Abstract
Background
Cyclophilin A (CypA) represents a potential target for antiretroviral therapy since inhibition of CypA suppresses human immunodeficiency virus type 1 (HIV-1) replication, although the mechanism through which CypA modulates HIV-1 infectivity still remains unclear. The interaction of HIV-1 viral protein R (Vpr) with the human peptidyl prolyl isomerase CypA is known to occur in vitro and in vivo. However, the nature of the interaction of CypA with Pro-35 of N-terminal Vpr has remained undefined.
Results
Characterization of the interactions of human CypA with N-terminal peptides of HIV-1 Vpr has been achieved using a combination of nuclear magnetic resonace (NMR) exchange spectroscopy and surface plasmon resonance spectroscopy (SPR). NMR data at atomic resolution indicate prolyl cis/trans isomerisation of the highly conserved proline residues Pro-5, -10, -14 and -35 of Vpr are catalyzed by human CypA and require only very low concentrations of the isomerase relative to that of the peptide substrates. Of the N-terminal peptides of Vpr only those containing Pro-35 bind to CypA in a biosensor assay. SPR studies of specific N-terminal peptides with decreasing numbers of residues revealed that a seven-residue motif centred at Pro-35 consisting of RHFPRIW, which under membrane-like solution conditions comprises the loop region connecting helix 1 and 2 of Vpr and the two terminal residues of helix 1, is sufficient to maintain strong specific binding.
Conclusions
Only N-terminal peptides of Vpr containing Pro-35, which appears to be vital for manifold functions of Vpr, bind to CypA in a biosensor assay. This indicates that Pro-35 is essential for a specific CypA-Vpr binding interaction, in contrast to the general prolyl cis/trans isomerisation observed for all proline residues of Vpr, which only involve transient enzyme-substrate interactions. Previously suggested models depicting CypA as a chaperone that plays a role in HIV-1 virulence are now supported by our data. In detail the SPR data of this interaction were compatible with a two-state binding interaction model that involves a conformational change during binding. This is in accord with the structural changes observed by NMR suggesting CypA catalyzes the prolyl cis/trans interconversion during binding to the RHFP35RIW motif of N-terminal Vpr.
Background
The 96 amino acid virion-associated multifunctional viral protein R (Vpr) [1,2] is encoded by primate lentiviruses, the human immunodeficiency viruses, types 1 and 2 (HIV-1/HIV-2), and simian immunodeficiency viruses (SIV). This accessory protein fulfils multiple functions in the viral life cycle including increase of viral replication in non-dividing host cells, induction of G2 cell-cycle arrest [3,4], apoptosis [5,6], and transduction through cell membranes [7]. Vpr facilitates transport of the pre-integration complex into the nucleus of non-dividing cells [8] and interacts with several cellular factors, including the human peptidyl prolyl isomerase CypA [9].
The interaction of HIV-1 Vpr with CypA is known to occur in vitro and in vivo [9-11]. In addition to the extensively studied interaction between CypA and HIV-1 capsid, that is crucial for viral replication [12,13], CypA was also reported to be significant for the de novo synthesis of Vpr, as the Vpr-mediated cell cycle arrest in HIV-1 infected T cells appeared to be eliminated in the absence of CypA activity [9]. However, more recently Ardon et al. [10] concluded that the interaction of Vpr with CypA is independent of the ability of Vpr to induce cell cycle arrest. Nonetheless, specific inhibitors of the prolyl cis/trans isomerase activity of CypA, such as cyclosporine A and SDZ-NIM811 inhibit HIV replication [11,14-18].
Previous structural studies of Vpr by 2D NMR spectroscopy in aqueous organic solvents provided evidence of proline cis/trans isomerism for the highly conserved N-terminal Pro-5, -10, -14 and -35 of Vpr [19]. In particular Pro-35 exhibited a relatively high proportion of the cis isomer under these solvent conditions (15% cis isomer content). This suggested prolyl cis/trans isomerisation may be important for the folding of the molecule. At pH 7 Vpr has a relatively random structure in aqueous solution but assumes a folded structure in a hydrophobic membranous environment [7,19]. This fact together with observation of considerable amounts of CypA in virions [20] prompted a study, using surface plasmon resonance (SPR) spectroscopy, of the interaction of Vpr with the prolyl cis/trans isomerase CypA [9]. A qualitative interaction was detected for N-terminal peptides containing Pro-35, indicating an essential role for this Pro residue, although this could not be quantified. Indeed the interaction of Vpr with CypA could not be confirmed under the solution conditions used. Hence, the nature of the interaction of CypA with Pro-35 of N-terminal Vpr has remained undefined.
The catalytic activity of CypA, as a peptidyl-prolyl cis/trans isomerase (PPiase), has previously been studied on short model peptides containing one Pro residue. Thus, based on interaction studies of eight short model peptides, each comprising four residues containing only one Pro residue preceded by a variety of different amino acids, Harrison and Stein [21] concluded that CypA exhibited a broad tolerance as a prolyl cis/trans isomerase of several substrates. Endrich et al. [22] reported that CypA interacts with three 14-37 residues HIV-1 Capsid-derived peptides, each containing 1-4 Pro residues and at least one Gly-Pro motif. However, exact information regarding the interaction of individual Pro residues within peptides containing more than one Pro unit was not accessible with the applied methodology.
The suitability of NMR spectroscopy for studying the catalytic activity of CypA has been demonstrated in a more limited number of reports. NMR spectroscopy is the only method wherefrom information about prolyl cis/trans isomerase interaction of CypA with individual Pro residues of a peptide, containing several Pro residues, is accessible at atomic resolution. However when determined, only selective interactions involving one of the Pro residues in the peptides [12,23] or the full length protein [13] containing more than one Pro residue have been reported.
In this paper the interactions of CypA with the N-terminal synthetic Vpr (sVpr) peptides sVpr1-20, sVpr21-40 and sVpr25-40, containing all the highly conserved Pro residues of Vpr, have been studied by NMR spectroscopy at atomic resolution. The NMR experiments were performed in aqueous phosphate buffer at physiological pH 7, where CypA retains its enzymatic activity and Vpr exists in an unstructured state [7,19]. Complementary experiments, using SPR spectroscopy and NMR spectroscopy, have provided an unambiguous distinction between the substrate requirements for the catalytic activity of CypA and those for a strong specific binding to N-terminal Vpr. For the first time this has also allowed quantification of this interaction.
Results
The combination and access to both highly pure recombinant CypA and synthetic N-terminal Vpr peptides, as well as sensitive NMR and SPR facilities, has allowed us to probe the interactions of Vpr and CypA in more detail than previously.
Characterization of sVpr1-20, sVpr25-40 and sVpr21-40 by NMR spectroscopy
Previous studies [7,19,24,25] have shown that Vpr has a relatively random structure in aqueous solution at pH 7, but has a propensity for three well-defined α-helical structural domains in aqueous solution at lower pH or in the presence of organic co-solvent (TFE-d2 or CD3CN), where the extent of secondary structure is dependent on the hydrophobicity of the solvent.
The N-terminal region of Vpr includes four highly conserved Pro residues at positions 5, 10, 14 and 35, respectively (Fig. 1) (more than 98% conserved according to references [19] and [26]). Among these, the presence of Pro-35 is vital for several functional interactions of Vpr. The tertiary structure of Vpr observed in aqueous and 30% aqueous acetonitrile solution at low pH is characterized by a hydrophobic core formed by the three α-helices [25] and partly reliant on the presence of the Pro-35 residue that acts as a helix breaker between helix 1 comprising residues 17-33, in which the p6 binding domain of Vpr is located [27], and helix 2 (comprising residues 38-50) of Vpr [28]. At pH 7 Vpr is essentially unstructured in aqueous solution and requires under these conditions a hydrophobic environment to achieve α-helical structure [7,19] (Additional file 1, Fig. S1).
