Abstract
Development of a functional neuronal network during embryogenesis begins with pioneer axons creating a scaffold along which later-outgrowing axons extend. The molecular mechanism used by these follower axons to navigate along pre-existing axons remains poorly understood. We isolated loss-of-function alleles of fmi-1, which caused strong axon navigation defects of pioneer and follower axons in the ventral nerve cord (VNC) of C. elegans. Notably follower axons, which exclusively depend on pioneer axons for correct navigation, frequently separated from the pioneer. fmi-1 is the sole C. elegans ortholog of Drosophila flamingo and vertebrate Celsr genes, and this phenotype defines a new role for this important molecule in follower axon navigation. FMI-1 has a unique and strikingly conserved structure with cadherin and C-terminal G-protein coupled receptor domains and could mediate cell-cell adhesion and signaling functions. We found that follower axon navigation depended on the extracellular but not on the intracellular domain, suggesting that FMI-1 mediates primarily adhesion between pioneer and follower axons. By contrast, pioneer axon navigation required the intracellular domain, suggesting that FMI-1 acts as receptor transducing a signal in this case. Our findings indicate that FMI-1 is a cell-type dependent axon guidance factor with different domain requirements for its different functions in pioneers and followers.
Keywords: Axon guidance, flamingo, C. elegans, Pioneer, Follower, Nervous system, Cadherin, Adhesion, GPCR
INTRODUCTION
The creation of a functional neuronal network during embryogenesis is a complex process. Pioneer axons initially generate a scaffold, along which later-outgrowing follower axons extend (R. M. Durbin, PhD Thesis, University of Cambridge, 1987; Kuwada, 1986; Mastick and Easter, 1996; Raper et al., 1983; Ross et al., 1992). In the grasshopper, the Ti axon functions as a pioneer in the developing limb. Ablation of Ti leads to failure of SGO follower axons to extend (Klose and Bentley, 1989). Deletion of four pioneer axons in the ventral nerve cord (VNC) of Drosophila causes severe defects in the establishment of longitudinal axon tracks (Hidalgo and Brand, 1997). In the mouse, cerebral cortex short-lived subplate neuron axons are the first to invade the thalamus, which guide later outgrowing cortical axons to their correct target (McConnell et al., 1989). However, follower axons do not always strictly depend on the presence of pioneer axons for correct axon navigation. In zebrafish, prevention of TPOC axon outgrowth leads to failure in later nucPC axon guidance, but in more than half of the lesioned embryos, nucPC axon navigation is unaffected (Chitnis and Kuwada, 1991). In some cases, pioneer axons are even dispensable for follower axon guidance (Cornel and Holt, 1992; Eisen et al., 1989; Keshishian and Bentley, 1983).
In C. elegans the VNC is the main nerve bundle running in the anterior-posterior direction. It consists of two axon tracks with the majority of axons running in the right bundle. VNC pioneer axons have been identified using electron microscopic reconstructions of staged embryos (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). The right axon tract is pioneered by the AVG axon (Fig. 1A). Elimination of this pioneer leads to a disorganization of the axon bundle and axons erroneously extending in the left axon track or crossing back and forth between right and left axon bundles (R. M. Durbin, PhD Thesis, University of Cambridge, 1987) (Hutter, 2003). The left axon tract is pioneered by the PVPR axon closely followed by the PVQL axon (Fig. 1A). In the absence of the PVPR axon, the PVQL axon joins the right VNC track, suggesting that the PVQL axon alone is unable to pioneer the left axon track (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). Furthermore, defects in PVPR pioneer axon navigation lead to identical PVQL follower navigation defects, suggesting a strong link between pioneer and follower axons (Hutter, 2003; Hutter et al., 2005). Similarly HSN axons, which extend postembryonically, rely on the presence of either the PVP or PVQ axons for correct navigation (Garriga et al., 1993). The molecular basis of pioneer-mediated navigation of follower axons is not well understood, but presumably requires specific adhesion between pioneer and follower axons.
Fig. 1.
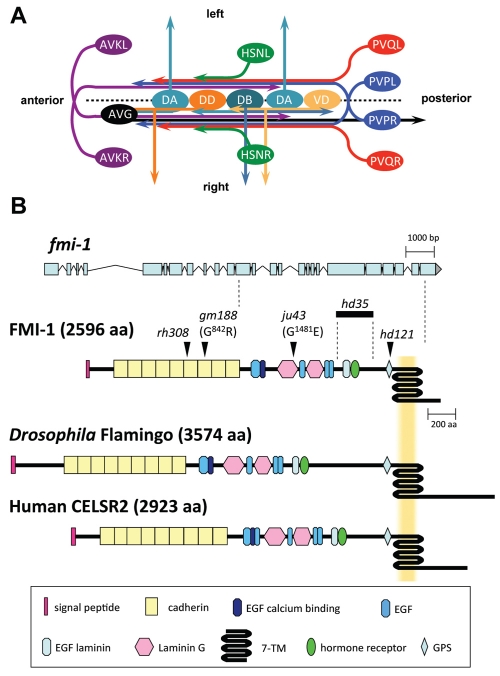
Development of the ventral nerve cord (VNC) in C. elegans, and gene model and protein domain organization of FMI-1. (A) The C. elegans VNC consists of two axon tracks with motoneuron cell bodies (light and dark orange, light and dark turquoise) lining the ventral midline (broken line). During embryogenesis, the AVG axon (black) pioneers the right VNC axon track. PVP axons (blue) decussate and extend on the contralateral side of the VNC. The PVPR axon pioneers the left VNC axon track. Both PVP axons are closely followed by PVQ axons (red). DA, DB and DD motoneurons (light and dark turquoise, dark orange, respectively) extend axons into the right axon track and send commissures circumferentially to the DNC. Later, AVK axons (purple) enter the VNC from the anterior. Postembryonically, VD motoneurons (light orange) differentiate and send processes into the VNC and towards the DNC. HSN axons (green) join the VNC at the vulva region. Command interneuron axons, which extend in the right axon track, are omitted for clarity. Ventral view. (B) The fmi-1 gene. The top part illustrates the exon-intron structure of fmi-1. The positions of alleles used in this study are mapped onto the protein structure (arrowheads, bold line). The protein domain organization of Drosophila Flamingo and human CELSR2 are shown for comparison.
The cadherin superfamily constitutes one of the largest families of cell-adhesion molecules (CAMs). Cadherins extensively mediate neuronal interactions (Takeichi, 2007) and are also implicated in selective axon-axon fasciculation (Treubert-Zimmermann et al., 2002; Wohrn et al., 1999). Cadherin domains mediate mainly homophilic cell-cell adhesion (Patel et al., 2003). `Classical' cadherins are characterized by a conserved catenin-binding side in their intracellular tail that provides a dynamic link to the cytoskeleton (Drees et al., 2005; Yamada et al., 2005). In C. elegans the sole `classical' cadherin, hmr-1, has one neuronal splice variant and is implicated in motoneuron commissure guidance (Broadbent and Pettitt, 2002). Eleven more C. elegans proteins containing cadherin modules belong to various subgroups of `non-classical' cadherins with several evolutionary conserved members, including a Flamingo ortholog, FMI-1 (Pettitt, 2005). Flamingo/Starry Night (FMI, STAN – FlyBase) was first described in Drosophila as a component of the planar cell polarity pathway (PCP), where it functions together with Frizzled (FZ), Disheveled, Diego and Van Gogh/Strabismus (VANG) (Chae et al., 1999; Chen et al., 2008; Das et al., 2002; Lu et al., 1999; Usui et al., 1999). In addition, Drosophila fmi mutants show target selection defects of photoreceptor axons (Chen and Clandinin, 2008; Lee et al., 2003), defects in dendrite development and maintenance (Gao et al., 1999; Gao et al., 2000; Kimura et al., 2006; Reuter et al., 2003), and defects in synapse formation (Bao et al., 2007), suggesting various functions for fmi during nervous system development. Vertebrate genomes contain up to three orthologous genes (Celsr1, Celsr2 and Celsr3) with distinct functions (Shima et al., 2007; Shima et al., 2004; Tissir et al., 2005; Zhou et al., 2008). How the various Flamingo/CELSR functions are mediated is poorly understood and seems to differ even within the same cell (Lee et al., 2003).
