Abstract
The motions of mitotic chromosomes are complex and show considerable variety across species. A wealth of evidence supports the idea that microtubule-dependent motor enzymes contribute to this variation and are important both for spindle formation and for the accurate completion of chromosome segregation. Motors that walk towards the spindle pole are, however, dispensable for at least some poleward movements of chromosomes in yeasts, suggesting that depolymerizing spindle microtubules can generate mitotic forces in vivo. Tubulin protofilaments that flare outward in association with microtubule shortening may be the origin of such forces, because they can move objects that are appropriately attached to a microtubule wall. For example, some kinetochore-associated proteins can couple experimental objects, such as microspheres, to shortening microtubules in vitro, moving them over many micrometers. Here, we review recent evidence about such phenomena, highlighting the force-generation mechanisms and different coupling strategies. We also consider bending filaments of the tubulin-like protein FtsZ, which form rings girding bacteria at their sites of cytokinesis. Mechanical similarities between these force-generation systems suggest a deep phylogenetic relationship between tubulin depolymerization in eukaryotic mitosis and FtsZ-mediated ring contraction in bacteria.
Keywords: Microtubule, FtsZ, Motility, Mitosis, Chromosome motion, Coupler, Bacterial cleavage
Introduction
The assembly of molecules into ordered arrays can do mechanical work. The freezing of water can crack rock, and the assembly of proteins into fibers can push on subcellular structures. For example, growing microtubules (MTs) can deform the surfaces of protozoans (Tilney, 1968), polymerizing deoxygenated, sickle-cell hemoglobin can distort red blood cells (Samuel et al., 1990), actin assembly can help advance the leading edge of migrating cells (Pollard and Borisy, 2003) and polymerizing actin homologs can segregate chromosomes for prokaryotic cell division (Kruse and Gerdes, 2005). The ability of disassembling structure to do work is, however, less well established. It was first suggested as a mechanism for chromosome motion in the 1940s (Oestergren, 1949). As the reality of spindle fibers became evident, models based on MT dynamics were developed to explain many aspects of chromosome motion (Inoue and Sato, 1967). The simplicity of polymerization-based motility enhanced the popularity of these models, but exactly how a chromosome holds onto a shortening spindle fiber was not spelled out. Moreover, the early observations supporting this hypothesis (Inoue et al., 1975) were made on cells containing a full complement of motor enzymes; so the observed motions could have been caused by minus-end-directed motors, with MT shortening serving as a regulator that permitted motion without causing it. Thus, for many years the evidence for a depolymerization motor in mitosis was largely circumstantial.
Kirschner and colleagues pioneered the investigation of chromosome–MT interactions in vitro (Mitchison and Kirschner, 1985). They showed that depolymerizing MTs remained bound to isolated, coverslip-attached mammalian chromosomes (Koshland et al., 1988). However, they presented no evidence that mechanical work was actually done as these MTs shortened, and the possibility of a contribution from ATP-dependent motors was not rigorously excluded. Subsequent work from our laboratory addressed these issues through a system in which isolated chromosomes moved by MT depolymerization against a flow of buffer while nucleotide concentrations were driven below nanomolar range by addition of apyrase. These results demonstrated that tubulin depolymerization could do mechanical work in vitro (Coue et al., 1991). Antibodies against CENP-E, a kinetochore-localized kinesin, blocked this depolymerization-dependent, nucleotide-independent movement (Lombillo et al., 1995a). Analogous motions were seen with inert microspheres coated by MT-dependent motor enzymes; for example, a plus-end-directed motor was induced to move backwards under the influence of tubulin depolymerization in either the absence or presence of ATP (Lombillo et al., 1995b). From this work it emerged that MT shortening is, indeed, a motor – but to see its action in vitro one needs an effective coupling between the cargo under study and the shortening MT.
In recent years, several features of this depolymerization-dependent motor have been characterized. Ideas about how MT shortening can do work are now well developed, although there is disagreement on how tubulin depolymerization is coupled to its cargo. This Commentary summarizes aspects of recent studies on tubulin depolymerization and juxtaposes them with studies on the bacterial tubulin-like protein FtsZ. The results suggest that the mechanism by which tubulin polymers can exert force as they shorten is ancient and related to the machinery for bacterial cell cleavage.
MT depolymerization is a motor in eukaryotic cells
Hoyt and colleagues showed that mitosis can work in the budding yeast Saccharomyces cerevisiae with only two MT-dependent motors: a twofold symmetric, homo-tetrameric kinesin 5 that helps spindles to form; and a second motor that promotes tubulin depolymerizaton (Cottingham et al., 1999). Mitosis limped along well enough to sustain colony growth, even when cells contained only kinesin 5 and a mutated depolymerizing motor, so long as a drug was added to enhance MT depolymerization. However, these studies did not include direct observation of chromosome motions. More recent work from our laboratory has shown that, in the fission yeast Saccharomyces pombe, the deletion of all three minus-end-directed MT-dependent motors (the ones that could pull a chromosome poleward) does not reduce the maximal speed of such motions (Fig. 1) (Grishchuk and McIntosh, 2006). This result has been confirmed in budding yeast (Tanaka et al., 2007) and expanded in fission yeasts (Franco et al., 2007; Gachet et al., 2008), leading to the conclusion that minus-end-directed motors are dispensable for chromosome-to-pole motion in two rather distantly related yeasts.
Fig. 1.
