.
To map the dwarfism gene in Huaai 11, a population consisting of 122 doubled-haploid (DH) lines was developed from a cross between a common feed barley cultivar Huadamai 6 and Huaai 11 using anther culture in this study. The DH population and parents were planted on the Experimental Farm of Huazhong Agricultural University, Wuhan, China. The field trials were conducted following a randomized complete block design with three replications in 2006, 2007 and 2008, respectively. Each of the DH and parental lines were grown in three rows in a plot of 0.6 × 1.5 m2.
Height of plants before ripening was measured in the field from soil surface to top of the main culm (with the spike). The height was calculated as the mean of the twelve plants (in three replicates, four plants from each replicate were measured).
Extraction of genomic DNA
The leaves from each doubled-haploid (DH) lines and parents were collected and frozen for DNA extraction. The cetyltrimethylammonium bromide (CTAB) method was used to extract genomic DNA from about 0.6-1.0 g of young leaf-tissue of each accession [25]. The quality of DNA was checked using 0.8% agrose gel electrophoresis, and the DNA concentration was measured using spectrophotometer. Twelve extremely tall lines and twelve short lines were selected from the DH population to construct two DNA pools.
SSR genotyping and mapping analysis
Three-hundred and six SSR markers distributed on all seven barley chromosomes were used to screen polymorphism between the two parental lines. The polymorphic SSR markers were further used to analyze the two DNA pools. The polymorphic markers between the two bulks were used to genotype the 122 individuals from the DH population. Genotyping data of 122 DH lines were given in Additional file 1(Table S1).
Polymerase chain reaction (PCR) was carried out in a final volume of 15 μL, containing 3 μL of the 20 ng/μL genomic DNA, 1.5 μL of 10× PCR buffer (with 15 mM Mg2+), 0.3 μL of 10 mM dNTP mixture, 2.0 μL of a 2.5 μM solution of the forward and reverse primers, and 0.6 units of Taq DNA polymerase (TakaRa Biotechnology, Dalian, China). DNA amplifications were performed in a thermocycler using the following touchdown PCR protocol: 1 cycle of 3 min at 94°C, followed by 15 cycles 94°C for 30 sec, 30 sec at 60°C (decreasing 1°C per cycle), 45 sec at 72°C. Another 25 cycles of 30 sec at 94°C, 30 sec at 50°C, 45 sec at 72°C. The reaction ended with a 5 min extension at 72°C. PCR product was separated on separated on 6% denaturing polyacrylamide gel and visualized using silver staining.
Linkage map was constructed using the software MAPMAKER [26] and the genetic distance (centimorgan, cM) was derived using Kosambi function [27].
Results
Inheritance of the dwarf gene in Huaai 11
Huaai 11 was discovered from the barley landrace Daofu Bai Qing Ke in 1993. Plant height of Huaai 11 is about 40 cm which is significant shorter than Huadamai 6 (85 cm) (Figure 1). Huaai 11 was crossed with nine tall varieties, Monker, Mpyt, Zhenongda 3, Zaoshu 3, Advance, Huadamai 1, Huadamai 6, Hyproly and Ris01508. All the F1 plants were tall. Distribution of plant height in all F2 populations are discontinuous, the distribution curves are dropped to valley at 60 cm, the plant equal or higher than 60 cm was classified as tall, while less than 60 cm as dwarf. Both the tall and dwarf plants appeared in all the F2 populations with a 3:1 segregation ratio for tall : dwarf (plant height ≥ 60 cm : plant height < 60 cm), suggesting that the dwarf trait in the Huaai 11 is controlled by single recessive gene. This gene is designated as btwd1.
Figure 1.
Photo of the dwarf Huaai 11 and cultivar Huadamai 6 plants.
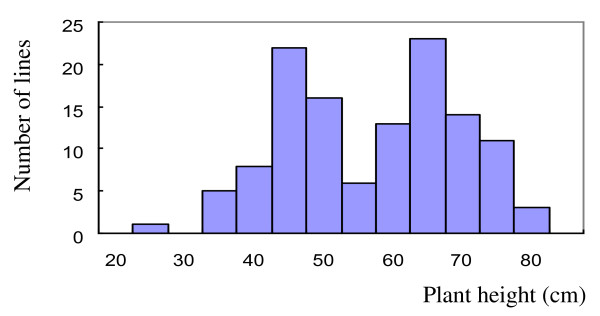
The DH population was constructed from the crosses between Huadamai 6 and Huaai 11 varieties. The averages of plant height of Huadamai 6 and Huaai 11 were 82.8 cm and 39.8 cm in three years, respectively. Figure 2 shows the distribution of plant height in the DH population in year 2007-2008. Plant height of the DH lines showed a bimodal distribution, ranging from 25 to 80 cm (Figure 2). The segregation ratio between tall and dwarf is 1:1.