In early VNC development, pioneer axons establish axon tracks along which follower axons extend. Here, we describe the role of C. elegans FMI-1 in pioneer and pioneer-mediated axon navigation. Mutations in fmi-1 cause strong axon navigation defects of VNC pioneer axons. In addition, follower axons, which exclusively depend on pioneer axons for correct navigation, frequently separated from the pioneer, defining a new role for this class of molecules. FMI-1 was expressed in neurons and localized to axons during and after nervous system development. FMI-1 probably functions as receptor in pioneer axon navigation. It has several functions in follower axon navigation, including a role in axon-axon adhesion between pioneer and follower axons.
MATERIALS AND METHODS
Nematode strains used and phenotypic description
The C. elegans wild-type strain CB4856 was used for SNP mapping of fmi-1(rh308). pha-1(e2123ts) III was used as selection marker for transgenic animals (Granato et al., 1994).
The following integrated GFP reporter constructs were used to characterize mutant phenotypes and to identify neurons expressing fmi-1::GFP reporter constructs: hdIs26[odr-2::CFP, sra-6::DsRed2] III; rhIs4[glr-1::GFP, dpy-20(+)] III; zdIs13[tph-1::GFP] IV; hdIs29[odr-2::CFP, sra-6::DsRed2] V; bwIs2[flp-1::GFP, rol-6(su1006)]; hdIs25[unc-129::CFP, unc-47::DsRed2]; and hdIs30[glr-1::DsRed2].
Other alleles used for phenotypic descriptions were: lin-17(n671) I; hmr-1b(hd37) I; mig-1(e1787) I; casy-1(tm718) II; cdh-1(hd44) III; prkl-1(ok3182) IV; cfz-2(ok1201) V; and vang-1(tm1422) X.
Strains were cultured at 20°C under standard conditions (Brenner, 1974).
Axonal defects in the VNC are defined here as axons crossing the ventral midline into the contralateral axon track, extending completely in the contralateral axon track or leaving the VNC. Defasciculation within the same axon track were not included.
Mapping and gene identification
In an EMS screen for axon guidance defects in command interneurons of C. elegans, we isolated the allele fmi-1(rh308) (Hutter et al., 2005). We mapped the allele to a 1.8 Mb region on chromosome V between the SNPs R11D1[1] and pKP5086. Injection of Fosmids (Geneservice, Cambridge, UK) narrowed the region to a 150 kb region on the right arm of chromsome V, which contained fmi-1 as candidate gene. Injection of the fmi-1-containing Fosmid WRM065dE09 into fmi-1(rh308) animals rescued PVQ axon guidance defects in three out of eight lines. Sequencing of the fmi-1-coding region in rh308 animals revealed an early nonsense mutation (Q725Ochre; V:13024281 C/T) in the sixth cadherin domain. In genetic screens, we also isolated the late nonsense allele hd121 (W2184Amber; V:13031487 G/A) and the missense mutation gm188 (G842R; V:13025328 G/A). The out-of-frame deletion hd35, starting in exon 20 and ending in exon 22 (V:13030417/13030418-13031319/13031320; context of deletion: GAAAAGCCATTTCTCAACTA-902 bp-AGTTGTTTATTCAATTGTAC), was isolated from our deletion library. The missense allele ju43 (G1481E; V:13029284 G/A) was a generous gift from Mei Zhen and Yishi Jin.
Cloning, expression analysis and domain analysis
fmi-1p::GFP was generated by PCR-based fusion (Hobert, 2002) of 2615 bp of upstream fmi-1 promoter region to GFP. fmi-1pintron was generated by PCR amplification of the fmi-1 promoter and part of the fmi-1 transcript (up to the 5. exon) and cloning into the BamHI site of pPD122.39 containing the pat-3 trans-membrane domain and GFP. To generate the translational reporter construct FMI-1::GFP, the GFP was inserted at the C terminus of the FMI-1 transcript in a genomic clone containing the entire upstream and coding part of FMI-1. The various deletion constructs were generated with FMI-1::GFP as starting point. Details of the cloning can be made available upon request.
For all constructs, transgenic animals were generated as described (Mello et al., 1991). The extra-chromosomal array containing fmi-1p::GFP was integrated and resulting strains were outcrossed three times. For each construct, at least two independent strains were imaged. GFP-positive cells were identified by location and cell morphology in comparison with reference images from Wormatlas (http://www.wormatlas.org/) and with the help of marker strains with known expression patterns.
Microscopy
Confocal images of fluorescent protein containing mixed stage worms were made using a Zeiss Axioplan II microscope (Carl-Zeiss AG, Germany). Stacks of confocal images with 0.3 to 0.5 μm distance between focal planes were recorded with a Quorum WaveFX spinning disc system (Quorum Technologies, Canada). Image acquisition and analysis was carried out using the Volocity software package (Perkin-Elmer, Waltham, MA). Maximum intensity projections of all focal planes were used to generate images for the figures. Figures and GFP/Nomarski overlays were assembled with Adobe Photoshop CS 8.0 (Adobe, San Jose, CA, USA).
Mosaic analysis and targeted expression
For the mosaic analysis, transgenic animals were created by injecting Fosmid WRM065dE09, sur-5::GFP (Yochem et al., 1998) and pha-1(+) into pha-1(e2123ts) hdIs26 III; fmi-1(rh308) V; zdIs13 animals. Animals from one rescuing line were first analyzed for PVP and PVQ axon guidance defects. Then presence of the rescuing extra-chromosomal array in PVP and PVQ neurons was determined via the presence of SUR-5::GFP, which localizes to the nucleus.
To express FMI-1 in specific neurons, the fmi-1 cDNA was cloned into vectors containing the promoters of fmi-1, odr-2 (Chou et al., 1996), sra-6 (Troemel et al., 1995) and tph-1 (Sze et al., 2000). A detailed description of the cloning is available upon request. These constructs were injected alone and in combination into pha-1(e2123ts) hdIs26 III; fmi-1(rh308) V; zdIs13 animals. Axon guidance defects of eight to ten independent strains were evaluated.