Fission yeast chromosomes move poleward in the absence of pole-directed motor enzymes. (A,B) Cells were deprived of MTs by growing a cold-sensitive mutant of β-tubulin at a restrictive temperature (18°C) for 6 hours; this caused the kinetochores to lose their pole-proximal association (Grishchuk and McIntosh, 2006). Upon reversion to permissive conditions (32°C), MTs grew from the centrosomes and encountered the now-dispersed chromosomes. Subsequent chromosome motions were monitored using fluorescence microscopy. (A) Three S. pombe cells from a 3- to 10-minute sample showing stages in chromosome attachment to the spindle. White arrows suggest an inferred progression. Chromosomes, purple; centrosomes, green; kinetochores, red. (B) Fluorescence imaging of a living cell in which both poles (green arrowheads) and one kinetochore (red arrowheads) were tagged with GFP. Imaging was done over a period of 10 minutes. SPB, spindle pole body. (C) Four-dimensional microscopy allowed quantification of the rate of chromosome-to-pole motion in different mutant strains of motor proteins. The maximal rate of chromosome motion was unchanged in those strains that lack the kinesin 14 family members Pkl1 and Klp2 as well as the dynein heavy chain (Dhc1) (pkl1Δklp2Δdhc1Δ; green), revealing that these motors are not at the root of poleward chromosome motion. (D,E) Although the absence of minus-end-directed motors had a relatively minor impact on mitotic progression in wild-type cells (D), the motors contributed to the accuracy and expediency of the poleward motion and bi-orientation when cells were challenged by chromosome scatter (E). Adapted from Grishchuk and McIntosh with permission (Grishchuk and McIntosh, 2006). Chromosomes, purple; centrosomes, green; kinetochores, red (for panels A,D,E).
In higher eukaryotes the associations of chromosomes with spindle fibers in early mitosis are often between kinetochores and MT walls; the resulting pole-directed chromosome motions are probably driven by dynein (Yang et al., 2007). With this exception, motor inhibition in most well-studied cells cause more problems for spindle assembly than for chromosome movement per se. When kinetochore motors are compromised at metaphase by injection of antibody (Sharp et al., 2000), mutation (Garcia et al., 2002; Mao et al., 2010; Grishchuk et al., 2007; Tanaka et al., 2007) or RNAi of components required for motor–kinetochore association (Wordeman et al., 2007; Yang et al., 2007) the results are generally changes in chromosome speed and/or an increase in the rate of chromosome loss, but the chromosomes continue to move. Thus, it is likely to be some engine other than a minus-end-directed motor that is the fundamental driver for chromosome-to-pole motion in vivo.
One could argue that these mitotic motions are based on actin or some unknown motile machinery, but two additional findings support the hypothesis that tubulin depolymerization is a motor for chromosome motion in vivo: the aforementioned fact that MT depolymerization can move chromosomes at physiological rates in vitro and the observation that inhibition of MT dynamics with low concentrations of Taxol blocks mitosis without increasing MT polymer mass (Jordan et al., 1993). Understanding the mechanical properties of tubulin depolymerization is, therefore, important for understanding cellular mechanisms for chromosome motion.
Finally, it is worth noting that MTs can lose subunits from either end. Depolymerization from the pole-associated minus end of MTs might have a role in both the flux of kinetochore MTs towards the spindle pole (Mitchison and Salmon, 1992) and the segregation of some chromosomes during anaphase (LaFountain et al., 2001). In this Commentary we look only at kinetochore-associated plus-end depolymerization, partly because this is the important mechanism for anaphase in both yeast and mammals, partly because this process is better described and partly because we believe that the mechanics of depolymerization are likely to be the same at both MT ends.
Transmitting the force of MT depolymerization
The first fully developed theoretical model for attaching a load to a depolymerizing MT was based on a hypothetical ‘sleeve’, which was proposed to surround each kinetochore-associated MT near its plus end, establishing many weak bonds with the MT wall (Hill, 1985). These weak bonds allowed the sleeve to diffuse on the MT so its position was not fixed. It could not, however, diffuse off the MT end, because that would require breaking all bonds. Tubulin depolymerization therefore biased the diffusion of the sleeve, allowing the energy released by MT shortening to do work on objects attached to the sleeve. More recent treatments of this hypothesis have pointed out several weaknesses, such as its inability to accommodate the expanded shape of a depolymerizing MT end (Efremov et al., 2007) or to withstand substantial counter forces (Joglekar and Hunt, 2002; Molodtsov et al., 2005a). The latter is of particular significance because spindles can exert large forces when a chromosome is stalled, either by experiment (Nicklas, 1997) or entanglement. Nonetheless, biased diffusion of a loosely bound structure is a physically realistic way of coupling MT shortening to the movement of a light load (Grishchuk et al., 2010), so it remains a popular idea in the field.
Marc Kirschner and colleagues brought the issue of MT structure into focus on the basis of their early findings that depolymerizing MTs formed curved tubulin oligomers, e.g. rings and helices (Kirschner et al., 1975). They proposed that in a MT wall the strands of tubulin, the protofilaments (PFs), curve as they depolymerize, making a conformational ‘wave’ that would allow a chromosome to ‘surf’ on the MT end as the polymer shortened (Koshland et al., 1988). These interpretations paved the way for a different view of depolymerization-dependent generation of force and coupling between cargo and MT.
The presence of bending PFs at the ends of depolymerizing MTs was demonstrated using electron microscopy (EM) of MTs that were rapidly frozen while depolymerizing in vitro, then imaged in the frozen-hydrated state (Mandelkow et al., 1991). Numerous subsequent studies have helped to characterize the changes in tubulin shape associated with GTP hydrolysis (Chretien et al., 1995; Elie-Caille et al., 2007; Muller-Reichert et al., 1998; Wang and Nogales, 2005), providing strong support for the idea that GDP–tubulin in MTs is strained within the MT wall. Stressed lateral bonds in the GTP-containing MT cap prevent PFs from adopting their more curved, minimum-energy shape. A relaxation of this stress during bending is now generally thought to be part of the pathway for GDP–tubulin depolymerization (for a simulation of MT depolymerization, see supplementary material Movie 1). Although some structural evidence suggests that a tubulin dimer is bent regardless of the nucleotide it binds (Rice et al., 2008), the balance of current evidence supports the idea that MT shortening works simply through the action of PFs that relax to a higher curvature as they depolymerize.