Figure 2.
Frequency distribution of plant height of the Huadamai 6/Huaai 11 DH population in 2007-2008.
Allelic relationship between btwd1 and other dwarfing genes in barley
Plant height of Huaai 11 (40 cm) is much shorter than that of several other dwarf varieties such as Himalaya (br, 65.7 cm), Aiganqi (uzu, 71.7 cm), India dwarf (sdw1, 61.3 cm) and Maris Mink (denso, 69.2 cm). To test whether Huaai 11 shares the same allele with those four dwarf varieties, the F1 of Huaai 11 × Monker was crossed with those four varieties. All progeny in the four crosses are tall (Table 3), suggesting that the gene btwd1controlling plant height in the Huaai 11 is nonallelic with the genes in these four dwarf varieties.
Table 3.
Segregation of plant height in the test crosses between F1 of Huaai 11 × Monker and four dwarf varieties
| Cross | Generation | No. of total plants | No. of dwarf plants (< 60 cm) | No. of high plants (≥60 cm) | χ2 (1:1) |
|---|---|---|---|---|---|
| (Huaai11/Monker)/Huaai 11 | BC1 | 135 | 62 | 73 | 0.90 |
| (Huaai11/Monker)/Aiganqi | TC1 | 119 | 0 | 119 | 0 |
| (Huaai 11/Monker)/Himalaya | TC1 | 157 | 0 | 157 | 0 |
| (Huaai 11/Monker)/Indian dwarf | TC1 | 117 | 0 | 117 | 0 |
| (Huaai 11/Monker)/Maris Mink | TC1 | 143 | 0 | 143 | 0 |
χ21, 0.05 = 3.841
Molecular mapping of the dwarf gene btwd1 in Huaai 11
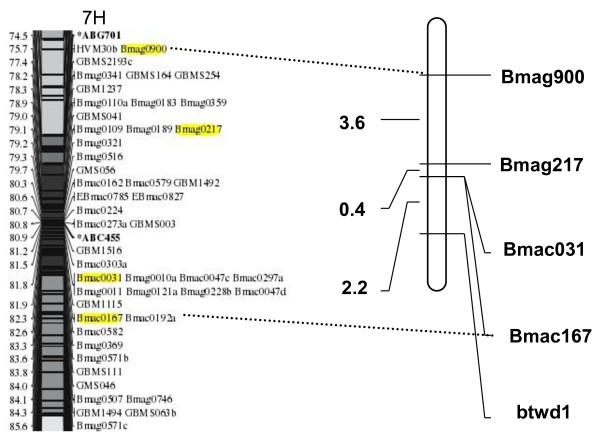
Out of the 306 SSR primers screened, ninety-six are polymorphic between parental lines Huadamai 6 and Huaai 11. The polymorphic SSR markers were used to test the two DNA pools for polymorphism. Four SSR markers, Bmac167, Bmac031, Bmag217 and Bmag900 located on chromosome 7H were polymorphic between the two DNA pools. Polymorphism was further confirmed using the individuals that were used to construct the two DNA pools, suggesting that the gene btwd1 controlling plant height of Huaai 11 was located on chromosome 7H. The four polymorphic markers were used to analyze the 122 individuals of DH population. Linkage analysis between SSR markers and plant height found that the dwarfing gene was located on the long arm of chromosome 7H, associated with the marker Bmac167 and Bmac031 at a genetic distance of 2.2 cM (Figure 3).
Figure 3.