RESULTS
Follower axon navigation in fmi-1 mutant animals is severely disrupted
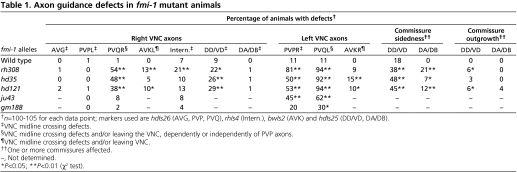
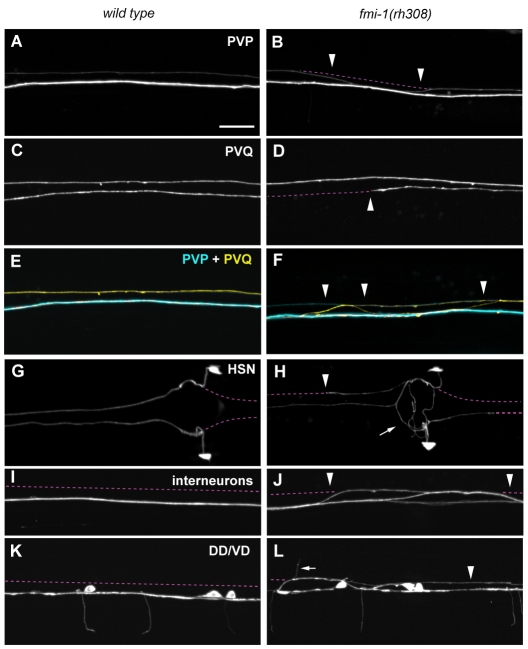
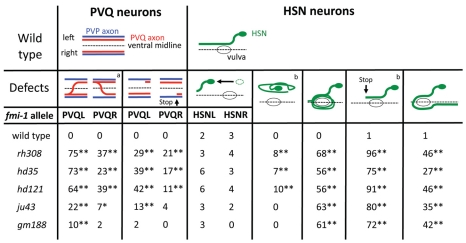
The molecular basis for pioneer-mediated axon navigation is largely unclear. We performed genetic screens to identify novel molecules involved in follower axon navigation in the C. elegans VNC. In these screens, we isolated alleles of the Flamingo-ortholog FMI-1. fmi-1(rh308) animals showed specific axonal defects in VNC neurons. In 81% of fmi-1(rh308) animals, the PVPR axon, which pioneers the left ventral cord axon tract (Fig. 1A), irregularly crossed the ventral midline and joined the right axon track (Table 1, Fig. 2B). The pioneer axon of the right axon tract, AVG, was not affected. Notably, follower axons exhibited strong axon navigation defects in fmi-1(rh308) animals. PVQ axons crossed the ventral midline, left the ventral cord and extended posteriorly (Table 1, Fig. 2F) – frequently in the absence of corresponding pioneer defects (Fig. 2F, Fig. 3). In many fmi-1-deficient animals PVQ axons crossed the ventral midline multiple times, typically combining pioneer-depended and pioneer-independent crossings. When midline crossing events were counted individually, 58% (n=223) of PVQL and 83% (n=82) of PVQR midline crossing defects occurred independently of PVPR axon crossings, indicating that the tight pioneer-follower relationship between PVP and PVQ axons was severely disrupted. In addition, follower axons displayed outgrowth defects. PVQ axons frequently stopped prematurely and did not reach the nerve ring (Fig. 2D, Fig. 3). Both PVQ axons in the left and right axon tract were affected, with the PVQL axon exhibiting more penetrant defects (Fig. 3).
Table 1.
Axon guidance defects in fmi-1 mutant animals
Fig. 2.
Axonal defects in fmi-1(rh308) animals. Axon guidance of VNC axons in wild-type (A,C,E,G,I,K) and fmi-1(rh308) (B,D,F,H,J,L) animals. (E,F) Overlays of AVG, PVP and PVQ axons. (A) Wild type; (B) PVPR axon crossed into contralateral axon track (arrowheads). (C) Wild type; (D) PVQR axon stopped prematurely (arrowhead). (E) Wild type; (F) PVQL axon (yellow) crossed into contralateral axon track independently of the PVPR axon (cyan) (arrowheads) and extended posteriorly. (G) Wild type; (H) HSN axons circled the vulva (arrow), extended posteriorly along the VNC and stopped prematurely (arrowhead). (I) Wild type; (J) interneurons crossed into the contralateral axon track (arrowheads). (K) Wild type; (L) DD/VD processes extended along the left axon track (arrowhead) and commissures projected along the left side of the animal into the dorsal cord (arrow). The broken line (purple) indicates the position of the left and right VNC axon tracks. Ventral views, anterior towards the left. Markers used: hdIs26 (PVP/PVQ), zdIs13 (HSN), rhIs4 (Interneurons) and hdIs25 (DD/VD). Scale bar: 10 μm.
Fig. 3.
Defects of PVQ and HSN follower axons are independent of PVP pioneer axon navigation. aPVQ axons independently crossing the ventral midline and/or leaving the VNC. bPercentage of axons with n=200-220, remaining values are percentage of animals with n=100-110; combined HSNL/R axonal defects are shown; *P<0.05, **P<0.01 (χ2 test) compared with wild type; all graphics are ventral views, anterior towards the left. Marker used are hdIs26 (PVP/PVQ) and zdIs13 (HSN).
The HSN neurons, a second pair of followers, are born in the posterior of the animal and migrate during embryogenesis to lateral positions in the midbody region. Post-embryonically, HSN neurons project axons ventrally towards the VNC. In the VNC, the axons turn anteriorly and extend into the nerve ring (Fig. 1A, Fig. 2G). In fmi-1 mutant animals, migration of HSN neurons was generally unaffected (Fig. 3). We observed only mild defects in HSN axon projections towards the VNC. HSN axon guidance along the VNC axon tracks, however, was severely disrupted. HSN axons appeared to circle the vulva in the majority of the animals (Fig. 2H, Fig. 3). This might have been due to ectopic branching of the HSN axons and/or axons growing in a circle. HSN axons also often projected to the posterior instead of the anterior after leaving the vulva region. We observed HSN axons frequently crossing the ventral midline outside the vulva region. HSN axons that extended along the VNC often stopped before they reached the nerve ring (Fig. 2H, Fig. 3).
AVK axons, which extend from the anterior nerve ring into the VNC (Fig. 1A), showed mild defects in all fmi-1 alleles tested (Table 1). Motoneuron cell bodies line the ventral midline of the VNC (Fig. 1A). These neurons extend axons along the right axon track and show a highly stereotypic commissure development. An invariable number of commissures navigate circumferentially towards the dorsal cord in a distinct left-right pattern (Fig. 1A, Fig. 2K). In fmi-1 mutant animals, GABAergic DD/VD motoneurons erroneously sent axons into the left VNC axon track and the invariant left-right choice of commissures of the DD/VD and the cholinergic DA/DB motoneurons was partially disrupted (Table 1, Fig. 2L). We also observed mild midline crossing defects of command interneuron axons, which normally extend in the right VNC track (Table 1, Fig. 2J). Taken together, our results suggest a crucial role for FMI-1 in pioneer axon navigation, as well as pioneer-mediated follower navigation.
FMI-1 is a highly conserved non-classical cadherin and adhesion GPCR
Flamingo orthologs are cadherin-like proteins with a unique domain composition. In addition to nine cadherin domains at the N terminus, the extracellular domain of FMI-1 contains six EGF and two laminin G domains, a configuration reminiscent of type-III classical cadherins (Fig. 1B). The GPS cleavage site and seven-pass transmembrane domain (7TM), however, resemble the C-terminal part of an adhesion G-protein coupled receptor (adhesion GPCR). The short intracellular domain does not contain any recognizable known domain. The domain organization is strikingly conserved among Flamingo-like proteins in different species (Fig. 1B). The Drosophila genome contains one homolog, flamingo/starry night, whereas vertebrate genomes have up to three homologs: Celsr1, Celsr2 and Celsr3. The extracellular domains show a high degree of similarity between species, whereas the intracellular domains vary in length and are generally not conserved. FMI-1 has characteristics of a non-classical cadherin as well as an adhesion GPCR and could function as cell-surface receptor and cell-cell adhesion molecule.
Axon guidance defects were most penetrant in animals containing the earliest nonsense mutation rh308 located in the sixth cadherin domain (Fig. 1B). An antibody directed to the intracellular domain of FMI-1 showed no staining in fmi-1(rh308) animals (E.H.N., L.W., M. Zhen, E. Pinedo Carpio, A. Goncharov, G.G., Y. Jin and B.D.A., unpublished), suggesting that rh308 is a null allele. hd121 is a late nonsense mutation in the GPS domain (Fig. 1B) and hd35 a small deletion leading to a frame-shift and premature stop codon before the GPS domain. All these alleles, including rh308, would lead to the production of a truncated unanchored protein fragment. Both alleles showed axonal defects similar to fmi-1(rh308) animals (Table 1; Fig. 3). Interestingly the missense allele ju43, a mutation within the laminin G domain, caused PVP navigation defects similar to the alleles hd121 and hd35, but did not cause penetrant PVQ follower defects. However, the gm188 allele, which carries a missense mutation in the seventh cadherin domain, had no effect on PVP navigation. Interestingly, HSN axonal defects were highly penetrant in all alleles tested, suggesting that HSN axons are more susceptible to changes in the fmi-1 protein (Fig. 3).