Figure 1.
Interactions of N-terminal Vpr with CypA. Schematic overview of the N-terminal sequence of HIV-1NL4-3 Vpr showing its interactions with CypA that include regions undergoing transient enzymatic prolyl cis/trans catalysis and the region of strong selective binding. In aqueous phosphate buffer at pH 7 N-terminal Vpr peptides, as well as full-length Vpr, are essentially unstructured but assume α-helical structure at lower pH and under hydrophobic conditions [7,19]. Notice that the region of selective strong binding of Vpr with CypA includes the entire loop region connecting regions that form α-helices 1 and 2.
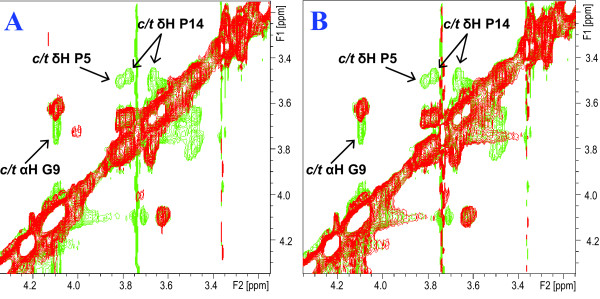
In an initial series of 2D NMR experiments (2D 1H TOCSY, ROESY and NOESY NMR spectra), the 1H chemical shifts of the all-trans isomers of sVpr1-20, sVpr21-40 and sVpr25-40 in aqueous phosphate buffer solution at pH 7 containing 10% v/v D2O were completely assigned (Additional file 1, Table S1, Additional file 1, Table S2, Additional file 1, Table S3, Additional file 1, Table S4, Additional file 1, Table S5, Additional file 1, Table S6 and Additional file 1, Table S7). In accordance with previous investigations in aqueous organic solvent [19] the NMR spectra of sVpr1-20 revealed two sets of 1H NMR signals originating from the same residues in the sequence (Fig. 2). In each case one set of signals was more intense than the second set, and the corresponding amidic protons showed significant chemical shift differences. The fact that the residues showing the largest shift differences were those either adjacent to or very close to each of the Pro residues indicated that the weaker sets of signals in each case arose from the cis isomers of the Pro residues. The identification of sets of signals for Ala-4, Gly-9 and Tyr-15 (Fig. 2A), in addition to the 1H NMR signals originating from the cis isomers of Pro-5 and 14 (Fig. 3), is a strong indication that all Pro residues are involved in such processes under the conditions used. Similarly, 1H NMR signals originating from the cis isomer of Pro-35 were observed in the 2D 1H NMR spectra of sVpr21-40 (Fig. 2B) and sVpr25-40.
Figure 2.
Presence of cis and trans proline isomers in N-terminal sVpr. A1: Fingerprint region of the 2D 1H-1H TOCSY spectrum showing the assigned spin systems of residues close to proline residues in sVpr(1-20) and A2: Superimposed expanded HN-HN regions of the 2D 1H-1H NOESY spectra of sVpr1-20 in H2O:D2O (9:1, v/v) at pH 7 prior to (red signals) and after addition of 50 μl (0.1 mg) CypA (green signals). Notice the appearance of exchange peaks originating from enhanced prolyl cis/trans interconversion rate after addition of CypA. B1: Expanded Hα-Hβ region of the 2D 1H-1H TOCSY spectrum of sVpr21-40 in H2O:D2O (9:1, v/v) at pH 7 showing the signals of trans and cis Pro-35. B2: Superimposed expanded Hα-Hβ regions of the 2D 1H-1H NOESY spectra of sVpr21-40 in H2O:D2O (9:1, v/v) at pH 7 prior to (red signals) and after addition of 100 μl (0.2 mg) CypA (green signals). Notice the appearance of exchange peaks originating from enhanced prolyl cis/trans interconversion rate after addition of CypA.
Figure 3.
Catalysis of isomerisation of Pro-5,10 and 14 of Vpr1-20 by CypA. A: Superimposed expanded Hα-Hβ region of the 2D 1H-1H NOESY spectra of sVpr1-20 in H2O:D2O (9:1, v/v) at pH 7 prior to (red signals) and after addition of 50 μl (0.1 mg) CypA (green signals). Notice the appearance of exchange peaks originating from enhanced prolyl cis/trans interconversion rate after addition of CypA. B: Superimposed expanded Hα-Hβ region of the 2D 1H-1H NOESY spectra of sVpr1-20 in H2O:D2O (9:1, v/v) at pH 7 after addition of 50 μl (0.1 mg) CypA (green signals) and after additional addition of 5 μl (0.1 mg) cyclosporine A (red signals). Notice that the prolyl cis/trans related exchange peaks detected after addition of CypA disappear after addition of cyclosporine A.
It is particularly noteworthy that exchange peaks between analogous 1H signals of the cis and trans isomer were not observed in the NOESY spectra of any of the pure peptides, which is a clear indication that only a slow cis/trans interconversion on the NMR time scale is taking place in the absence of enzymatic catalysis. As a consequence of the relatively large energy barrier, uncatalyzed isomerisation is a rather slow process with an interconversion time on the order of several minutes [29,30].
CypA catalyzes the prolyl cis/trans isomerisation of Pro-35 of sVpr21-40 and sVpr25-40
NMR spectroscopy provides information about prolyl cis/trans isomerase interaction of CypA with Pro-containing peptides and proteins at atomic resolution. Originally, prolyl cis/trans isomerase activity of CypA with relatively short model peptides, containing one Pro residue, was revealed by line shape analysis of 1 D spectra [31-33]. Exchange spectroscopy including 2D 1H-1H NOESY and ROESY NMR experiments have been applied to determine prolyl cis/trans isomerase interactions of CypA with unlabelled model peptides [23,34,35]. Selective interaction of Pro-90 of HIV-1 Capsid was revealed by 3D 15N-edited NOESY-HSQC of 15N-labelled N-terminal (1-146) and full-length HIV-1 Capsid, in addition to a fragment of the HIV-1 Gag polyprotein containing the full Matrix protein and the N-terminal domain of Capsid [12,13].
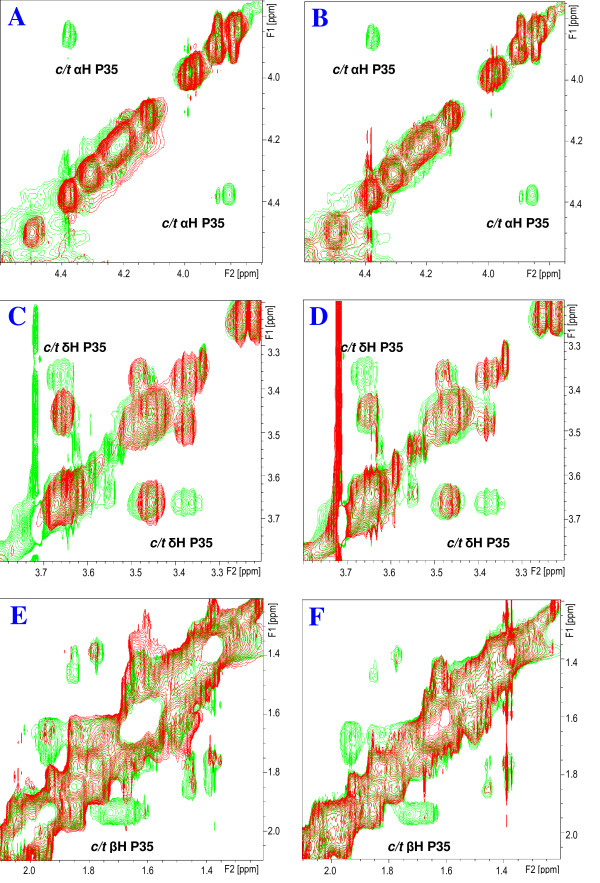
When the complete series of NMR experiments allowing full assignment of the 1H chemical shifts of sVpr21-40 were recorded, CypA was added to the peptide solution at a molar ratio sVpr21-40-CypA 140:1, and analogous NMR experiments were then recorded. In the presence of catalytic amounts of CypA, strong exchange peaks between related signals of the cis Pro-35 and trans-Pro-35 isomer of sVpr21-40 were observed (Fig. 4A, C and 4E). In particular, strong exchange peaks between cis-Hα/trans-Hα and cis-Hδ/trans-Hδ of Pro-35 were observed in the NOESY spectra after addition of catalytic amounts of CypA.