Bending MT protofilaments can generate force
Direct in-vitro assays have demonstrated that PFs that bend at the plus end of a shortening MT can, indeed, generate force (Grishchuk et al., 2005). We attached a microsphere to a MT wall using beads coated with streptavidin and MTs assembled from biotinylated tubulin. First, polymers were initiated from coverslip-bound nucleators, elongated with biotinylated tubulin and then capped with biotin-free, Rhodamine-labeled tubulin assembled in the presence of GMPCPP, a slowly hydrolyzed analogue of GTP that makes MTs unusually stable (Fig. 2A). Soluble tubulin could now be rinsed out, avidin-coated beads washed in, and many of them attached to the tethered, biotinylated MTs. We used laser tweezers to trap individual beads and then a pulse of bright light to bleach the Rhodamine – which dispersed the stable tubulin cap – inducing the bead-associated MT to depolymerize. When the end of the shortening MT reached the bead, the bead experienced a brief jerk that was visible with a quadrant photo-diode (Fig. 2B). This confirmed the reality of curling PFs at the end of a shortening MT and allowed a rather direct measure of the force these conformational changes could generate.
Fig. 2.
Tubulin depolymerization exerts force on inert objects coupled tightly to a MT wall. (A,B) Experimental setup to measure the depolymerization force. MTs initiated from coverslip-bound nucleators grow with their plus ends distal. A microsphere coated with streptavidin binds to biotinylated MT segments (blue) and can be captured in a laser trap (red cone) as well as monitored with a quadrant photodiode (not shown). Although Brownian movement renders the position of the bead variable (see photodiode data in B), MT depolymerization pulls the bead briefly, moving it away from the center of the trap until the bead-associated tubulin is fully depolymerized. The bead then relaxes to the center of the trap. (C) Relative size of bead (0.25 μm radius in this drawing) and the MT. The lateral mode of the attachment of the bead, the large distance between the MT attachment site (where the force is exerted) and the center of the bead (where the force is measured with a laser trap) cause the measured force to be less than that developed by the bending PFs. (D) Consistent with the interpretation given in C, beads of smaller diameter report a larger force (compare black horizontal bars). Yellow vertical bars indicate the range of force predicted theoretically. Adapted from Grishchuk et al. (Grishchuk et al., 2008a).
The observed force was small – on average ~0.2 pN, with a maximum of 0.5 pN (for comparison, it takes about 6 pN to stall a normal motor enzyme). We have argued, however, that the force actually generated by bending PFs is much larger than the measured value, given the geometry of the system. Our wall-attached beads had a radius of 250–1000 nm, whereas the MT radius is ~12 nm; bending PFs are usually less than 50 nm long. If – as the chemistry suggests – the bead is bound to the MT wall, the bending PFs would act at the surface of the bead, whereas the laser trap acts at its center (Fig. 2C). Thus, the trap has a huge mechanical advantage over the bending PF(s) (Grishchuk et al., 2005). This idea is supported by the fact that smaller beads report larger forces from MT depolymerization (Fig. 2D). One must admit, however, that the exact geometry of bead–MT interaction is unknown, either in our work or in that of others who do similar studies, so there is some question about the right way to interpret the observations. When our interpretation is used, one bending PF generates about the same force as an ATP-dependent motor enzyme; when one considers that there are 13 PFs in each MT, a depolymerizing MT that is correctly coupled to a chromosome should exert ~10× the force of a kinesin motor. This force approaches the theoretical maximum defined by the energy of GTP hydrolysis, and it is on the same order as the probable force per MT observed by Bruce Nicklas when he stalled chromosome motion in anaphase (for a review, see Nicklas, 1997). No wonder the ATP-dependent action of kinetochore motors is dispensable for chromosome-to-pole motion.
Coupling objects to shortening MTs
The depolymerization-dependent motion of streptavidin-coated beads is short; this kind of static coupling cannot produce processive motion and is a bad model for chromosome motion in vivo. How, then, do cells attach objects to MTs so they can follow depolymerization over biologically useful distances? It has long been recognized that cargos bound to MT walls could be pulled poleward if depolymerization were at the poles (Margolis and Wilson, 1981), but if depolymerization were at the kinetochore, the problem would be more acute. How can a tip-attached cargo move if subunits are leaving from the tip itself, and how can the cargo stay bound if it is attached at the site where subunits are leaving? The sleeve in Terrell Hill's model attaches to the wall at a place where it can include the tip (Hill, 1985). Both Koshland et al. as well as Inoue and Salmon proposed lateral attachments near the MT end, and the latter paper added the suggestion of fibrillar binding directly to the tip (Koshland et al., 1988; Inoue and Salmon, 1995). Our early evidence showed that bead-coupled motor enzymes, which probably attached to the MT wall, could follow a depolymerizing end (Lombillo et al., 1995b), but the results admitted the possibility that these beads rolled as they moved (Grishchuk et al., 2008b; Peskin and Oster, 1995) – a poor model for mitosis. Work at that time with non-motor MT-binding proteins provided no evidence for processive motions, and the problem lay dormant for several years.
Ring-shaped couplers
MT–cargo coupling was opened to rigorous investigation following the discovery that a kinetochore protein complex from yeast – the Dam1 or DASH complex (Cheeseman et al., 2001) – can bind to and form rings around MTs in vitro (Miranda et al., 2005; Wang et al., 2007; Westermann et al., 2005). Initially, fluorescence microscopy suggested that these complexes diffuse along a MT surface and follow depolymerization over many micrometers (Westermann et al., 2006). Additional work with laser tweezers showed that beads coated with the Dam1 complex attached to MT tips in such a way that, when pulled in the direction of MT polymerization, they would ride with the growing MT end (Asbury et al., 2006). When the same MT started to depolymerize, the Dam1-coated beads followed the shortening end. This shortening could even continue against a small load, albeit for only a few hundred nanometers (Franck et al., 2007).