Pioneer and follower axons express fmi-1
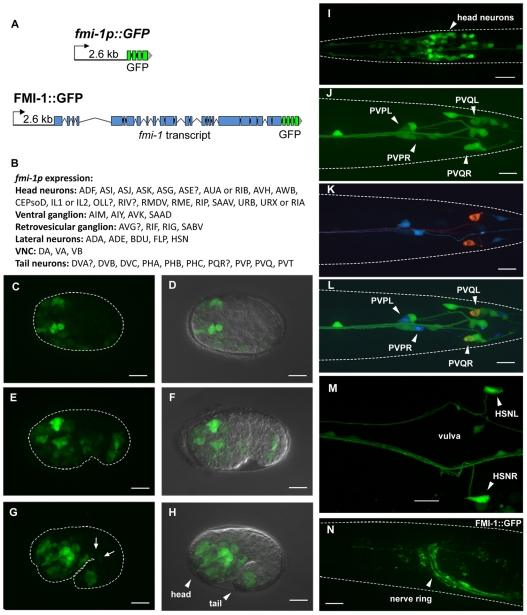
We used transgenic animals expressing GFP reporter constructs to determine the expression pattern of fmi-1. The fmi-1p construct includes a 2.6 kb upstream region fused to the cDNA for GFP (Fig. 4A). GFP expression could first be observed in the embryo during gastrulation in neuronal precursors (Fig. 4C,D). At the comma and 1.5-fold stage, expression was apparent in neurons in the head, the tail and along the ventral cord (Fig. 4E-H). Expression was maintained into the adult stage (Fig. 4I-M). Using cell type-specific markers, expression of the fmi-1p construct was confirmed in many neurons, including PVP and PVQ (Fig. 4J-L). HSN neurons started to express fmi-1 post-embryonically before axon outgrowth and maintained expression in adults (Fig. 4M). The fmi-1 gene contains a large intron near the 5′ end of the gene, which might contain additional regulatory elements (Fig. 1B). We generated a second construct (fmi-1pintron) extending into the fifth exon with a transmembrane version of GFP to prevent secretion of the reporter. Differences in the expression pattern between the two constructs were minor. However, expression in SDQ, PVD and PDE neurons was seen only with the fmi-1pintron construct, suggesting that additional elements controlling expression are located in introns.
Fig. 4.
fmi-1 expression and localization. (A) Transcriptional and translational reporter constructs. (B) Cells expressing fmi-1p::GFP. Expression of the fmi-1p construct was detected embryonically (C-H) and postembryonically (I-N). (C,D) Gastrulation-stage embryo. (E,F) Comma-stage embryo. (G,H) 1.5-fold-stage embryo with expression in motoneurons (G, arrows). D, F and H are overlays of Nomarski and GFP channels. (I) L1 larva head region. (J-L) fmi-1p expression (green) in PVP (cyan) and PVQ (red) neurons. (M) Adult animal, vulva region. (N) FMI-1::GFP expression in the adult. Broken white lines indicate the outline of animals where possible. (C-H,N) Lateral views; (I-M) Ventral views. In all pictures, anterior is towards the left. Scale bars: 10 μm.
To determine the subcellular localization of the fmi-1 protein, we created a construct (FMI-1::GFP) containing the genomic region of fmi-1 with a GFP cDNA fused to its C terminus (Fig. 4A). FMI-1::GFP was functional and was able to rescue PVP, PVQ and HSN defects in fmi-1(rh308) animals. FMI-1::GFP localized diffusely to major axon tracts such as the nerve ring and the ventral cord (Fig. 4N). In the embryo, during stages of axon outgrowth, FMI-1::GFP was found in axons in the nerve ring, the tail and along dendrites of sensory neurons.
FMI-1 acts partially cell autonomously in follower axons
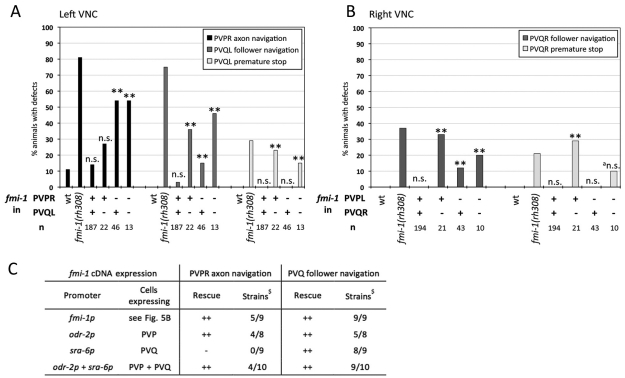
We performed a mosaic analysis to determine whether FMI-1 is required cell-autonomously in PVP and PVQ axons. Pioneer axon guidance defects in the left axon track were completely rescued when FMI-1 was present in the pioneer (PVPR) alone (Fig. 5A) and the loss of FMI-1 in PVPR correlated with increased pioneer axon guidance defects. These results suggest that FMI-1 is required cell-autonomously in PVPR for correct pioneer navigation. PVQL follower axon guidance was restored completely when FMI-1 was present in both pioneer (PVPR) and follower (Fig. 5A). fmi-1 expression in PVQL alone was also sufficient for a nearly complete rescue of the PVQL axon navigation defects. The premature stopping of PVQ axons could not be rescued at all unless FMI-1 was present in PVQ itself (Fig. 5A,B). Similarly, all defects of PVQR in the right ventral cord were only rescued when FMI-1 was present in PVQR, suggesting that FMI-1 also acts largely cell autonomously in this follower (Fig. 5B).
Fig. 5.
Mosaic analysis and targeted expression of fmi-1 in fmi-1(rh308) animals. (A,B) Mosaic analysis; (C) targeted expression. (A,B) Presence (+) or absence (–) of the rescuing array containing fmi-1(+) in PVP and PVQ neurons was determined by presence of nuclear localized GFP. Defects in wild-type (wt) and fmi-1(rh308) animals are shown for comparison. Mosaic analysis results are for (A) left axon track and (B) right axon track neurons. **P<0.01 and not significant (n.s.) with P>0.05 compared with wild type (χ2 test). The marker used was hdIs26. (C) Results of targeted fmi-1 cDNA expression in PVP and PVQ neurons of fmi-1(rh308) animals. Categories: –, no rescue with P>0.05 compared with fmi-1(rh308); ++, rescue with P>0.05 compared with best fmi-1p rescue (χ2 test); $Independent best rescuing strains out of total of analyzed strains; n=27-110. The marker used was hdIs26.
To confirm the results of the mosaic analysis, we expressed the fmi-1 cDNA specifically in pioneer and follower neurons in fmi-1(rh308) animals. Expression of fmi-1 in PVP pioneers under the control of the odr-2 promoter was able to rescue both pioneer and follower axon guidance defects (Fig. 5C). fmi-1 expression in PVQ neurons alone, under control of the sra-6 promoter, rescued PVQ follower but not PVPR pioneer defects. fmi-1 expression in both PVP and PVQ axons rescued pioneer and follower defects as effectively as the fmi-1 promoter itself. These results support the idea of a cell autonomous function in the pioneer and partially non-cell autonomous function in the follower.