Figure 4.
Catalysis of isomerisation of Pro-35 of Vpr21-40 by CypA. A, C, E: Superimposed expanded Hα-Hβ regions of the 2D 1H-1H NOESY spectra of sVpr21-40 in H2O:D2O (9:1, v/v) at pH 7 prior to (red signals) and after addition of 100 μl (0.2 mg) CypA (green signals). Notice the appearance of exchange peaks originating from enhanced prolyl cis/trans interconversion rate after addition of CypA. B, D, F: Superimposed expanded Hα-Hβ region of the 2D 1H-1H NOESY spectra of sVpr21-40 in H2O:D2O (9:1, v/v) at pH 7 after addition of 100 μl (0.2 mg) CypA (green signals) and after additional addition of 5 μl (0.1 mg) cyclosporine A (red signals). Notice that the prolyl cis/trans related exchange peaks detected after addition of CypA disappear after addition of cyclosporine A.
To confirm that CypA interacts with Pro-35 of sVpr21-40 and Pro-35 of sVpr25-40 as a prolyl cis/trans isomerase, an excess of cyclosporine A (CsA), a selective inhibitor of prolyl cis/trans isomerase activity of CypA was added, and a complete series of NMR spectra were again recorded. The NOESY and ROESY NMR spectra of sVpr21-40, after the sequential addition of CypA and cyclosporine A, revealed that the prolyl cis/trans exchange crosspeaks disappeared after addition of cyclosporine A, and the NMR spectra closely resembled those of pure sVpr21-40 (Fig. 4B, D and 4F). Similar results were observed for sVpr25-40. Thus, CypA behaves as a prolyl cis/trans isomerase, causing an increase in the interconversion rate of Pro-35 of Vpr.
CypA catalyzes the prolyl cis/trans interconversion of Pro-5, -10 and -14 of sVpr1-20
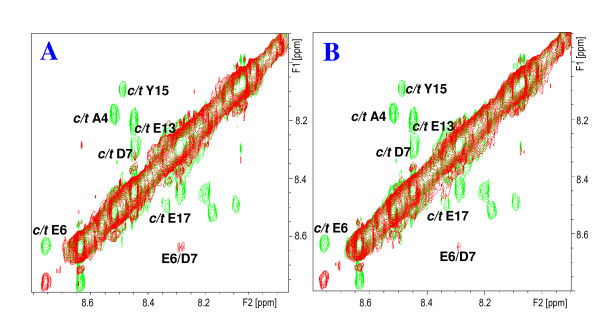
The NMR spectra of sVpr1-20 in aqueous phosphate buffer at pH 7 revealed cis/trans proline isomerism of all Pro residues of this peptide through the observation of separate signals for the all-trans and various cis isomers originating from a slow equilibrium under non-catalytic conditions.
In order to reveal a potential interaction of CypA with the highly conserved Pro-5, -10 and -14 of Vpr, complete series of NMR experiments allowing full assignment of the 1H chemical shifts of sVpr1-20 were recorded, followed by addition of catalytic amounts of CypA (molar ratio sVpr1-20-CypA 224:1; molar ratio sVpr1-20 proline substrate-CypA 672:1). After addition of catalytic amounts of CypA, particularly the HN-HN region of the 2D 1H-1H NOESY NMR spectrum of sVpr1-20 showed strong cis/trans exchange peaks between related protons of the all-trans isomer and various cis isomers originating from cis Pro-5, cis Pro-10 and cis Pro-14, respectively (Fig. 5A). This was further confirmed by the observation of exchange peaks for Hα of Gly-9 and those of Hδ of Pro-5 and Pro-14 (Fig. 3A). As described above for sVpr21-40 containing Pro-35, cyclosporine A was added and a further series of NMR spectra was recorded that showed the disappearance of the prolyl cis/trans exchange crosspeaks (Fig. 3B and 5B). Although Pro-5, -10 and -14 of Vpr are highly conserved [31], any interactions involving these residues have not previously been identified.
Figures 5.
Catalysis of isomerisation of Pro-5,10 and 14 of Vpr1-20 by CypA. A: Superimposed expanded HN-HN regions of the 2D 1H-1H NOESY spectra of sVpr1-20 in H2O:D2O (9:1, v/v) at pH 7 prior to (red signals) and after addition of 50 μl (0.1 mg) CypA (green signals). Notice the appearance of exchange peaks originating from enhanced prolyl cis/trans interconversion rate after addition of CypA. B: Superimposed expanded HN-HN regions of the 2D 1H-1H NOESY spectra of sVpr1-20 in H2O:D2O (9:1, v/v) at pH 7 after addition of 50 μl (0.1 mg) CypA (green signals) and after additional addition of 5 μl (0.1 mg) cyclosporine A (red signals). Notice that the prolyl cis/trans related exchange peaks detected after addition of CypA disappear after addition of cyclosporine A.
Thus we have direct experimental evidence that all four conserved Pro residues in Vpr undergo cis/trans isomerism in aqueous solution at pH 7 that is catalyzed by CypA. Only small amounts of enzyme are required and the NMR method is sufficiently sensitive to detect these effects in ratios of substrate to enzyme as high as 672:1. The applied relative proportions of enzyme to substrate were considerably lower than the relative proportions of CypA (enzyme to substrate 1:8-1:300) used in previous studies with NMR techniques [12,13,23,31-35].
Determination of cis/trans interconversion rate by ROESY NMR experiments
To obtain the rate constant of the prolyl cis/trans interconversion, a series of ROESY experiments with variable mixing times were performed (Additional file 1, Table S8 and Additional file 1, Fig. S2). For each mixing time, volume integration of the well-resolved prolyl cis/trans exchange crosspeaks originating from cisHαPro-35 -transHαPro-35 were performed, and based thereupon, the reaction rate constant was calculated. The mean reaction rate was calculated using the data from ROESY experiments with mixing times 200-500 ms. Mean reaction rate constant for the prolyl cis/trans interconversion was determined to be 0.19 ± 0.01 s-1
Characterization of CypA-Vpr interaction using surface plasmon resonance spectroscopy
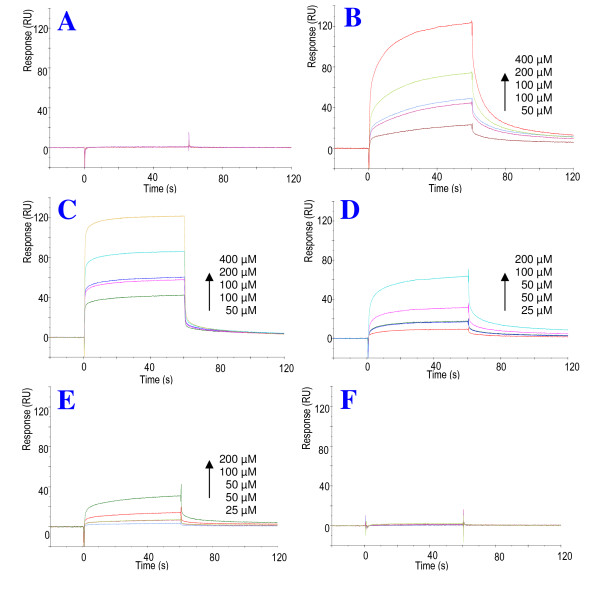
Previous Biacore studies [9] concluded that Pro-35 was a determinant for the ability of N-terminal Vpr peptides to interact with CypA. sVpr1-20 or any mutant N-terminal Vpr peptide lacking Pro-35 did not interact with CypA in the Biacore assay. Pure enzymatic interactions such as the observed catalysis of prolyl cis/trans isomerisation of N-terminal Vpr peptides by CypA will not be detected by SPR because of the short lifetime of the enzyme-substrate (ES) complexes (between 10-7 and 10-4 sec according to references [36] and [37]). We confirmed that the N-terminal peptides sVpr1-40, sVpr21-40 and the mutant sVpr1-40 P5,10,14N bind to CypA, while sVpr1-20 and the mutant sVpr1-40 P35N failed to bind (Fig. 6). Thus, the binding region of N-terminal Vpr peptides for the observed interaction with CypA subsequently involves Pro-35.