Structural studies of MTs coated with Dam1 revealed individual rings and extended helices, permitting detailed analysis of Dam1 complexes bound to MT walls (Miranda et al., 2007; Wang et al., 2007). The positively charged C-terminus of the Dam1 protein itself, probably in combination with parts of other proteins in this complex, projected in towards the MT wall where interaction with the negatively charged C termini of α- and/or β-tubulin was possible. Because early observations of fluorescent Dam1 complexes bound to MTs in vitro showed rather rapid one-dimensional diffusion (Westermann et al., 2006), and EM showed predominantly ring-shaped aggregates of Dam1 on MTs, the work was interpreted to mean that Dam1 rings could slide easily over the MT surface. This suggests that they move with a shortening MT end through biased diffusion (supplementary material Movie 2), providing an elegant mechanism for chromosome–MT coupling and provoking a resuscitation of the ideas of Terrell Hill – with the sleeve recast as a ring and a conformational wave as the ‘limiter’ of ring diffusion (Davis and Wordeman, 2007; Efremov et al., 2007).
Dam1 forms more than just rings
Subsequent work from our group has used EM to characterize Dam1 oligomers as they associate with MTs bound through biotin-avidin interaction to an electron-transparent film. When samples were aldehyde-fixed before negative staining, we found many oligomers that were curved but not ring-shaped (Fig. 3A). Moreover, we and others have used fluorescence microscopy to quantify the number of Dam1 complexes in oligomers associated with MT walls, and have shown that many of them are small and diffused quickly (Fig. 3B) (Gestaut et al., 2008; Grishchuk et al., 2008b). Bigger oligomers, e.g., those containing ≥16 subunits, bind the MT so strongly that their diffusion is negligible (Grishchuk, 2008b), suggesting that earlier work – which showed Dam1 diffusion – was carried out on small Dam1 oligomers not rings. Apparently, the early EM evidence that MT-associated Dam1 formed only rings was misleading. Perhaps the smaller oligomers were washed off the MT wall by the negative stain and were too disordered to appear distinct when using cryoEM.
Fig. 3.
The Dam1 complex forms rings and smaller oligomers that can serve as processive couplers to MT depolymerization. (A) EM images (right panels) of a negatively stained MT that is either attached to an immobilizing film, followed by incubation without (top) and with soluble Dam1 (middle) or that is not attached and incubated together with soluble Dam1 (bottom). (Left panels) Schematic of the same experimental setup. (B,C) Fluorescent studies reveal that all Dam1 oligomers (but not stacks of multiple rings) exhibit some diffusion and move with a shortening MT end; but their rates of diffusion (B) and tracking shortening MTs (C) decrease with increasing oligomer size (error bars in B are ± s.e.m.; curve in C is a hyperbolic fit to the data and vertical bar at each point shows the 95% confidence level). (D) Dam1-coated beads follow the shortening MT ends by two distinct mechanisms, depending on the presence of soluble Dam1. (E) Large oligomers allow a shortening MT to push on a bead with a force that is approximately five times greater than that seen with a biotin–avidin linkage. Shown are unprocessed quadrant photodiode records from a Dam1-coated bead in the presence of soluble Dam1 and a streptavidin-coated bead (inset). Adapted from Grishchuk et al. (Grishchuk et al., 2008b).
Although this issue is still open to dispute, our interpretation of current evidence is that, at saturating concentrations, Dam1 self-associates to form helices around MTs but, at more physiological Dam1:MT ratios, Dam1 forms curved oligomers that vary in size. Small oligomers, probably even monomers, can become MT associated, show 1-D diffusion and even track the shortening MT ends. Ring-sized oligomers are, however, unlikely to diffuse because they slow the rate of MT disassembly (Fig. 3C), and several rings stacked together will even cause depolymerization to pause until some of these complexes disassemble (Grishchuk et al., 2008b).
Strikingly, both small oligomers and rings of Dam1 can couple objects to MTs in ways that support motion with MT shortening. Microbeads coated with Dam1 move processively with MT shortening, and they do so freely enough not to slow MT depolymerization (Asbury et al., 2006); in our hands they even increase the rate at which MTs shorten relative to the depolymerization rate of free MTs. However, these Dam1-coated beads roll as they move, so they cannot be coupled by a sliding ring, suggesting again a poor model for chromosome motion (Grishchuk et al., 2008b). When Dam1 is bead-bound and in solution, the beads do not roll and the speed at which MTs shorten is lower, just as is the case for ring-sized Dam1 complexes without beads attached (Fig. 3D). These beads are pulled by the MT with a force approximately fivefold greater than that observed with the biotin-avidin linkage (Fig. 3E), consistent with there being a ring of Dam1 that surrounds the MT, allowing many PFs to push in parallel (Grishchuk et al., 2008a).
Biased diffusion or something else?
The steady motion of ring-shaped Dam1 oligomers with shortening MT ends, whereas similarly sized oligomers show negligible diffusion on a stable MT wall, suggests that the ring moves because of a diffusion-independent mechanism, prompting us to call this kind of motion a ‘forced walk’ (supplementary material Movie 3) (Efremov et al., 2007). This latter mechanism, which combines tight binding and depolymerization-dependent motion, has many advantageous properties for kinetochore-MT coupling: processive motion, strong binding and thus a low probability of payload loss during polymer shortening, and good control on MT dynamics (supplementary material Movie 4). This might be particularly important in cells, such as budding yeast, that have only one MT associated with each kinetochore. Thus, Dam1 has caused much excitement among students of mitosis.
For all these reasons it was disappointing to learn – by using bioinformatics – that the subunits of the Dam1 complex are not widely conserved: the relevant proteins have been found only in fungi. Moreover, although these subunits are present in fission yeast, they are not essential (as shown for Dam1 deficient S. pombe cells) (Sanchez-Perez et al., 2005). There must, therefore, be other kinds of couplers that can harness the energy available from MT shortening to the motion of biological cargos. Considerable attention has now been addressed to other kinetochore proteins that are MT associating and more widely conserved – be they motor enzymes or not.