Different domains of FMI-1 are required for pioneer and follower navigation
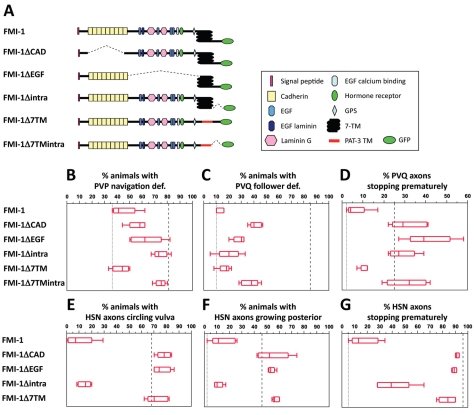
FMI-1 contains domains typically found in receptors, as well as domains characteristic for cell-adhesion molecules. Thus, it is conceivable that FMI-1 acts as receptor for guidance cues in the pioneer and as adhesion molecule in the follower. To test this hypothesis, we created FMI-1 variants lacking certain protein domains and tested their ability to rescue pioneer and follower axon defects in fmi-1(rh308) animals. To identify domains that might mediate adhesion or contain ligand-binding sites, we divided the extracellular domain into a cadherin and non-cadherin part, which we deleted individually (FMI-1ΔCAD, ΔEGF) (Fig. 6A). To test the importance of the 7TM domain, we replaced it with an unrelated single TM domain (FMI-1Δ7TM). Similarly we deleted the intracellular domain either individually (FMI-1Δintra) or together with the 7TM domain (FMI-1Δ7TMintra) to uncover parts required for downstream signaling.
Fig. 6.
Domain analysis of FMI-1. (A) For each FMI-1 construct, five transgenic strains were generated and their phenotypes scored. (B-G) Defects in fmi-1(rh308) animals (dashed line) and in best fmi-1 full-length rescue (dotted line) are shown for comparison. Boxes outline 50% of the data set around the median, indicated as a line within the box. The upper and lower ends of the box indicate the 25th and 75th quartiles. Whiskers mark the maximum and minimum values. (B) Rescue of PVP axon guidance. (C) Rescue of PVQ follower axon navigation. (D) Rescue of PVQ premature stop. (E) Rescue of HSN circling phenotype. (F) Rescue of HSN posterior extension. (G) Rescue of HSN premature stop. (B,C) n=39-100; (D) n=92-202; (E,F) n=35-61; (G) n=80-122. Marker used were hdIs26 (PVP,PVQ) and zdIs13 (HSN).
First, we examined the effect of each construct on pioneer axon navigation (Fig. 6B). Somewhat surprisingly, all constructs removing various parts of the extracellular domain (FMI-1ΔCAD, ΔEGF, Δ7TM) were able to at least partially rescue PVP navigation defects. This suggests that more than one ligand binding side exists or that FMI-1 acts as co-receptor and does not bind a specific ligand itself. The FMI-1Δ7TM construct rescued to the same extent as the complete protein, indicating that the 7TM domain is dispensable for PVP axon navigation. However, guidance of the PVP axon was not rescued with constructs that lack the intracellular domain, which seems absolutely essential here.
Analysis of PVQ defects revealed different domain requirements to rescue follower defects. Constructs without various non-cadherin domains (FMI-1ΔEGF, Δ7TM, Δintra) rescued PVQ follower defects as well as full-length FMI-1 (Fig. 6C). Even the construct lacking the cadherin domains (FMI-1ΔCAD) similarly rescued these defects, suggesting that FMI-1 does not simply act as homophilic adhesion molecule here. The 7TM and intracellular domain were dispensable for follower axon guidance. The premature stopping of PVQ axons could not be rescued with any of the deletion constructs except FMI-1Δ7TM (Fig. 6D), pointing to the requirement of multiple domains for proper axon outgrowth. Similarly, HSN outgrowth and navigation defects could only be effectively rescued with FMI-1Δintra (Fig. 6E-G). Partial rescue of the HSN premature stopping defect was also obtained with FMI-1Δ7TM (Fig. 6G).
The extracellular domain is necessary for axonal localization of FMI-1
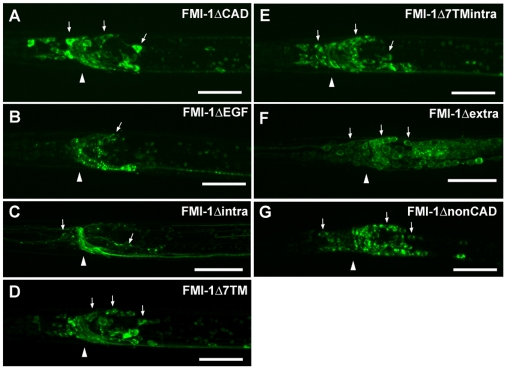
All FMI-1 constructs were GFP tagged, enabling us to examine the subcellular localization of each construct (Fig. 7). FMI-1ΔEGF and FMI-1Δintra showed a normal distribution of the protein in axons only, suggesting that the non-cadherin part of the extracellular domain and the intracellular domain are not crucial for proper subcellular localization. FMI-1ΔCAD, Δ7TM and Δ7TMintra showed increased GFP signal in cell bodies in addition to strong staining in axons. Two more constructs, one with the entire extracellular domain deleted (FMI-1Δextra) and one with only membrane-bound cadherin domains (FMI-1ΔnonCAD) showed almost no axonal staining, suggesting that the extracellular domain is required for proper axonal localization. These constructs were not able to rescue any of the defects. However, as this could be due to their mislocalization, they were excluded from the domain analysis.
Fig. 7.
Localization of FMI-1 domain analysis constructs. (A-G) GFP images of head region with arrows indicating neuronal cell bodies and an arrowhead indicating the position of the nerve ring. Localization of construct (A) FMI-1ΔCAD, (B) FMI-1ΔEGF, (C) FMI-1Δintra, (D) FMI-1Δ7TM, (E) FMI-1Δ7TMintra, (F) FMI-1Δextra and (G) FMI-1ΔnonCAD in head region; lateral views, anterior towards the left. Scale bars: 10 μm.
lin-17/frizzled and fmi-1 act together in follower axon navigation
To identify putative components of the FMI-1 signaling pathway, we analyzed C. elegans orthologs of PCP pathway genes for defects in VNC axon guidance. No defects were detected in cfz-2/frizzled-, mig-1/frizzled-, vang-1/vangogh-, prkl-1/prickle- or cdh-1/dachsous-deficient animals (data not shown). lin-17/frizzled(n671) animals, however, exhibited strong PVPR axon guidance defects (60%, n=100). Pioneer axon guidance defects were significantly enhanced in lin-17; fmi-1 double mutants (97%, n=100), suggesting that LIN-17/Frizzled functions in parallel to FMI-1 rather than in the same pathway. lin-17 deficient worms also exhibited mild defects in PVQL follower axon navigation (23%, n=100), but not in axon extension. We observed no enhancement of PVQL follower defects in lin-17(n671); fmi-1(rh308) double mutants (70%, n=100), suggesting that LIN-17/Frizzled functions together with FMI-1 in follower axon navigation.
During follower axon navigation, FMI-1 might also interact with other cadherins. We tested casy-1/Calsytenin- and hmr-1b-deficient animals for follower axon guidance defects but were unable to detect any (data not shown). To test whether the partial non-cell autonomous rescue of PVQL defects depends on the presence of other cadherins, we created fmi-1 double mutants with casy-1/Calsyntenin and hmr-1b. We expressed fmi-1 cDNA either in PVP or PVQ neurons in these double mutants to test whether FMI-1 could still rescue the follower defects. The rescuing ability of the cDNA was unchanged in either double mutant (data not shown), suggesting that neither hmr-1b nor casy-1 interact with fmi-1 here.