Partial linkage map of chromosome 7H with the dwarfing gene btwd1. The chromosome 7H map was cited from Varshney et al. [35] for comparison
Discussion
Utilization of dwarfing genes in barley breeding programs has greatly increased barley yields, particularly in Asia and Europe [28]. In barley, more than 30 types of dwarfs or semidwarfs have been reported. However, because pleiotropic effects of dwarf genes cause some other undesired agronomic traits such as reduced weight and yield [29,30], a few have been exploited in barley breeding. The plant architecture of newly discovered dwarf germplasm Huaai 11 consisted of desirable agronomic traits such as shortened stature and early maturity. It is a new source of dwarfs for broadening the genetic base of dwarfism. The segregation ratio (3:1) of tall : dwarf in the all crosses in two years suggested that dwarfism in Huaai 11 is controlled by a single recessive gene, btwd1.
The allelism test of Huaai 11 with four dwarf varieties, Himalaya (br), Aiganqi (uzu), India dwarf (sdw1) and Maris Mink (denso) showed that all progeny derived from test crosses between the F1 of Huaai 11 × Monker and those four varieties are tall (Table 3). Since the dwarfism of Huaai 11 is controlled by a Mendelian recessive allele, the F1 of Huaai 11 × Monker was tall as the tall parent. If the Huaai 11 is allelic with one of the four dwarf varieties tested here, the progeny will be expected to have tall : dwarf ratio of 1:1. No dwarf plants from the four crosses were observed, suggesting that the gene btwd1 controlling plant height in the Huaai 11 is nonallelic with the gene br, uzu, sdw1 and denso.
In barley, most of the dwarf and semi-dwarf genes were mapped on the chromosome 2H, 3H and 4H. Tsuchiya reported that two loci, the br1 locus on the short arm of chromosome 7 (7H) and br2 on the short arm of chromosome 4 (4H) [31,32]. Recently, Yu et al. mapped a semi-dwarfing QTL PH-7 in the ZAU7 on chromosome 7HL [28]. In our study, the dwarfing gene btwd1 was mapped on the long arm of chromosome 7H. The chromosome location of btwd1 is different from the uzu (3HL) [9,33,34], sdw1/denso (3HL) [18,19], which further demonstrated that the btwd1 in Huaai 11 is nonallelic with br2, uzu sdw1/denso genes. The br1 was mapped on 7HS, while the btwd1 was mapped on 7HL, indicating that they belong to different locus, and are non-allelic. The QTL PH-7 was located between the markers bPb4541 and bPb3107 on chromosome 7HL [28]. Examining the 7H linkage maps of Varshney et al. [35] and Yu et al. [28] revealed that QTL PH-7 is close to the distal of the 7HL, far away from centromere, while the btwd1 in Huaai 11 is linked with Bmac167 that is close to the centromere of 7H, suggesting that the btwd1 in Huaai 11 is different from QTL PH-7.
Most barley cultivars developed in China are dwarfs and semi-dwarfs since 1950 [2]. The study on origin of dwarfing genes used in barley breeding in China indicated that 68.4% of dwarf and semi-dwarf barley cultivars were derivatives of Chibadamai, Xiaoshanlixiahuang, Cangzhouluodamai, Aiganqi, Zhepi 1 and Yanfu Aizao 3 [2]. Allelism test indicated that Chibadamai, Xiaoshanlixiahuang, Cangzhouluodamai, and Aiganqi carried the same dwarfing gene uzu on 3HL [9,33,34]. Zhepi 1 and Yanfu Aizao 3 likely are mutations at the sdw1 locus [2]. The dwarfing gene btwd1 in Huaai 11 is non-allelic with the uzu and sdw1 which have widely been used in China, and provides barley breeders with a new gene in China. Quite often, when breeders use the dwarfing gene to reduce plant height, due to the confounded effects of other loci controlling plant height and environmental effects, not all plant carrying the dwarfing gene can be easily identified [28]. Thus, the linked SSR markers identified in the present study can provide a useful marker-assisted selection tool to transfer the dwarfing gene btwd1 into elite barley germplasm.
Conclusions
Our study indicated that the dwarfing gene btwd1 in Huaai 11 is non-allelic with the uzu and sdw1 which have widely been used in China, and provides barley breeders with a new gene in China. Linkage analysis located the gene btwd1 gene (btwd1) onto the long arm of chromosome 7H, and it is closely linked to Bmac031 and Bmac167 with genetic distance of 2.2 cM.
Authors' contributions
XFR is the major executive person in this study, including phenotyping of plant height, marker genotyping and statistical analysis. WWG assisted in phenotyping and marker genotyping. DFS and GLS conceived the design of this study, coordinated the experiments, and wrote the manuscript. CDL produced the Huadamai 6 and Huaai 11 DH population using the anther-culture method. All authors have read and approved the final manuscript.