In search of a universal chromosome–MT coupling
The most conserved of all MT-binding kinetochore proteins is the Ndc80 complex, a fibrous hetero-tetramer that associates with the MT wall at one end and proteins of the inner kinetochore at the other (Ciferri et al., 2005; Wei et al., 2007). Microbeads coated with nematode Ndc80 complex, oriented with its MT-binding end protruding, can follow the end of a depolymerizing MT, albeit with lesser processivity than Dam1-coated beads (McIntosh et al., 2008) (Fig. 4A). Also, beads coated with either human or yeast Ndc80 will track shortening MTs (Powers et al., 2009), although it is not yet known whether these beads roll as they move. The Ndc80 complex diffuses quite quickly on the MT surface (Fig. 4B), so objects coupled to MTs by this molecule may, in principle, move through biased diffusion (Powers et al., 2009). Alternatively, this complex could provide fibrillar coupling to PF power strokes (McIntosh et al., 2008), as discussed below. Ndc80 may be widely used for coupling mitotic kinetochores to spindle fibers, but the details of its biophysics remain to be characterized.
Fig. 4.
Bead tracking assays to identify kinetochore–MT couplers. (A) Microbeads coated with the Ndc80 complex attach to tethered MTs (left panel) and follow their ends as they shorten (right panel); time-lapse images of a moving bead, adapted from McIntosh et al. with permission (McIntosh et al., 2008). (B) Fluorescent Ndc80 molecules diffuse on a MT wall in a low-ionic-strength buffer, as illustrated with a kymograph. Reproduced with permission from Powers et al. (Powers et al., 2009). (C) Comparison of the coupling properties of different kinetochore proteins (for more details see supplementary material Table S1). By the two criteria of percent processive beads (red bars) and mean interaction time (green bars and ± s.e.m.), the Dam1 complex provides the most efficient bead transport, followed by its presumptive ortholog, the Ska1-containing complex.
Kinetochores contain many additional proteins, some of which bind MTs, e.g. components of the KNL1-Mis12-complex–Ndc80-complex (KMN) network (Cheeseman et al., 2006). We have tried this complex as a coupler, both in toto and in parts, but none of these mixtures has yet provided coupling as processive as Dam1 (Fig. 4C and supplementary material Table S1). The more recently discovered Ska1 complex offers another option as it, too, is kinetochore associated (Gaitanos et al., 2009; Hanisch et al., 2006) and exhibits MT binding (Welburn et al., 2009). Ska1 can couple beads to MTs and produce depolymerization-dependent movements over a distance of several micrometers; it is better than Ndc80 but, again, not as processive as Dam1. There is, however, no quantitative assessment of just how processive a coupler must be to serve well in anaphase. Certainly, some anaphase chromosomes move many micrometers, but most kinetochores bind multiple MTs; cooperative MT action might confer processivity, even when the attachment to one polymer is limited.
Kinetochore-localized motor enzymes might contribute additional kinetochore–MT coupling. Kinesin 8 from fission yeast is a plus-end-directed, kinetochore-associated motor that can couple beads to MTs; in the absence of ATP it delivers quite processive, minus-end-directed motion in vitro (Grissom et al., 2009) (supplementary material Table S1). Indirect evidence also implies that CENP-E is capable of a mechanically analogous activity for mammalian chromosomes (Lombillo et al., 1995a), so motor enzymes working in an ATP-independent manner must also be considered as potential couplers. Together, these studies pose the question: what does the connection between a kinetochore and a spindle MT really look like?
Structural studies of kinetochore–MT interaction
Light microscopy does not, a priori, offer sufficient resolution to reveal the molecular structure of a kinetochore–MT interface, but recent work with fluorescently labeled kinetochore proteins has provided insights into both their abundance (Joglekar et al., 2008) and their order along the centromere-to-pole axis (Joglekar et al., 2009; Schittenhelm et al., 2007; Wan et al., 2009). These studies have defined important constraints on the molecular organization of kinetochores but they have not yet told us which kinetochore components hold onto MTs as they grow and shrink in mitosis.
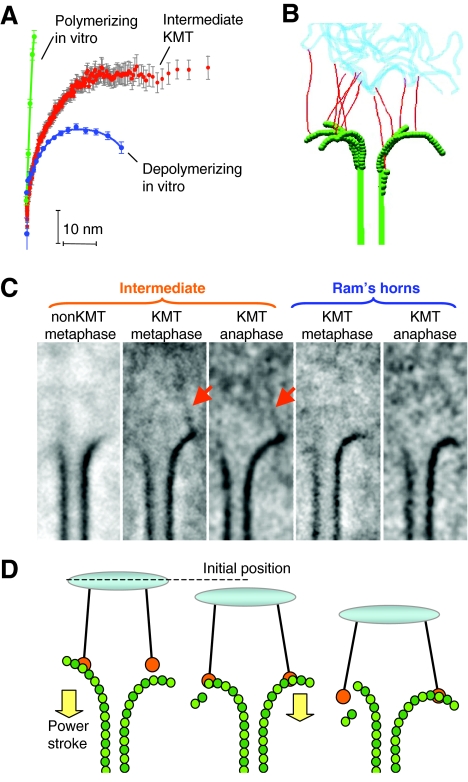
Structural studies using conventional EM have long shown a darkly staining outer kinetochore plate into which the plus ends of MTs insert (Brinkley and Stubblefield, 1966). More recent work with cells prepared by rapid freezing and freeze-substitution fixation has confirmed that kinetochore-associated MTs end in a fibrous network (Dong et al., 2007), however, the outer plate has become less distinct as fixation methods have improved (McEwen et al., 1998). In yeast (Grishchuk et al., 2007; O'Toole et al., 1999) and nematode blastomeres (O'Toole et al., 2003) there is no detectable outer plate; kinetochore-associated MTs simply end near the chromatin in a web of fibrous material. Strikingly, almost half of the PFs at the end of kinetochore-associated MTs show a distinct shape: their curvature is intermediate between that found on PFs at the ends of shrinking and growing polymers of purified tubulin (Fig. 5A).
Fig. 5.