DISCUSSION
FMI-1 mediates axon track formation in the VNC
Establishment of a functional neuronal network begins with pioneer axons creating a scaffold of axon tracks (R. M. Durbin, PhD Thesis, University of Cambridge, 1987) (Kuwada, 1986; Mastick and Easter, 1996; Raper et al., 1983; Ross et al., 1992). In many organisms, later outgrowing axons use these pioneer axons to navigate to their correct target areas (R. M. Durbin, PhD Thesis, University of Cambridge, 1987) (Chitnis and Kuwada, 1991; Garriga et al., 1993; Hidalgo and Brand, 1997; Hutter, 2003; Klose and Bentley, 1989; McConnell et al., 1989). However, the molecular basis for pioneer-mediated navigation is largely unclear. In C. elegans, sequential outgrowth of pioneer and follower axons leads to the establishment of axon tracks in the VNC (R. M. Durbin, PhD Thesis, University of Cambridge, 1987), providing a model with which to study axon-axon interactions at the single cell level and to identify genes controlling pioneer-mediated navigation of follower axons. In genetic screens for defects in follower axon navigation in the VNC, we isolated alleles of the Flamingo ortholog FMI-1. fmi-1 mutant animals are characterized by strong defects in the navigation of VNC pioneer and follower axons. In C. elegans, PVPR pioneer axon navigation defects observed in fmi-1-deficient animals are also reported for several other genes, e.g. the basement membrane components UNC-6/NETRIN and NID-1/NIDOGEN, the IgCAMs WRK-1 and SAX-3/ROBO, the Fat-like cadherin CDH-4, and the SYNDECAN ortholog SDN-1 (Boulin et al., 2006; Hutter, 2003; Rhiner et al., 2005; Schmitz et al., 2008; Schwarz et al., 2009). fmi-1-deficient animals, as well as lin-17/frizzled, cdh-4 and nid-1 mutants, show highly penetrant PVPR defects, suggesting that in this complex network of guidance cues, these molecules might play a central role for correct axon navigation and midline avoidance of the PVPR pioneer axon. flamingo mutants in Drosophila also display defects in VNC axon track development (Sweeney et al., 2002; Usui et al., 1999), in sensory neuron advance (Steinel and Whitington, 2009) and in photoreceptor axon target choice (Chen and Clandinin, 2008; Lee et al., 2003). In mouse, CELSR3 also functions in axon navigation (Tissir et al., 2005; Zhou et al., 2008).
During VNC development, PVQ axons closely follow PVP axons on the left and right side of the VNC (R. M. Durbin, PhD Thesis, University of Cambridge, 1987) (Hutter, 2003). Ablation of the pioneer of the left track (PVPR) causes the PVQL axon to irregularly join the right VNC axon track, whereas ablation of the PVQL axon has no effect on PVPR axon navigation (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). Furthermore, PVPR axonal defects observed in various mutants lead to identical PVQL axon guidance defects (Hutter, 2003). These results suggest that correct PVQL axon navigation is highly dependent on interactions with the pioneer axon PVPR. In fmi-1 mutant animals, PVQ axons initially correctly navigated towards the VNC, in part mediated by UNC-6/NETRIN attraction (Hutter, 2003; Wadsworth et al., 1996), but then failed to maintain contact with the PVP axons. FMI-1 seems to be a key mediator of the previously described interaction between the PVPR/PVQL axon pair in the left axon track. In the right axon track, several axons (the AVG pioneer axon and DD motoneuron axons) are already present, when PVPL grows into the VNC (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). Ablation of PVPL has no effect on PVQR axon navigation (R. M. Durbin, PhD Thesis, University of Cambridge, 1987), which still extends in the right track, possibly now following other axons present here. However, PVQR also partially navigated independently of PVPL in fmi-1 mutants, suggesting that FMI-1 has a similar role in both axon tracts with respect to mediating PVP-dependent navigation of PVQ axons.
The AVKR axon, which extends along the left VNC track towards the posterior, is also thought to depend on the presence of the PVPR/PVQL axon pair (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). After PVPR ablation, no processes were found in the left axon track. However, it remained unclear whether the AVKR axon joined the right VNC track or failed to extend (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). We found strong PVPR pioneer axon navigation defects in fmi-1(rh308) animals, but only mild AVKR axon guidance defects. This suggests that AVKR axon navigation differs from navigation of PVQ axons and neither depends on FMI-1 nor on the PVPR axon. Alternatively, AVKR might have different requirements for extension (potentially pioneer dependent) and navigation (potentially pioneer independent).
Postembryonically, a second pair of follower axons, the HSN axons, fasciculate with the PVP and PVQ axons and extend into the nerve ring (Asakura et al., 2007; Garriga et al., 1993). In many fmi-1 mutant animals, HSN axons failed to fasciculate with PVP and PVQ axons. These defects differed from defects seen in animals with ablated PVPR and PVQL neurons, where HSNL axons join the right VNC axon track (Garriga et al., 1993), suggesting that these were not just consequences of missing or misguided pioneer axons. In fmi-1-deficient animals, HSN axons appeared continuously attracted by guidance cues emanating from the vulva precursor cells (VPC), possibly UNC-6/NETRIN (Asakura et al., 2007). HSN axons also interact with the vulva epithelium via heterophilic binding of CAMs of the immunoglobulin domain superfamily in order to establish correct synaptic connections (Chao and Shen, 2008; Shen and Bargmann, 2003; Shen et al., 2004). FMI-1 might regulate UNC-6/NETRIN signaling and/or adhesive interactions with the vulva epithelium to allow HSN axons to leave the vulva region. In summary, FMI-1 is a key regulator of follower axon navigation along pre-existing pioneer axons, leading to the establishment of VNC axon tracks.
FMI-1 acts cell-autonomously and non cell-autonomously in a neuron-type-dependent manner
How does FMI-1 mediate pioneer and follower axon navigation? fmi-1 is expressed in pioneer and follower axons, suggesting that it could act cell-autonomously. We found that in the pioneer axon FMI-1 indeed predominantly functions cell-autonomously. Similar results were reported for Flamingo in Drosophila sensory neuron axon extension (Steinel and Whitington, 2009) and in PCP pathway signaling (Usui et al., 1999). Follower axon guidance was partially rescued when FMI-1 was present in PVP but not in PVQ. This raises the possibility that FMI-1 might mediate heterophilic interactions, possibly with other cadherins. This has been demonstrated for the cadherins Fat and Dachsous in Drosophila (Matakatsu and Blair, 2004). Alternatively, FMI-1 could regulate adhesion through other cell-surface molecules. In Xenopus morphogenesis paraxial protocadherin, for example, has no adhesive function itself but regulates cell adhesion by downregulating C-cadherin adhesion (Chen and Gumbiner, 2006). With respect to PVQ axon extension, FMI-1 functions cell-autonomously and could therefore also function as a receptor in the PVQ axons.
FMI-1 might act as modulator of cell-cell adhesion and as receptor for correct axon navigation and extension
To distinguish whether FMI-1 acts as receptor or adhesion molecule, we tested the different parts of the protein for their ability to rescue the various fmi-1 phenotypes. Pioneer axon guidance depended on the intracellular domain, suggesting that FMI-1 might have a receptor-like function in the pioneer and mediate downstream signaling events. Interestingly, the various parts of the FMI-1 ectodomain were all able to partially rescue the pioneer defects. The 7TM domain, however, was completely dispensable for PVPR axon navigation, indicating that FMI-1 does not act as GPCR here. Potentially FMI-1 has more than one ligand binding site or is recruited into a receptor complex. Flamingo is known to interact with Frizzled and Van Gogh/Strabismus in Drosophila PCP signaling (Chen et al., 2008). Mutations in fz and vang, however, show no axonal defects in flies (Clandinin and Zipursky, 2000; Senti et al., 2003; Steinel and Whitington, 2009). Furthermore, the intracellular domain of Flamingo is dispensable for rescue of PCP signaling in flamingo deficient flies (Chen et al., 2008), in contrast to its requirement in FMI-1-mediated pioneer axon navigation. We failed to detect genetic interactions between PCP pathway genes and fmi-1 in pioneer navigation, suggesting that FMI-1 function in the PVPR axon does not depend on PCP pathway components.