Figure 6.
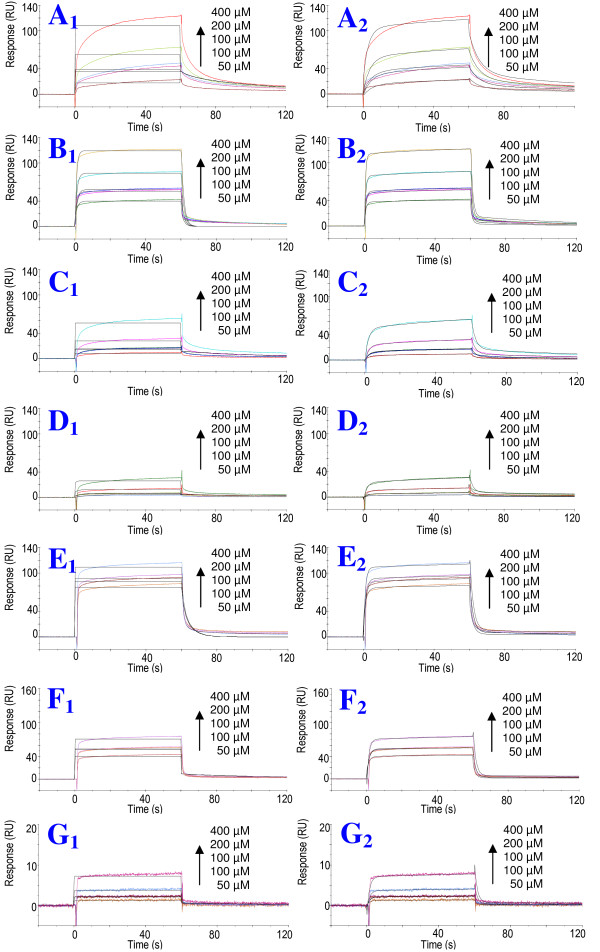
Characterization of CypA binding to N-terminal peptides of sVpr. Synthetic Vpr1-20(A), Vpr21-40 (B), Vpr25-40 (C), Vpr1-40(D) and synthetic mutants carrying Pro to Asn exchanges at amino acid positions 5, 10 and 14(sVpr1-40P5,10,14N) (E) and at position 35 (sVpr1-40 P35N) (F) were tested for binding to immobilized recombinant CypA using a SPR biosensor system. A, B and C were injected at concentrations ranging from 0-400 μM over CM5 chip immobilized with 918 RU CypA, while D, E and F were injected at concentrations ranging from 0-200 μM. The curves A, B, C, D and E were best fit to a two state reaction model (Fig. 7). ka1, ka2, kd1, kd2 and KD were calculated for respective sensograms (Table 1).
Kinetic analysis of the Biacore binding data showed deviations from a pseudo-first order 1:1 binding model. The binding curves of CypA with sVpr25-40, sVpr21-40, sVpr1-40 and the short peptides sVpr30-40 and sVpr32-38 were best described by a two-state binding interaction model (Fig. 7, Additional file 1, Fig. S3). The minor deviation observed between the experimental and modelled curve for sVpr21-40 at the highest concentration, i.e. 400 μM, may be due to aggregation of the peptide at this concentration. The two-state binding model, also called the conformational change model, is based on a 1:1 binding of an analyte to the immobilized ligand followed by a conformational change in the complex. This model should be regarded as indicative, rather than as direct evidence, for a conformational change upon binding [38]. However, our NMR data confirm that CypA acts as a prolyl cis/trans isomerase of the highly conserved Pro residues of N-terminal Vpr including Pro-35, hence a conformational change upon binding is probable. Thus, CypA catalyzes the prolyl cis/trans interconversion during binding to the sVpr peptides. The interconversion rate constant derived from the ROESY NMR experiments was determined to be 0.19 ± 0.01 s-1. Thus, a peptide-CypA complex would undergo approximately 12 prolyl cis/trans interconversions during the 60 sec. association phase of the Biacore experiment. From the two-state binding model the association rate (ka1), dissociation rate (kd1), forward rate (ka2) and backward rate (kd2) constants were calculated, and subsequently the magnitude of the dissociation constants (KD) could be calculated (1/((ka1/kd1)*(ka2/kd2)) (Table 1).
Figure 7.
Optimized fit of SPR sensograms to binding models. The sensograms were fit to the binding models 1:1 (Langmuir) (A1-G1) and two-state reaction (conformational change) model (A2-G2). Black lines show the best curve fit in each instance. The curves originating from the interactions of sVpr21-40 (A), sVpr25-40 (B), sVpr1-40(C), sVpr1-40 P5,10,14N (D), sVpr30-40 (E), sVpr32-38 (F) and sVpr33-37(G) with CypA were considered to fit to two state reaction model (sensogram A2-G2) best.
Table 1.
Estimated kinetic constants for binding of HIV-1 N-terminal Vpr peptides to CypA.
| Peptide | Amino acid sequence | Kinetic constants by two state reaction model | ||
|---|---|---|---|---|
| KD (M) | ka1 (1/Ms) ka2 (1/s) |
kd(1/s) | ||
| sVpr21-40 | ELLEELKSEAVRHFP35 RIWLH | 2.01 × 10-4 | ka1 = 466 ka2 = 0.0084 |
kd1 = 0.164 kd2 = 0.011 |
| sVpr25-40 | ELKSEAVRHFP35 RIWLH | 4.04 × 10-4 | ka1 = 2004 Ka2 = 0.0028 |
kd1 = 0.9415 kd2 = 0.0173 |
| sVpr1-40 | MEQAP5EDQGP10QREP14YNEWTTLELLEELKSEAVRHFP35 RIWLH | 2.84 × 10-4 | ka1 = 752 ka2 = 0.0073 |
kd1 = 0.37 kd2 = 0.0099 |
| sVpr1-40 P5,10,14 N | MEQAN5EDQGN10QREN14YNEWT TLELLEELKSEAVRHFP35 RIWLH |
0.00615* | ka1 = 46* ka2 = 0.0047 |
kd1 = 0.56* kd2 = 0.0048 |
| sVpr30-40 | AVRHFP35 RIWLH | 1.67 × 10-4 | ka1 = 2467 ka2 = 0.00145 |
kd1 = 0.527 kd2 = 0.0053 |
| sVpr32-38 | RHFP35 RIW | 4.87 × 10-4 | ka1 = 924 ka2 = 0.0011 |
kd1 = 0.67 kd2 = 0.0023 |
| sVpr33-37 | HFP35RI | 0.00357 | ka1 = 120.4 ka2 = 0.00138 |
kd1 = 0.66 kd2 = 0.0025 |
*ka1 beyond the values measurable by the Biacore instrument.