Fibrillar structures between kinetochores and spindle MTs, and their possible role in force-transducing coupling. (A) The average shape of PFs from kinetochore-associated MTs (KMTs) is intermediate between those of polymerizing and depolymerizing MTs. (B) This shape may be caused by fibrillar attachments between the bending PFs and chromosomes, which serve as a load; drawing based on electron tomography (fibrils, red; MTs, green; chromatin, blue). (C) Fibrils connect these intermediate protofilaments with chromatin in averaged images of kinetochore-associated MTs from cultured mammalian cells (red arrows). The more-bent PFs, called ‘Ram's horns’ lack these connections in averaged images. Adapted from McIntosh et al. with permission (McIntosh et al., 2008). (D) Consecutive stages in this hypothetical coupling; non-diffusing fibrils pull on the attached cargo because of power-strokes from bending PFs.
Careful examination of such ends in a mammalian cell has identified slender fibrils that connect bending PFs directly with the underlying chromatin (McIntosh et al., 2008) (Fig. 5B). Individual fibrils are at the limit of what can reliably be detected using cellular EM, but we found about one to two such structures per PF in mitotic mammalian cells. Fibrillar connections are also seen in averaged images of many kinetochore MT ends, in which the PFs have been aligned and then averaged (Fig. 5C). Because the fibrils were not used for alignment, their presence in an average supports their reality, slender as they are. Work now ongoing in our laboratory has confirmed the existence of similar structures in the alga Chlamydomonas, blastomeres of the nematode Caenorhabditis elegans, and in both fission and budding yeasts. The macromolecules that comprise these fibrils are not yet known, but these structural results reinforce our interest in fibrous couplers. The fibrils might include Ndc80, KNL1, long kinetochore-associated motors or other, not-yet-recognized kinetochore proteins. Identification and characterization of the molecules that comprise these structures will go a long way towards clarifying how cells couple mitotic cargos to MT depolymerization.
Models for fibril–MT coupling
Rings provide an intuitively straightforward mechanism for coupling a load to MT depolymerization, but MT-binding fibrils are initially less satisfying. One can imagine a mechanism on the basis of weak MT binding, in which one end of a fibril diffuses rapidly over the MT surface while the other retains a firm attachment to its load. MT shortening might then bias the diffusion of the MT-associated end (Powers et al., 2009). This mechanism should work well for small loads, but it cannot maintain attachment when the load is high; the result is either cargo loss or an inhibition of MT depolymerization (Grishchuk et al., 2010). These behaviors call into question the efficacy of weakly bound fibrils as components at the heart of chromosome motion. An alternative is, however, available in the PF power-stroke-dependent mechanism mentioned above. If fibrils bind strongly both to their load and to polymerized tubulin, then PF bending will pull on the fibrils and, thus, on the load (Fig. 5D). If, in addition, the affinity of the fibril for soluble tubulin is low, then the dissociation of a fibril-associated tubulin dimer from the distal end of a bending PF will release that fibril, allowing it to be recycled. This process would permit a release of kinetochore-attached fibrils from fully bent PFs and their subsequent engagement with those that are less bent or just about to bend, providing a continuous pull on the load (supplementary material Movie 5). Such a mechanism offers both efficient and processive coupling, even with high loads and pausing MTs (McIntosh et al., 2008). Our calculations show that processive motion of a 40 pN load is readily achieved by this mechanism. This load is within the range of the ~80 pN maximum defined by the energy release of GTP hydrolysis, which sets an upper limit on the work a depolymerizing MT can do. However, if binding between fibril and PF is not strong enough, the PF may peel away from its load and the motion would not be processive (supplementary material Movie 6).
Chromosomal oscillations during prometaphase and metaphase imply that one kinetochore can experience a higher force than its sister and move towards the pole it faces. When this happens, the sister kinetochore must permit tubulin addition so that associated MTs can elongate and the spindle-fiber attachment can be retained (Cassimeris et al., 1988). Thus, the kinetochore–MT coupling in its broad sense must include mechanisms that allow a cargo to follow an elongating MT as well as one that is shortening. A fibril-based model to couple a large load to a growing MT end is not yet available.
Bending tubulin protofilaments may be an ancient mechanism for biological motility
The ability of tubulin PFs to transition from a straight conformation in the MT wall to considerably curved (>20° per monomer in a Ram's horn) appears to have been around for a long time: ‘flared’ ends are found on MTs in all groups of eukaryotes examined so far. It is too early to say whether such ends are commonly involved in the generation of force, but the properties of one well-studied tubulin homolog suggest that force generation through bending protofilaments is widespread and ancient. The FtsZ protein found in most Eubacteria, some Archaea and in chloroplasts contributes to cleavage in prokaryotic cells.
FtsZ is a GTP-binding member of the tubulin family that assembles into linear, non-tubular filaments, both in vitro and in vivo (Erickson and Stoffler, 1996; Li et al., 2007; Lutkenhaus, 1993). It is a homolog of eukaryotic tubulin (Desai and Mitchison, 1998; Nogales et al., 1998) and essential for bacterial cytokinesis, where it bundles to form the Z-ring that girds the cell at its equator prior to and during cytokinesis. The Z-ring is attached by associating proteins to the inner surface of the plasma membrane. After bacterial chromosomes have separated, the ring decreases in diameter as the plasma membrane pinches in (Fig. 6A). An allele of FtsZ, engineered to have a membrane anchor, has been purified and introduced into liposomes in vitro. Over time, these rings constricted and indented the liposomes, demonstrating that FtsZ is sufficient to induce membrane bending (Osawa et al., 2008) (Fig. 6B,C). Polymers of FtsZ, as seen using EM and atomic force microscopy, are clearly curved in vitro (Erickson and Stoffler, 1996; Hamon et al., 2009) and in vivo (Fig. 6D), and there are indications that their minimum energy shape acquires greater curvature in association with the hydrolysis of protein-bound GTP (Erickson, 2009). This idea is somewhat confounded by the observation that FtsZ with reduced GTPase activity can still support bacterial cell cleavage (Redick et al., 2005), but it is clear that a change in the curvature of linear FtsZ oligomers can generate force on membranes to which they are attached (Osawa et al., 2009). Thus, a tubulin homolog in bacteria is a morphogenetic motor that uses changes in curvature to shape the plasma membrane in a manner dependent on cell cycle stage.