Supplementary Material
Table S1. Genotyping data of 122 DH lines (*A: Plant height < 60 cm; B: Plant height ≥60 cm in btwd1).
Contributor Information
Xifeng Ren, Email: renxifeng1979@yahoo.com.cn.
Dongfa Sun, Email: sundongfa1@mail.hzau.edu.cn.
Weiwei Guan, Email: guan-910@163.com.
Genlou Sun, Email: Genlou.sun@smu.ca.
Chengdao Li, Email: chengdao.li@agric.wa.gov.au.
Acknowledgements
This project is supported in part by the National Natural Science Foundation of China (30630047) and China 863 project (2006AA10Z1C3).
References
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol. 2004;54:533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang WX. Tracing sources of dwarfing genes in barley breeding in China. Euphytica. 2003;131:285–292. doi: 10.1023/A:1023955131523. [DOI] [Google Scholar]
- Sears RG, Kronstad WE, Metzger RJ. Inheritance of dwarf and semidwarf plant height in barley. Crop Sci. 1981;21:828–833. doi: 10.2135/cropsci1981.0011183X002100060008x. [DOI] [Google Scholar]
- Franckowiak JD. Coordinator's report on the semi-dwarf barley collection. Barley Genet Newslett. 1987;17:114–115. [Google Scholar]
- Franckowiak JD, Pecio A. Coordinator's report: Semi-dwarf gene: A listing of genetic stocks. Barley Genet Newslett. 1992;21:116–126. [Google Scholar]
- Hoskins PH, Poehlman JM. Pleiotropic effects of uzu and spike-density genes in a barley cross. J Hered. 1971;62:153–156. [Google Scholar]
- Tsuchiya T. Allelism testing in barley. II. Allelic relationships of three uzu genes. Crop Sci. 1976;16:496–499. doi: 10.2135/cropsci1976.0011183X001600040013x. [DOI] [Google Scholar]
- Zhang J. Changes in plant height of varieties and analysis of dwarf sources with progress in barley breeding in China. Barley Sci. 1994;4:11–13. [Google Scholar]
- Zhang J. Inheritance of agronomic traits from the Chinese barley dwarfing gene donors Xiaoshan Lixiahuang and Cangzhou Luodamai. Plant Breed. 2000;119:523–524. doi: 10.1046/j.1439-0523.2000.00543.x. [DOI] [Google Scholar]
- Saisho D, Tanno K, Chono M, Honda I, Kitano H, Takeda K. Spontaneous brassinolide-insensitive barley mutants 'uzu' adapted to east Asia. Breeding Sci. 2004;54:409–416. doi: 10.1270/jsbbs.54.409. [DOI] [Google Scholar]
- Gymer PT. An attempt to locate the sdw gene for prostrate growth habit or the perils and pitfalls of classical genetics. Barley Genet Newslett. 1992;22:19–22. [Google Scholar]
- Hellewell KB, Rasmusson DC, Meagher MG. Enhancing yield of semidwarf barley. Crop Sci. 2000;40:352–358. doi: 10.2135/cropsci2000.402352x. [DOI] [Google Scholar]
- Jia QJ, Zhang JJ, Westcott S, Zhang XQ, Bellgard M, Lance R, Li CD. GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct Integrat Genomics. 2009;9:255–262. doi: 10.1007/s10142-009-0120-4. [DOI] [PubMed] [Google Scholar]
- Rasmusson DC. A plant breeder's experience with ideotype breeding. Field Crops Res. 1991;26:191–200. doi: 10.1016/0378-4290(91)90035-T. [DOI] [Google Scholar]
- Mickelson HR, Rasmusson DC. Genes for short stature in barley. Crop Sci. 1994;34:1180–1183. doi: 10.2135/cropsci1994.0011183X003400050007x. [DOI] [Google Scholar]
- Ivandic V, Malyshev S, Korzun V, Graner A, Borner A. Comparative mapping of a gibberellic acid-insensitive dwarfing gene (Dwf2) on chromosome 4HS in barley. Theor Appl Genet. 1999;98:728–731. doi: 10.1007/s001220051127. [DOI] [Google Scholar]
- Zhang J, Li Z, Zhang CH. Analysis on the dwarfing genes in Zhepi 1 and Aizao 3: two dwarfing gene donors in barley breeding in China. Agric Sci China. 2006;5:643–647. [Google Scholar]