The calculated dissociation constants were all of the same order of magnitude for the N-terminal Vpr peptides sVpr1-40, sVpr21-40, sVpr25-40, sVpr30-40 and sVpr32-38 containing Pro-35 (Table 1). Reliable dissociation constants for sVpr1-40 P5,10,14N, which also contains Pro-35 and binds to CypA (Fig. 6E), could not be determined with high confidence. This may be due to relatively poor solubility of the latter peptide in aqueous buffer solution at pH 7.
The two facts, that sVpr1-20 did not bind to CypA in the Biacore assay (Fig. 6) although CypA was shown by our NMR studies to interact with these Pro residues of the peptide as a prolyl cis/trans isomerase (Fig. 3, 5) and that the presence of Pro-35 on the other hand is required for binding, indicate the observed specific binding of N-terminal Vpr peptides to CypA require a binding region that is structurally dependent on Pro-35.
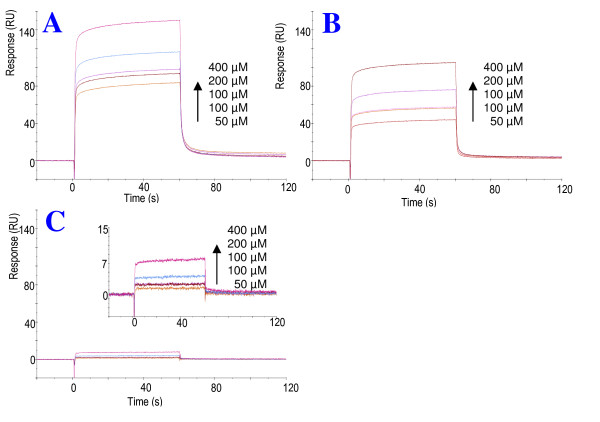
To identify and determine the specific binding region of N-terminal Vpr to CypA, SPR studies of the shorter Vpr peptides sVpr30-40, sVpr32-38 and sVpr33-37 were performed. The sensograms revealed that sVpr30-40and sVpr32-38 maintain the strong binding to CypA similar to that of longer N-terminal Vpr peptides (Fig. 8). In contrast, the shortest peptide sVpr33-37 binds considerably weaker. The remarkably weaker response of the binding curves of sVpr33-37 to CypA compared with the longer peptides, demonstrates that the shortest peptide sequence maintaining strong binding is the heptapeptide sVpr32-38. Thus the seven-residue motif RHFPRIW centred at Pro-35 defines the region for strong specific binding to CypA. In keeping with our findings, Zander et al. [9] and Ardon et al. [10] reported that CypA co-immunoprecipitates with wild-type Vpr, while mutation of Pro-35 caused loss of this phenomenon.
Figure 8.
Characterization of CypA binding region of the N-terminus of Vpr. Synthetic Vpr30-40 (A), Vpr32-38 (B), Vpr33-37(C, D) were tested for binding to immobilized recombinant CypA using SPR biosensor system. Individual peptides were injected at concentration ranging from 0-400 μM over CM5 chip immobilized with 980 RU CypA. The curves were best fit to a two state model, and ka1, ka2, kd1, kd2 and KD were calculated for the respective sensograms (Table 1.).
Discussion
As a continuation of our previous discovery of the interaction of CypA with Vpr, these interactions have in this work been characterized in detail at atomic resolution. Direct experimental evidence that all four conserved Pro residues in Vpr undergo cis/trans isomerism in aqueous solution at pH 7 that is catalyzed by CypA was achieved. Only small amounts of enzyme are required and the NMR method is sufficiently sensitive to detect these effects in ratios of substrate to enzyme as high as 672:1.
The apparent differentiation between the results originating from interaction studies performed by NMR spectroscopy and SPR indicates a different additional mode of interaction observed in the latter case. The fact that only N-terminal Vpr peptides containing Pro-35 bind to CypA in the Biacore assay and strong binding is maintained in the heptapeptide sVpr32-38, although CypA also catalyzes prolyl cis/trans interconversions of Pro-5, 10 and 14 of sVpr1-20 as shown by NMR spectroscopy at atomic resolution, is convincing evidence that CypA only binds specifically to peptides containing the seven-residue motif RHFPRIW centred at Pro-35 of N-terminal Vpr. With the exception of Ile-37, residues comprising the binding region of N-terminal Vpr to CypA are highly conserved [26]. This region includes the loop region connecting helices 1 and 2 of Vpr, in addition to Arg-32 and His-33 that terminates the well-defined helix 1 of Vpr.
The situation found here is analogous to the NMR study reported by Bosco et al. [12] where CypA not only interacts as a chaperone with Pro-90 of N-terminal HIV-1 Capsid but also selectively catalyzes the prolyl cis/trans interconversion of the Gly-89 -Pro-90 bond. Crystal structure of the complex of CypA with the N-terminal HIV-1 Capsid peptide (residues 1-151) revealed that the binding region of HIV-1 Capsid to CypA encompasses the nine residues 85 to 93 (PVHAGPIAP) belonging to a loop region connecting helices 4 and 5, respectively, of HIV-1 Capsid [39]. Based on X-ray crystallography, Vajdos et al. [40] reported that the HIV-1 Capsid-derived hexapeptide HAGP90IA maintained the strong interaction with CypA, thus limiting the binding region of HIV-1 Capsid to include these six residues.
Piotukh et al. [41] used phage display to delineate the requirements for stable binding of CypA to linear peptide motifs containing the consensus motif FGPXLp that is present in many human proteins. This binding motif is quite different from the sequence RHFPRIW found here that defines the binding region of N-terminal Vpr to CypA and also differs from the binding region of HIV-1 Capsid (HAGPIA), indicating that the Pro-dependent binding pocket of CypA is tolerant of substrates with several different sequences.
Pro-35 in Vpr is located in a loop region separating helix 1 and helix 2 [25]. Structural data for an N-terminal mutant of Vpr in which Pro-35 is exchanged for Asn indicate a merge of helices 1 and 2 leading to a significant conformational change in the hydrophobic core of Vpr [11]. This will thus disrupt the hydrophobic interactions of helices 1 and 2 with helix 3 that were observed by long-range NOEs [25]. The tertiary structure of wt Vpr consists of three well-defined α-helices consisting of residues 17-33 (helix 1), 38-50 (helix 2) and 56-77 (helix 3), respectively, folded around a hydrophobic core constituted of Leu, Ile, Val and aromatic residues [25]. According to Morellet et al. [25], the hydrophilic accessible domains of Vpr exposed to the solvent molecules comprise the acidic residues Glu-21, Glu-22, Glu-24, Glu-25, Asn-28 and Glu-29 of helix 1, the basic residues Arg-62, Arg-73, Arg-77, Gln-65 and Gln-66 of helix 3 and the basic residues Arg-80, Arg-87, Arg-88, Arg-90 and Lys-95 of the flexible C-terminal domain, respectively. The fact that the mutant Vpr R80A failed to co-immunoprecipitate with CypA [10] may indicate that the binding of full-length Vpr to CypA is also dependent on the maintenance of the tertiary structure of Vpr. Clearly there are at least two highly conserved residues in the molecule that are crucial for maintaining the interaction of CypA with Vpr, namely Pro-35 and Arg-80.
Previous literature has indicated that CypA is relatively tolerant as a prolyl cis/trans isomerase to a variety of different substrates [21]; however, these studies have been performed exclusively on short peptides comprised of four amino acids and containing only one Pro residue. To our knowledge catalysis by CypA of prolyl cis/trans interconversions of more than one Pro residue in a peptide containing several Pro residues has not previously been reported. However, exclusively selective prolyl cis/trans isomerase catalysis mediated by CypA on only one Pro unit have been reported for peptides containing more than one Pro residue [12,13,23].