Fig. 6.
Organization and function of FstZ filaments in eubacteria. (A) Fusion of FtsZ protein with GFP forms a band at the mid-region of a bacterial cell. This band decreases in diameter as the cell cleaves (image kindly provided by J. Lutkenhaus, University of Kansas, Kansas City, KS). (B) Purified FtsZ–YFP chimera, engineered to include a membrane-binding domain. This protein induces indentations (arrows) in tubular liposomes in vitro, demonstrating its sufficiency for bending membranes. (C) The same liposome imaged before and after a 6-minute interval, scale bar 5 μm. Adapted from Osawa et al. with permission (Osawa et al., 2008). (D) Slices cut from cryo-electron tomograms of cleaving Caulobacter cells that are overexpressing FtsZ. Straight and curved segments of FtsZ filaments are visible in these 6.7-nm slices. Abrupt kinks are sometimes seen (black arrows) as well as direct connections of straight filaments to the membrane (white arrowheads); scale bar 50 nm. Adapted from Li et al. with permission (Li et al., 2007). (E) Summary of curvature-dependent cellular functions of bending tubulin-like proteins (blue) and their attaching links (red) to a cargo (black).
The mechanical analogy between bending filaments of FtsZ and bending PFs of tubulin is obvious. Both are linear assemblies of tubulins, and each alters its curvature in response to a change in its biochemical environment. Both sets of bending filaments are coupled to a payload to do their job: the plasma membrane for FtsZ, and the kinetochore for tubulin. The process of FtsZ coupling to the membrane is quite well understood, but that of tubulin coupling to the kinetochore is still under investigation. Nonetheless, these two systems are mechanically similar (Fig. 6E), suggesting that an ancestral protein also functioned as a bending engine used for mechanical purposes by an ancient cell.
The ability of proteins to alter their shape in response to ligand binding is certainly not unique to members of the tubulin family. All motor enzymes use this trick as part of their mechanochemical cycles, and many other enzymes display analogous conformational changes. Members of the tubulin family are unique in their ability to modify their shape while associated with other cellular components. These changes empower dynamic tubulins to act on big cellular objects, such as a chromosome or a cell membrane, altering its position or shape. The ability to affect cellular structure over long distances through local interactions and structural changes is a hallmark of cytoskeletal polymers.
Perspective
Both FtsZ and dimeric tubulin can assemble into linear strands whose minimum energy curvature is under cellular control. On the one hand, tubulin curvature in MTs can be seen only at the end of the polymer because PF bending is inhibited by interactions with neighboring tubulins in the cylindrical MT wall. The tubular wall is, on the other hand, useful for allowing a long-range connection between the MT end that is fastened to its initiator and the other end, which might be many micrometers away. The distal end, behaving a little like the growth cone of an axon, can explore the surrounding volume, probing for objects to which it binds (Kirschner and Mitchison, 1986). If the binding includes an appropriate coupler, subsequent tubulin depolymerization will allow PF bending to pull, providing an effective motile system that can search, capture and gather. The membrane-coupled fibers of FtsZ, however, can change their curvature in response to an altered cellular environment, affecting the shape of the membrane to which they are bound. Given the mechanical and phylogenetic relatedness of these proteins across a wide evolutionary gap, the biological engines they make up are probably among the oldest now available for study.
Both systems have clearly been modified as time has passed and selection has acted. For example, the efficacy of motility based on MT depolymerization has been enhanced by the addition of ATP-dependent motors that drive depolymerization and other proteins, such as mitotic motors and checkpoints that greatly increase mitotic accuracy – factors of great biological significance. The regulation of PF bending in today's cells might be defined both by tubulin depolymerases and MT tip-associated proteins. Nonetheless, the root of mitosis as we know it is probably found in the bending PFs that spindle MTs share with tubulin homologs in distantly related organisms.
Supplementary Material
Acknowledgments
The work from our laboratory reviewed here was supported in part by grants GM033287 and RR000592 to J.R.M., and RFBI-09-04-12077 and RFBI-09-04-05085 to F.I.A. The authors thank members of their laboratories for many fruitful discussions, M. Molodtsov and A. Efremov for theoretical modeling of depolymerization-dependent processes, A. Efremov, A. Zheleznjakov, E. Salova and M. Molodtsov for their contributions to the supplementary movies, and I. Cheeseman for kinetochore proteins used in some of the bead studies and for his critical reading of the manuscript. Deposited in PMC for release after 12 months.
Footnotes
This article is part of a Minifocus on microtubule dynamics. For further reading, please see related articles: ‘Microtubule plus-end tracking proteins (+TIPs)’ by Anna Akhmanova and Michel O. Steinmetz (J. Cell Sci. 123, 3415-3419), ‘Kinesins at a glance’ by Sharyn A. Endow et al., (J. Cell Sci. 123, 3420-3424), ‘Towards a quantitative understanding of mitotic spindle assembly and mechanics’ by Alex Mogilner and Erin Craig (J. Cell Sci. 123, 3435-3445) and ‘Post-translational modifications of microtubules’ by Dorota Wloga and Jacek Gaertig (J. Cell Sci. 123, 3447-3455).