- Barua UM, Chalmers KJ, Thomas WTB, Hackett CA, Lea V, Jack P, Forester BP, Waugh R, Powell W. Molecular mapping of genes determining height, time to heading, and growth habit in barley (Hordeum vulgare) Genome. 1993;36:1080–1087. doi: 10.1139/g93-143. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Romero C, Simpson E, Snape JW. Assignment of the denso dwarfing gene to the long arm of chromosome 3(3H) of barley by use of RFLP markers. Plant Breed. 1993;111:198–203. doi: 10.1111/j.1439-0523.1993.tb00630.x. [DOI] [Google Scholar]
- Thomas WTB, Powell W, Wood W. The chromosomal location of the dwarfing gene present in the spring barley variety Golden Promise. Heredity. 1984;53:177–183. doi: 10.1038/hdy.1984.73. [DOI] [Google Scholar]
- Pakniyat H, Handley LL, Thomas WTB, Connolly T, Macaulay M, Caligari PDS, Forster BP. Comparison of shoot dry weight, Na+ content and δ13 C values of ari-e and other semi-dwarf barley mutants under salt-stress. Euphytica. 1997;94:7–14. doi: 10.1023/A:1002956802041. [DOI] [Google Scholar]
- Borner A, Korzun V. Genetical studies of two barley mutants differing in their GA response. Barley Genet Newslett. 1996;25:27–29. [Google Scholar]
- Falk DE. New dominant dwarfing gene (Dwf2) in barley. Barley Genet Newslett. 1994;24:87–89. [Google Scholar]
- Borner A, Korzun V, Malyshev S, Ivandic V, Graner A. Molecular mapping of two dwarfing genes differing in their GA response on chromosome 2H of barley. Theor Appl Genet. 1999;99:670–675. doi: 10.1007/s001220051283. [DOI] [PubMed] [Google Scholar]
- Stein N, Herren G, Keller B. A new DNA extraction method for high-throughput marker analysis in a large-genome species such as Triticum aestivum. Plant Breed. 2001;120:354–356. doi: 10.1046/j.1439-0523.2001.00615.x. [DOI] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Liu RH, Meng JL. MapDraw: A microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003;25:317–321. [PubMed] [Google Scholar]
- Yu GT, Horsley RD, Zhang BX, Franckowiak JD. A new semi-dwarfing gene identified by molecular mapping of quantitative trait loci in barley. Theor Appl Genet. 2010;120:853–861. doi: 10.1007/s00122-009-1216-x. [DOI] [PubMed] [Google Scholar]
- Foster AE, Thompson AP. In: 5th Proceedings of international barley genetics symposium. Yasuda S, Kanishi T, editor. Sanyo Press Co., Okayama; 1987. Effects of a semidwarfing gene from Jotun on agronomic and quality traits of barley; pp. 979–982. [Google Scholar]
- Rasmusson DC, Phillips RL. Plant breeding progress and genetic diversity from de novo variation and elevated epistasis. Crop Sci. 1997;37:303–310. doi: 10.2135/cropsci1997.0011183X003700020001x. [DOI] [Google Scholar]
- Tsuchiya T. Allelism testing of genes between brachytic and eractoides mutants. Barley Genet Newslett. 1976;6:79–81. [Google Scholar]
- Li M, Pan Y, Li AS, Kudrna D, Kleinhofs A. Fine mapping of a semi-dwarf gene brachytic 1 in barley. Acta Genet Sin. 2002;29:565–570. [PubMed] [Google Scholar]
- Zhang J. Allelism tests for the dwarf genes in the three main dwarf sources of barley. Acta Agronom Sin. 1998;24:42–46. [Google Scholar]
- Zhang J. SSR maker tagging of the dwarfing gene uz in barley (Hordeum velgare L.) Acta Agronom Sin. 2003;29:637–640. [Google Scholar]
- Varshney RK, Marcel TC, Ramsay L, Russell J, Roder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A. A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet. 2007;114:1090–1103. doi: 10.1007/s00122-007-0503-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genotyping data of 122 DH lines (*A: Plant height < 60 cm; B: Plant height ≥60 cm in btwd1).