The role of FMI-1 in follower axon guidance and extension is even more complex. FMI-1 function in pioneer-mediated follower axon navigation did not rely on FMI-1 signaling properties and seems to differ between PVQ and HSN axons. This observation is not too surprising, as both axons extend at different times during development (R. M. Durbin, PhD Thesis, University of Cambridge, 1987). FMI-1 function in follower navigation did not depend on specific extracellular domains, suggesting that FMI-1 itself does not mediate homophilic adhesion here. In cell culture, Drosophila Flamingo and murine CELSR2 were shown to mediate homophilic adhesion via their extracellular domain and cadherin repeats, respectively (Shima et al., 2004; Usui et al., 1999). The Flamingo ectodomain is thought to act bi-functionally in dendrite development, with homophilic binding of cadherin repeats mediating contact repulsion, and the hormone receptor domain negatively regulating growth through a putative ligand (Kimura et al., 2006). Similar bi-functionality was observed for CELSR2 in dendritic maintenance (Shima et al., 2004). In primary hippocampal neurons, homophilic binding of cadherin repeats to murine CELSR2 and CELSR3 triggers Ca2+ signaling and modulates neurite growth through the 7TM and intracellular domain, suggesting a link between homophilic cell adhesion and intracellular signaling (Shima et al., 2007). Drosophila Flamingo also probably mediates homophilic interactions between photoreceptor cell axons for correct axon target selection (Chen and Clandinin, 2008). However, loss of Flamingo causes no obvious defects in photoreceptor axon track formation (Lee et al., 2003), suggesting that Flamingo and FMI-1 functions differ here. FMI-1 might recruit or regulate other receptors or CAMs to mediate follower axon navigation. Interestingly, we found that lin-17/frizzled genetically interacted with fmi-1 in follower axon navigation. However, we failed to find similar interactions with other PCP genes, suggesting that this pathway probably differs from the PCP pathway. Interestingly, Fzd3/frizzled and Celsr3-deficient mice have similar nervous system defects, and both genes are co-expressed, also suggesting a possible interaction between both proteins in the vertebrate nervous system (Tissir et al., 2005). fmi-1 function in the PVQ axon is complex and might include homophilic binding in addition to recruitment of receptors and/or modulation of cell-cell adhesion.
HSN axon navigation depended on the presence of the extracellular domain and the 7TM domain, and missense mutations in the extracellular domain had dramatic effects on HSN axon guidance, suggesting that FMI-1-mediated cell adhesion might play a more important role in HSN axon navigation. But this result does not rule out that FMI-1 might also recruit other adhesion molecules or receptors in HSN axons. However, based on the domain requirements, FMI-1 function in HSN axon guidance seems to differ from its role in PVQ axon navigation.
For the extension (premature stop) of follower axons, FMI-1 probably acts as a receptor for an unidentified ligand, as both the extra- and intracellular domains were required in PVQ axon extension. Similar axon extension defects in HSN axons required the same domains, suggesting a similar mechanism of action for the same aspect of axon outgrowth in different neurons.
All adhesion GPCRs, including FMI-1, contain a GPS cleavage side in their ectodomain (Krasnoperov et al., 2002; Krasnoperov et al., 1997), which could lead to a complete shedding of the extracellular domain and explain non cell-autonomous functions of FMI-1. However, FMI-1 functions mainly cell-autonomously, and in the domain analysis we found no specific requirement for the GPS domain in either pioneer or follower axon navigation.
In summary, FMI-1 function in C. elegans axon outgrowth and navigation is complex. In pioneer axons, FMI-1 might act as receptor for a putative ligand, or more likely is part of a receptor complex. In follower axons, FMI-1 might regulate adhesion between pioneer and follower axons, as well as transduce a signal to promote continued outgrowth. Our results suggest that pioneer-mediated navigation of follower axons is not simply based on homophilic adhesion mediated by FMI-1, but it seems to require a modulation of axon-axon interactions that probably involve other receptors and also CAMs.
Acknowledgments
We thank Mei Zhen and Yishi Jin for providing us with the ju43 allele. We also thank Thomas Unsöld, Mei Zhen and Yishi Jin for comments on the manuscript, and all our lab members for critical discussion of the experiments. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The C. elegans Gene Knockout Consortium and the National Bioresource Project for the nematode isolated some of the strains used in this study. This work was supported by grant MOP 93719 from the Canadian Institutes of Health Research to H.H. and by NIH grant NS32057 to G.G. H.H. is supported by the Michael Smith Foundation for Health Research through a senior scholar award. B.D.A. was supported, in part, by P20 RR016475 from the INBRE Program of the NCRR and RC1 GM091086 from the NIH. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Asakura T., Ogura K., Goshima Y. (2007). UNC-6 expression by the vulval precursor cells of Caenorhabditis elegans is required for the complex axon guidance of the HSN neurons. Dev. Biol. 304, 800-810 [DOI] [PubMed] [Google Scholar]
- Bao H., Berlanga M. L., Xue M., Hapip S. M., Daniels R. W., Mendenhall J. M., Alcantara A. A., Zhang B. (2007). The atypical cadherin flamingo regulates synaptogenesis and helps prevent axonal and synaptic degeneration in Drosophila. Mol. Cell. Neurosci. 34, 662-678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T., Pocock R., Hobert O. (2006). A novel Eph receptor-interacting IgSF protein provides C. elegans motoneurons with midline guidepost function. Curr. Biol. 16, 1871-1883 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent I. D., Pettitt J. (2002). The C. elegans hmr-1 gene can encode a neuronal classic cadherin involved in the regulation of axon fasciculation. Curr. Biol. 12, 59-63 [DOI] [PubMed] [Google Scholar]
- Chae J., Kim M. J., Goo J. H., Collier S., Gubb D., Charlton J., Adler P. N., Park W. J. (1999). The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126, 5421-5429 [DOI] [PubMed] [Google Scholar]
- Chao D. L., Shen K. (2008). Functional dissection of SYG-1 and SYG-2, cell adhesion molecules required for selective synaptogenesis in C. elegans. Mol. Cell. Neurosci. 39, 248-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. L., Clandinin T. R. (2008). The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 58, 26-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. S., Antic D., Matis M., Logan C. Y., Povelones M., Anderson G. A., Nusse R., Axelrod J. D. (2008). Asymmetric homotypic interactions of the atypical cadherin flamingo mediate intercellular polarity signaling. Cell 133, 1093-1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Gumbiner B. M. (2006). Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J. Cell Biol. 174, 301-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A. B., Kuwada J. Y. (1991). Elimination of a brain tract increases errors in pathfinding by follower growth cones in the zebrafish embryo. Neuron 7, 277-285 [DOI] [PubMed] [Google Scholar]
- Chou J. H., Troemel E. R., Sengupta P., Colbert H. A., Tong L., Tobin D. M., Roayaie K., Crump J. G., Dwyer N. D., Bargmann C. I. (1996). Olfactory recognition and discrimination in Caenorhabditis elegans. Cold Spring Harbor Symp. Quant. Biol. 61, 157-164 [PubMed] [Google Scholar]
- Clandinin T. R., Zipursky S. L. (2000). Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron 28, 427-436 [DOI] [PubMed] [Google Scholar]
- Cornel E., Holt C. (1992). Precocious pathfinding: retinal axons can navigate in an axonless brain. Neuron 9, 1001-1011 [DOI] [PubMed] [Google Scholar]
- Das G., Reynolds-Kenneally J., Mlodzik M. (2002). The atypical cadherin Flamingo links Frizzled and Notch signaling in planar polarity establishment in the Drosophila eye. Dev. Cell 2, 655-666 [DOI] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W. J., Weis W. I. (2005). Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell 123, 903-915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. S., Pike S. H., Debu B. (1989). The growth cones of identified motoneurons in embryonic zebrafish select appropriate pathways in the absence of specific cellular interactions. Neuron 2, 1097-1104 [DOI] [PubMed] [Google Scholar]