The binding curves from Biacore analyses of the specific interaction of Pro-35 were consistent with a two-state binding interaction model involving a conformational change during the binding. This is in accord with the catalysis of prolyl cis/trans isomerism observed by NMR in solution, indicating that the RHFPRIW motif of N-terminal Vpr binds the catalytic site of CypA, which maintains the ability to catalyze the prolyl cis/trans interconversion during binding.
The fact that CypA specifically binds to the inter-helical loop region included in the RHFPRIW binding motif centred at Pro-35, under solution conditions where Vpr has a relatively random structure, suggests a mechanism in which CypA may act as a folding chaperone. Whether this plays a role in the de novo synthesis of Vpr, in which CypA appears to be involved, remains to be determined [9]. Clearly the functional consequences of the interactions of CypA still remain to be determined. Further studies will necessitate the evaluation of these interactions in the context of the full length Vpr molecule in the various environments that it encounters in vivo.
Conclusions
Binding of CypA to a defined inter-helical loop region of HIV-1 Vpr is highly specific, which is analogous to the previously characterised HIV-1 Capsid-CypA interaction. The loop region centred at Pro-35 of Vpr comprising the binding motif to CypA maintains the integrity of the N-terminal and middle helices of Vpr and is functionally important for incorporation of Vpr into virus particles and replication of R5-tropic HIV-1 in human lymphoid tissue [28]. CypA acts as a catalyst of prolyl cis/trans interconversions of Pro 5, 10, 14 and 35 of N-terminal HIV-1 Vpr, which may indicate a folding chaperone role of CypA in HIV replication compatible with the fact that inhibitors of CypA, which inhibit CypA-Vpr binding [9,10] and CypA-mediated prolyl cis/trans catalysis, also suppress HIV-1 replication in cell culture.
Methods
Peptide synthesis
The synthesis, purification and molecular characterization of synthetic Vpr (sVpr) fragments, sVpr1-40, sVpr1-40 P35N, sVpr1-40 P5,10,14N, sVpr1-20, sVpr21-40, sVpr25-40, sVpr30-40, sVpr32-38 and sVpr33-37 were performed as described in detail elsewhere [7,19] and all showed the expected high quality HPLC peaks and correct molecular masses by MALDI-MS and positive ion ESI-MS.
Cyclosporine A and CypA
Cyclosporine A was purchased from Sigma (C3662; CAS number 59865-13-3). The procedure on which the purification of recombinant human CypA is based has previously been described by others [42]. The E. coli strain BL21(DE3) (Invitrogen) was transformed with an pET11c expression vector encoding human CypA [43]. A 4-liter culture of E. coli BL21(DE3) was grown at 37°C in a LB medium containing ampicillin to an A595 of 0.8 and isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. The incubation was continued for additional 8 h. The cells were harvested (3000 g, 15 min), washed with Tris buffer (1 mM DTT, 20 mM TrisHCl, pH 7, 8) and 12 g cell pellet was resuspended in 150 ml of the same buffer. Lysis was performed on ice using a BL12 sonifier (Branson). After separation of the cell debris by centrifugation (48000 g, 15 min) nucleic acids were precipitated by protamine sulphate (0.4% final concentration). The crude cell extract was dialyzed against the Tris buffer overnight and loaded onto a DEAE-Sepharose column (HiPrep16/10, GE Healthcare) equilibrated with 4 column volumes (CV) of Tris-buffer and the flow-through fractions were collected, containing essentially pure CypA. These fractions were combined and dialyzed against a 30 mM MES buffer pH 6.0 overnight. For final purification and concentration of CypA, the dialyzed solution was loaded on a MonoS cation exchange column (5/50 GL, GE Healthcare) which was equilibrated with 4 CV of the MES buffer. The final elution step of the bound CypA was performed with a 0-1 M NaCl gradient and yielded 30 mg of highly concentrated and pure CypA fractions, which were analysed by Coomassie staining and Maldi MS to check the purity and the correct molecular weight of the protein (Additional file 1, Fig. S4).
Mass Spectrometry
Matrix assisted laser desorption ionization mass spectra (MALDI-MS) were recorded on a Voyager-DE PRO BioSpectrometry Workstation from Applied Biosystems. Samples were dissolved in 50% aqueous acetonitrile and α-cyano-4-hydroxycinnamic acid was used as matrix. Positive ion electrospray ionization mass spectra (ESI-MS) were recorded on a micromass Q-Tof-2 mass spectrometer. Samples were dissolved in 70% aqueous methanol and infused into the electrospray chamber with a needle voltage of 0.9 kV at a flow rate of 40 nl/min.
Nuclear Magnetic Resonance (NMR) Spectroscopy
2D 1H Total correlation spectroscopy (TOCSY), Nuclear Overhauser enhancement spectroscopy (NOESY) and Rotating frame Overhauser enhancement spectroscopy (ROESY) NMR experiments were performed at 600.13 MHz on a Bruker Avance 600 MHz instrument equipped with an UltraShield Plus magnet and a triple resonance cryoprobe with gradient unit. Individual samples were dissolved in 600 μl 50 mM aqueous phosphate buffer pH 7.0 containing 10% D2O (v/v), at concentrations between 1-2 mM. The 2D NMR experiments were performed at 300 K without spinning with mixing times of 110 ms for the TOCSY experiments, 250 ms for the NOESY experiments and 500 ms for the ROESY experiments, respectively. Efficient suppression of the water signal was achieved with application of excitation sculpting in the 1 D 1H and the 2D 1H TOCSY and NOESY NMR experiments [44] and presat in the 2D 1H ROESY experiments [45]. 1H signal assignments of the NMR spectra were achieved by identification of the individual spin systems in the 2D 1H TOCSY spectra, combined with observations of sequence-specific short-distance crosspeaks (Hα-HN i, i+1) in the 2D 1H-1H NOESY spectra [9,46]. Readily recognisable spin systems were used as starting points for correlation of the individual spin systems observed in the TOCSY and NOESY spectra with individual residues in the peptide sequences. Acquisition of data, processing and spectral analysis were performed with Bruker Topspin 1.3 software. Assigned NMR data of the N-terminal Vpr peptides have been deposited at the BMRB (Accession number 17003).
Interactions of CypA with sVpr1-20 and sVpr21-40 peptides
After acquisition of 1 D 1H and 2D 1H TOCSY, NOESY and ROESY NMR spectra of pure sVpr1-20 and sVpr21-40, 50-100 μl 0.11 mM CypA dissolved in aqueous Tris buffer at pH 7 was added to the individual peptide solutions, followed by acquisition of identical series of NMR spectra (1 D 1H and 2D 1H TOCSY, NOESY and ROESY) to those of the pure peptides. Exchange peaks occurring in the spectra after addition of CypA were identified by supposition of analogous NOESY and/or ROESY spectra prior to and after addition of CypA using Bruker Topspin 1.3 software.
Determination of rate constant for Pro cis/trans interconversion by ROESY NMR
A series of ROESY experiments of Vpr32-38 (sample concentration 4.2 mM) with addition of 50 μl 0.11 mM CypA (sample concentration of CypA 8.5 μM; total volume 650 μl); relative proportions of substrate to enzyme 494:1, maintaining the same experimental condition as described above but with variable mixing time (500 ms, 450 ms, 400 ms, 350 ms, 300 ms, 250 ms 200 ms 150 ms, 100 ms and 50 ms, respectively) were performed. Volume integration of exchange peaks were carried out with Sparky software [47] and rate constants for each mixing time were calculated as described in Keller et al. [48].