- Gao F. B., Brenman J. E., Jan L. Y., Jan Y. N. (1999). Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 13, 2549-2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. B., Kohwi M., Brenman J. E., Jan L. Y., Jan Y. N. (2000). Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron 28, 91-101 [DOI] [PubMed] [Google Scholar]
- Garriga G., Desai C., Horvitz H. R. (1993). Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development 117, 1071-1087 [DOI] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R. (1994). pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22, 1762-1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A., Brand A. H. (1997). Targeted neuronal ablation: the role of pioneer neurons in guidance and fasciculation in the CNS of Drosophila. Development 124, 3253-3262 [DOI] [PubMed] [Google Scholar]
- Hobert O. (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728-730 [DOI] [PubMed] [Google Scholar]
- Hutter H. (2003). Extracellular cues and pioneers act together to guide axons in the ventral cord of C. elegans. Development 130, 5307-5318 [DOI] [PubMed] [Google Scholar]
- Hutter H., Wacker I., Schmid C., Hedgecock E. M. (2005). Novel genes controlling ventral cord asymmetry and navigation of pioneer axons in C. elegans. Dev. Biol. 284, 260-272 [DOI] [PubMed] [Google Scholar]
- Keshishian H., Bentley D. (1983). Embryogenesis of peripheral nerve pathways in grasshopper legs. III. Development without pioneer neurons. Dev. Biol. 96, 116-124 [DOI] [PubMed] [Google Scholar]
- Kimura H., Usui T., Tsubouchi A., Uemura T. (2006). Potential dual molecular interaction of the Drosophila 7-pass transmembrane cadherin Flamingo in dendritic morphogenesis. J. Cell Sci. 119, 1118-1129 [DOI] [PubMed] [Google Scholar]
- Klose M., Bentley D. (1989). Transient pioneer neurons are essential for formation of an embryonic peripheral nerve. Science 245, 982-984 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V. G., Bittner M. A., Beavis R., Kuang Y., Salnikow K. V., Chepurny O. G., Little A. R., Plotnikov A. N., Wu D., Holz R. W., et al. (1997). alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18, 925-937 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V., Lu Y., Buryanovsky L., Neubert T. A., Ichtchenko K., Petrenko A. G. (2002). Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J. Biol. Chem. 277, 46518-46526 [DOI] [PubMed] [Google Scholar]
- Kuwada J. Y. (1986). Cell recognition by neuronal growth cones in a simple vertebrate embryo. Science 233, 740-746 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Clandinin T. R., Lee C. H., Chen P. L., Meinertzhagen I. A., Zipursky S. L. (2003). The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat. Neurosci. 6, 557-563 [DOI] [PubMed] [Google Scholar]
- Lu B., Usui T., Uemura T., Jan L., Jan Y. N. (1999). Flamingo controls the planar polarity of sensory bristles and asymmetric division of sensory organ precursors in Drosophila. Curr. Biol. 9, 1247-1250 [DOI] [PubMed] [Google Scholar]
- Mastick G. S., Easter S. S., Jr (1996). Initial organization of neurons and tracts in the embryonic mouse fore- and midbrain. Dev. Biol. 173, 79-94 [DOI] [PubMed] [Google Scholar]
- Matakatsu H., Blair S. S. (2004). Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development 131, 3785-3794 [DOI] [PubMed] [Google Scholar]
- McConnell S. K., Ghosh A., Shatz C. J. (1989). Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science 245, 978-982 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. D., Chen C. P., Bahna F., Honig B., Shapiro L. (2003). Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr. Opin. Struct. Biol. 13, 690-698 [DOI] [PubMed] [Google Scholar]
- Pettitt J. (2005). The cadherin superfamily. WormBook, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper J. A., Bastiani M. J., Goodman C. S. (1983). Guidance of neuronal growth cones: selective fasciculation in the grasshopper embryo. Cold Spring Harbor Symp. Quant. Biol. 48, 587-598 [DOI] [PubMed] [Google Scholar]
- Reuter J. E., Nardine T. M., Penton A., Billuart P., Scott E. K., Usui T., Uemura T., Luo L. (2003). A mosaic genetic screen for genes necessary for Drosophila mushroom body neuronal morphogenesis. Development 130, 1203-1213 [DOI] [PubMed] [Google Scholar]
- Rhiner C., Gysi S., Frohli E., Hengartner M. O., Hajnal A. (2005). Syndecan regulates cell migration and axon guidance in C. elegans. Development 132, 4621-4633 [DOI] [PubMed] [Google Scholar]
- Ross L. S., Parrett T., Easter S. S., Jr (1992). Axonogenesis and morphogenesis in the embryonic zebrafish brain. J. Neurosci. 12, 467-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C., Wacker I., Hutter H. (2008). The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev. Biol. 316, 249-259 [DOI] [PubMed] [Google Scholar]
- Schwarz V., Pan J., Voltmer-Irsch S., Hutter H. (2009). IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Dev. 4, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K. A., Usui T., Boucke K., Greber U., Uemura T., Dickson B. J. (2003). Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr. Biol. 13, 828-832 [DOI] [PubMed] [Google Scholar]
- Shen K., Bargmann C. I. (2003). The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112, 619-630 [DOI] [PubMed] [Google Scholar]
- Shen K., Fetter R. D., Bargmann C. I. (2004). Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell 116, 869-881 [DOI] [PubMed] [Google Scholar]
- Shima Y., Kengaku M., Hirano T., Takeichi M., Uemura T. (2004). Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev. Cell 7, 205-216 [DOI] [PubMed] [Google Scholar]
- Shima Y., Kawaguchi S. Y., Kosaka K., Nakayama M., Hoshino M., Nabeshima Y., Hirano T., Uemura T. (2007). Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat. Neurosci. 10, 963-969 [DOI] [PubMed] [Google Scholar]
- Steinel M. C., Whitington P. M. (2009). The atypical cadherin Flamingo is required for sensory axon advance beyond intermediate target cells. Dev. Biol. 327, 447-457 [DOI] [PubMed] [Google Scholar]
- Sweeney N. T., Li W., Gao F. B. (2002). Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev. Biol. 247, 76-88 [DOI] [PubMed] [Google Scholar]
- Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G. (2000). Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560-564 [DOI] [PubMed] [Google Scholar]
- Takeichi M. (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11-20 [DOI] [PubMed] [Google Scholar]
- Tissir F., Bar I., Jossin Y., De Backer O., Goffinet A. M. (2005). Protocadherin Celsr3 is crucial in axonal tract development. Nat. Neurosci. 8, 451-457 [DOI] [PubMed] [Google Scholar]
- Treubert-Zimmermann U., Heyers D., Redies C. (2002). Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J. Neurosci. 22, 7617-7626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Chou J. H., Dwyer N. D., Colbert H. A., Bargmann C. I. (1995). Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83, 207-218 [DOI] [PubMed] [Google Scholar]
- Usui T., Shima Y., Shimada Y., Hirano S., Burgess R. W., Schwarz T. L., Takeichi M., Uemura T. (1999). Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98, 585-595 [DOI] [PubMed] [Google Scholar]
- Wadsworth W. G., Bhatt H., Hedgecock E. M. (1996). Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron 16, 35-46 [DOI] [PubMed] [Google Scholar]
- Wohrn J. C., Nakagawa S., Ast M., Takeichi M., Redies C. (1999). Combinatorial expression of cadherins in the tectum and the sorting of neurites in the tectofugal pathways of the chicken embryo. Neuroscience 90, 985-1000 [DOI] [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J. (2005). Deconstructing the cadherin-catenin-actin complex. Cell 123, 889-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. (1998). A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149, 1323-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Bar I., Achouri Y., Campbell K., De Backer O., Hebert J. M., Jones K., Kessaris N., de Rouvroit C. L., O'Leary D., et al. (2008). Early forebrain wiring: genetic dissection using conditional Celsr3 mutant mice. Science 320, 946-949 [DOI] [PMC free article] [PubMed] [Google Scholar]