Specific inhibition of prolyl cis/trans isomerase interactions of CypA with sVpr1-20 and sVpr21-40 by addition of cyclosporine A
The catalytic prolyl cis/trans isomerase interactions of CypA with sVpr1-20 and sVpr21-40 was inhibited by addition of excessive amounts of cyclosporine A dissolved in 5 μl deuterated dimethylsulfoxide (DMSO-d6). Addition of 6 μl pure DMSO did not inhibit the catalytic prolyl cis/trans isomerase interactions of CypA with a model peptide showing that CypA keeps its enzyme activity in 1% DMSO solution. The disappearance of NMR exchange peaks, originating from the catalytic prolyl cis/trans isomerase interactions of CypA with sVpr1-20 and sVpr21-40, after addition of cyclosporine A was revealed by supposition of analogous NOESY and ROESY spectra prior to and after addition of cyclosporine A, using Bruker Topspin 1.3 software.
Biacore Spectroscopy
SPR measurements were performed at 25°C on a Biacore T100 instrument (Biacore AB, Uppsala, Sweden) equipped with a CM5 research-grade sensor chip. CypA was immobilized in FC 2 and FC 4 to 180 and 918 RU (response units) respectively, using standard amine-coupling chemistry in two flow cells. FC 1 and FC 3 were treated according to FC 2 and FC 4 except for CypA immobilisation and functioned as reference cells. The synthetic Vpr peptides sVpr1-20, sVpr21-40, sVpr25-40, sVpr30-40, sVpr32-38 and sVpr33-37 were dissolved at four concentrations ranging from 50 to 400 μM in the running buffer (HBS-P buffer pH 7.4). Due to restricted solubility sVpr1-40, sVpr1-40 P35N and sVpr1-40 P5,10,14N were dissolved at lower concentrations ranging from 25-200 μM in the same buffer. The samples were injected over the flow cells at a flow rate of 30 μl/min. Data were collected at 2.5 Hz during the 60 s association and 120 s dissociation phase, and were automatically corrected for bulk buffer effects and unspecific binding of Vpr peptides to the chip matrix.
Analysis of biosensor data
Affinity, association and dissociation rate constants were obtained from sensograms by the Biacore T100 evaluation software version 2.0.1 in accordance with the global curve fit model. Sensorgram data for the four different concentrations were fitted to several binding models including 1:1 (Langmuir) binding model (A+B ↔ AB), two-state reaction (conformational change) model (A+B ↔ AB ↔ AB*) and heterogeneous ligand model (interaction one: A+B1 ↔ AB1; interaction two: A+B2 ↔ AB2). Kinetic constants were calculated for the best fitted model.
Authors' contributions
All authors read and approved the final manuscript. SMS, TRR, US, VW and TF participated in planning the experimental work. SMS, TRR, VW and TF planned and performed the structural and functional NMR studies. RR, PH and PH synthesized the peptides. DM produced CypA. SPR measurements were performed by SMS, OH, AJR, KB and TF. VW, SMS and TF wrote the manuscript.
Supplementary Material
Tables of 1H Chemical shifts of N-terminal Vpr peptides. MALDI-TOF Mass spectrum of CypA. Chemical shift differences of the Hα-protons between the experimental values and those for random coil residues, for sVpr1-20 and sVpr21-40. Calculated reaction rate constants for the prolyl cis/trans interconversion of Pro-35 of sVpr32-38. Optimised fit residuals of SPR sensograms. Peak intensities of cis/trans crosspeaks versus diagonal signal intensities influenced by mixing times observed in the ROESY spectra of sVpr32-38.
Contributor Information
Sara M Solbak, Email: Sara.Solbak@kj.uib.no.
Tove R Reksten, Email: Tove.Reksten@gades.uib.no.
Victor Wray, Email: victor.wray@helmholtz-hzi.de.
Karsten Bruns, Email: bugielski@gmx.de.
Ole Horvli, Email: Ole.Horvli@mbi.uib.no.
Arnt J Raae, Email: arnt.raae@mbi.uib.no.
Petra Henklein, Email: petra.henklein@charite.de.
Peter Henklein, Email: peter.henklein@peptides.de.
Rene Röder, Email: rene.roeder@charite.de.
David Mitzner, Email: D.Mitzner@ViroLogik.com.
Ulrich Schubert, Email: ulrich.schubert@viro.med.uni-erlangen.de.
Torgils Fossen , Email: Torgils.Fossen@kj.uib.no.
Acknowledgements
The authors are grateful to Dr. Manfred Nimtz for recording the mass spectra of CypA and the N-terminal Vpr peptides.
References
- Paxton W, Connor RI, Landau NR. Incorporation of Vpr into Human Immunodeficiency Virus Type 1 Virions: Requirement for the p6 Region of gag and Mutational Analysis. J Virol. 1993;67:7229–37. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MP, Schubert U, Williams SA, de Noronha CMC, Kreisberg JF, Henklein P, Greene WC. HIV-1 Vpr Displays Natural Protein-Transducing Properties: Implications for Viral Pathogenesis. Virology. 2002;302:95–105. doi: 10.1006/viro.2002.1576. [DOI] [PubMed] [Google Scholar]
- Levy DN, Fernandes LS, Williams WV, Weiner DB. Induction of cell differentiation by human immunodeficiency virus 1 vpr. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-Y. [DOI] [PubMed] [Google Scholar]
- Rogel ME, Wu LI, Emerman M. The Human Immunodeficiency Virus Type 1 vpr Gene Prevents Cell Proliferation during Chronic Infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Poon B, Jowett JBM, Chen ISY. Human Immunodeficiency Virus Type 1 Vpr Induces Apoptosis following Cell Cycle Arrest. J Virol. 1997;71:5579–92. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams WV, Green DR, Weiner DB. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor κB. Nature Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- Henklein P, Bruns K, Sherman MP, Tessmer U, Licha K, Kopp J, de Noronha CMC, Greene WC, Wray V, Schubert U. Functional and structural characterization of synthetic HIV-1 Vpr that transduces cells, localizes to the nucleus, and induces G(2) cell cycle arrest. J Biol Chem. 2000;275:32016–32026. doi: 10.1074/jbc.M004044200. [DOI] [PubMed] [Google Scholar]
- de Noronha CMC, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, Greene WC. Dynamic Distruption in Nuclear Envelope Architecture and Integrity Induced by HIV-1 Vpr. Science. 2001;294:1105–1108. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- Zander K, Sherman MP, Tessmer U, Bruns K, Wray V, Prechtel AT, Schubert E, Henklein P, Luban J, Neidleman J, Greene WC, Schubert U. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J Biol Chem. 2003;278:43202–43213. doi: 10.1074/jbc.M305414200. [DOI] [PubMed] [Google Scholar]
- Ardon O, Zimmermann ES, Andersen JL, DeHart JL, Blackett J, Planelles V. Induction of G2 Arrest and Binding to Cyclophilin A Are Independent Phenotypes of Human Immunodeficiency Virus Type 1 Vpr. J Virol. 2006;80:3694–3700. doi: 10.1128/JVI.80.8.3694-3700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Wray V, Schubert U. Role of cyclophilin A in HIV replication. Future Virol. 2007;2:65–78. doi: 10.2217/17460794.2.1.65. [DOI] [Google Scholar]
- Bosco DA, Eisenmesser EZ, Pochapsky S, Sundsquist WI, Kern D. Catalysis of cis/trans isomerisation in native HIV-1 capsid by human CypA. PNAS. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Kern D. Catalysis and Binding of Cyclophilin A with Different HIV-1 Capsid Constructs. Biochemistry. 2004;43:6110–6119. doi: 10.1021/bi049841z. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Göttlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Rosenwirth B, Billich A, Datema R, Donatsch P, Hammerschmid F, Harrison R, Hiestand P, Jaksche H, Mayer P, Peichl P, Quesniaux V, Schatz F, Schuurman HJ, Traber R, Wenger R, Wolff B, Zenke G, Zurini M. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppresive cyclosporine analog. Antimicrob Agents Chemother. 1994;38:1763–1772. doi: 10.1128/aac.38.8.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billich A, Hammerschmid F, Peichl P, Wenger R, Zenke G, Quesniaux V, Rosenwirth B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporine A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV Protein-cyclophilin A interactions. J Virol. 1995;69:2451–2461. doi: 10.1128/jvi.69.4.2451-2461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke EK, Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology. 1996;222:279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- Dorfman T, Weimann A, Borsetti A, Walsh CT, Göttlinger HG. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns K, Fossen T, Wray V, Henklein P, Tessmer U, Schubert U. Structural Characterization of the HIV-1 Vpr N Terminus. J Biol Chem. 2003;278:43188–43201. doi: 10.1074/jbc.M305413200. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HEH, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Harrison RK, Stein RL. Substrate Specificities of the Peptidyl Prolyl Cis- Trans Isomerase Activity of Cyclophilin and FK-506 Binding Protein: Evidence for the Existence of a Family of Distinct Enzymes. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. [DOI] [PubMed] [Google Scholar]
- Endrich MM, Gehrig P, Gehrig H. Maturation-induced Conformational Changes of HIV-1 Capsid Protein and Identification of Two High Affinity Sites for Cyclophilins in the C-termainal Domain. J Biol Chem. 1999;274:5326–5332. doi: 10.1074/jbc.274.9.5326. [DOI] [PubMed] [Google Scholar]
- Kern D, Drakenberg T, Wikström M, Forsèn S, Bang H, Fischer G. The cis/trans interconversion of the calcium regulating hormone calcitonin is catalyzed by cyclophilin. FEBS. 1993;323:198–202. doi: 10.1016/0014-5793(93)81338-Z. [DOI] [PubMed] [Google Scholar]
- Wecker K, Morellet N, Bouaziz S, Roques BP. NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol - Comparison with the Vpr N-terminal (1-51) and C-terminal (52-96) domains. Eur J Biochem. 2002;269:3779–3788. doi: 10.1046/j.1432-1033.2002.03067.x. [DOI] [PubMed] [Google Scholar]
- Morellet N, Bouaziz S, Petitjean P, Roques BP. NMR structure of the HIV-1 regulatory protein Vpr. J Mol Biol. 2003;327:215–227. doi: 10.1016/S0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Ayyavoo V, Mahalingam S, Kannan A, Boyd A, Datta D, Kalyanaraman V, Cristillo A, Collman RG, Morellet N, Sawaya BE, Murali R. A comprehensive analysis of the naturally occurring polymorphisms in HIV-1 Vpr: Potential impact on CTL epitopes. Virology journal. 2008;5:99. doi: 10.1186/1743-422X-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins Y, Pornillos O, Rich RL, Myszka DG, Sundquist WI, Malim MH. Biochemical Analyses of the Interactions between Human Immunodeficiency Virus Type 1 Vpr and p6Gag. J Virol. 2001;75:10537–42. doi: 10.1128/JVI.75.21.10537-10542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Studtrucker N, Sörgel S, Münch J, Rücker E, Kirchhoff F, Schick B, Henklein P, Fossen T, Bruns K, Sharma A, Wray V, Schubert U. Proline 35 of human immunodeficiency virus type 1 (HIV-1) Vpr regulates the integrity of the N-terminal helix and the incorporation of Vpr into virus particles and supports the replication of R5-tropic HIV-1 in human lymphoid tissue ex vivo. J Virol. 2007;81:9572–9576. doi: 10.1128/JVI.02803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grathwohl C, Wüthrich K. NMR Studies of the Rates of Proline Cis-Trans Isomerisation in Oligopeptides. Biopolymers. 1981;20:2623–2633. doi: 10.1002/bip.1981.360201209. [DOI] [Google Scholar]
- Nicholson LK, Lu KP. Prolyl cis-trans isomerisation as a molecular timer in Crk signaling. Mol Cell. 2007;25:483–485. doi: 10.1016/j.molcel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Hsu VL, Handschumacher RE, Armitage IM. Peptidyl-Prolyl Cis-Trans Isomerase Activity of Cyclophilin Studied by One-Dimensional 'H Nuclear Magnetic Resonance Spectroscopy. J Am Chem Soc. 1990;112:6145–6141. doi: 10.1021/ja00174a064. [DOI] [Google Scholar]
- Hübner D, Drakenberg T, Forsén S, Fischer G. Peptidyl-prolyl cis-trans isomerase activity as studied by dynamic proton NMR spectroscopy. FEBS. 1991;284:79–81. doi: 10.1016/0014-5793(91)80766-V. [DOI] [PubMed] [Google Scholar]
- Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Kinetic Analysis of Cyclophilin-Catalyzed Prolyl CisRrans Isomerisation by Dynamic NMR Spectroscopy. Biochemistry. 1995;34:13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- Reimer U, Drewello M, Jakob M, Fischer G, Schutkowski M. Conformational state of a 25-mer peptide from the cyclophilin-binding loop of the HIV type 1 capsid protein. Biochem J. 1997;326:181–185. doi: 10.1042/bj3260181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCornack MA, Kakalis LT, Caserta C, Handschumacher RE, Armitage IM. HIV protease substrate conformation: modulation by cyclophilin A. FEBS Lett. 1997;414:88. doi: 10.1016/S0014-5793(97)00974-5. [DOI] [PubMed] [Google Scholar]
- Reuben J. Substrate anchoring and the catalytic power of enzymes. PNAS. 1971;68:563–565. doi: 10.1073/pnas.68.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme Dynamics During Catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- Rich RL, Myszka DG. Survey of the year 2006 commercial optical biosensor literature. J Mol Recognit. 2007;20:300–366. doi: 10.1002/jmr.862. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/S0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Vajdos FF, Yoo S, Houseweart M, Sundquist WI, Hill CP. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Prot Sci. 1997;6:2297–2307. doi: 10.1002/pro.5560061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotukh K, Gu W, Kofler M, Labudde D, Helms V, Freund C. Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J Biol Chem. 2005;280:23668–23674. doi: 10.1074/jbc.M503405200. [DOI] [PubMed] [Google Scholar]
- Liu J, Albers MW, Chen CM, Schreiber SL, Walsh CT. Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc Natl Acad Sci USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grättinger M, Hohenberg H, Thomas D, Wilk T, Müller B, Kräusslich HG. In Vitro Assembly Properties of Wild-Type and Cyclophilin-Binding Defective Human Immunodeficiency Virus Capsid Proteins in the Presence and Absence of Cyclophilin A. Virology. 1999;257:247–260. doi: 10.1006/viro.1999.9668. [DOI] [PubMed] [Google Scholar]
- Hwang TL, Shaka AJ. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J Magn Reson. 1995;112:275–279. doi: 10.1006/jmra.1995.1047. [DOI] [Google Scholar]
- Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley and Sons, Inc. New York; 1986. [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California, San Francisco; http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- Keller M, Sager C, Dumy P, Schutkowski M, Fischer GS Mutter M. Enhancing the Proline Effect: Pseudo-Prolines for Tailoring Cis/Trans Isomerisation. J Am Chem Soc. 1998;120:2714–2720. doi: 10.1021/ja973966s. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables of 1H Chemical shifts of N-terminal Vpr peptides. MALDI-TOF Mass spectrum of CypA. Chemical shift differences of the Hα-protons between the experimental values and those for random coil residues, for sVpr1-20 and sVpr21-40. Calculated reaction rate constants for the prolyl cis/trans interconversion of Pro-35 of sVpr32-38. Optimised fit residuals of SPR sensograms. Peak intensities of cis/trans crosspeaks versus diagonal signal intensities influenced by mixing times observed in the ROESY spectra of sVpr32-38.