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/20/3425/DC1
References
- Asbury C. L., Gestaut D. R., Powers A. F., Franck A. D., Davis T. N. (2006). The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 103, 9873-9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B. R., Stubblefield E. (1966). The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma 19, 28-43 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Inoue S., Salmon E. D. (1988). Microtubule dynamics in the chromosomal spindle fiber: analysis by fluorescence and high-resolution polarization microscopy. Cell Motil. Cytoskeleton 10, 185-196 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Brew C., Wolyniak M., Desai A., Anderson S., Muster N., Yates J. R., Huffaker T. C., Drubin D. G., Barnes G. (2001). Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155, 1137-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983-997 [DOI] [PubMed] [Google Scholar]
- Chretien D., Fuller S. D., Karsenti E. (1995). Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 129, 1311-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K. J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E. D., Musacchio A. (2005). Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280, 29088-29095 [DOI] [PubMed] [Google Scholar]
- Cottingham F. R., Gheber L., Miller D. L., Hoyt M. A. (1999). Novel roles for saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147, 335-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M., Lombillo V. A., McIntosh J. R. (1991). Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 112, 1165-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Wordeman L. (2007). Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends Cell. Biol. 17, 377-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. (1998). Tubulin and FtsZ structures: functional and therapeutic implications. BioEssays 20, 523-527 [DOI] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K. J., Meng X., Khodjakov A., McEwen B. F. (2007). The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9, 516-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A., Grishchuk E. L., McIntosh J. R., Ataullakhanov F. I. (2007). In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc. Natl. Acad. Sci. USA 104, 19017-19022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elie-Caille C., Severin F., Helenius J., Howard J., Muller D. J., Hyman A. A. (2007). Straight GDP-tubulin protofilaments form in the presence of taxol. Curr. Biol. 17, 1765-1770 [DOI] [PubMed] [Google Scholar]
- Erickson H. P. (2009). Modeling the physics of FtsZ assembly and force generation. Proc. Natl. Acad. Sci. USA 106, 9238-9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P., Stoffler D. (1996). Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J. Cell Biol. 135, 5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A. D., Powers A. F., Gestaut D. R., Gonen T., Davis T. N., Asbury C. L. (2007). Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9, 832-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A., Meadows J. C., Millar J. B. (2007). The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J. Cell Sci. 120, 3345-3351 [DOI] [PubMed] [Google Scholar]
- Gachet Y., Reyes C., Courtheoux T., Goldstone S., Gay G., Serrurier C., Tournier S. (2008). Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol. Biol. Cell 19, 1646-1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T. N., Santamaria A., Jeyaprakash A. A., Wang B., Conti E., Nigg E. A. (2009). Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28, 1442-1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. (2002). Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21, 6015-6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D. R., Graczyk B., Cooper J., Widlund P. O., Zelter A., Wordeman L., Asbury C. L., Davis T. N. (2008). Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10, 407-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., McIntosh J. R. (2006). Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25, 4888-4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Molodtsov M. I., Ataullakhanov F. I., McIntosh J. R. (2005). Force production by disassembling microtubules. Nature 438, 384-388 [DOI] [PubMed] [Google Scholar]
- Grishchuk E. L., Spiridonov I. S., McIntosh J. R. (2007). Mitotic chromosome biorientation in fission yeast is enhanced by dynein and a minus-end-directed, kinesin-like protein. Mol. Biol. Cell 18, 2216-2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Efremov A. K., Volkov V. A., Spiridonov I. S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J. R., Ataullakhanov F. I. (2008a). The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA 105, 15423-15428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Spiridonov I. S., Volkov V. A., Efremov A., Westermann S., Drubin D., Barnes G., Ataullakhanov F. I., McIntosh J. R. (2008b). Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA 105, 6918-6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., McIntosh J. R., Molodtsov M., Ataullakhanov F. I. (2010). Force generation by dynamic microtubule polymers. In Biophysics, Vol. 7 (ed. Goldman Y. E., Ostap E. M.). Elsevier; (in press) [Google Scholar]
- Grissom P. M., Fiedler T., Grishchuk E. L., Nicastro D., West R. R., McIntosh J. R. (2009). Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol. Biol. Cell 20, 963-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon L., Panda D., Savarin P., Joshi V., Bernhard J., Mucher E., Mechulam A., Curmi P. A., Pastre D. (2009). Mica surface promotes the assembly of cytoskeletal proteins. Langmuir 25, 3331-3335 [DOI] [PubMed] [Google Scholar]
- Hanisch A., Sillje H. H., Nigg E. A. (2006). Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25, 5504-5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. L. (1985). Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA 82, 4404-4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Salmon E. D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Sato H. (1967). Cell motility by the labile association of molecules: the nature of the mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 50, 259-292 [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Fuseler J., Salmon E. D., Ellis G. W. (1975). Functional organization of mitotic microtubules. Physical chemistry of the in vivo equilibrium system. Biophys. J. 15, 725-744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Hunt A. J. (2002). A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys. J. 83, 42-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Salmon E. D., Bloom K. S. (2008). Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 85, 127-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Bloom K., Salmon E. D. (2009). In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19, 694-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Toso R. J., Thrower D., Wilson L. (1993). Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. USA 90, 9552-9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329-342 [DOI] [PubMed] [Google Scholar]
- Kirschner M. W., Honig L. S., Williams R. C. (1975). Quantitative electron microscopy of microtubule assembly in vitro. J. Mol. Biol. 99, 263-276 [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Mitchison T. J., Kirschner M. W. (1988). Poleward chromosome movement driven by microtubule depolymerization in vitro. Nature 331, 499-504 [DOI] [PubMed] [Google Scholar]
- Kruse T., Gerdes K. (2005). Bacterial DNA segregation by the actin-like MreB protein. Trends Cell. Biol. 15, 343-345 [DOI] [PubMed] [Google Scholar]
- LaFountain J. R., Jr, Oldenbourg R., Cole R. W., Rieder C. L. (2001). Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell 12, 4054-4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Trimble M. J., Brun Y. V., Jensen G. J. (2007). The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694-4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo V. A., Nislow C., Yen T. J., Gelfand V. I., McIntosh J. R. (1995a). Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 128, 107-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo V. A., Stewart R. J., McIntosh J. R. (1995b). Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 373, 161-164 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. (1993). FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9, 403-409 [DOI] [PubMed] [Google Scholar]
- Mandelkow E. M., Mandelkow E., Milligan R. A. (1991). Microtubule dynamics and microtubule caps: a time-resolved cryo- electron microscopy study. J. Cell Biol. 114, 977-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Varma D., Vallee R. 2010. Emerging functions of force-producing kinetochore motors. Cell Cycle 9, 715-719 [DOI] [PubMed] [Google Scholar]
- Margolis R. L., Wilson L. (1981). Microtubule treadmills-possible molecular machinery. Nature 293, 705-711 [DOI] [PubMed] [Google Scholar]
- McEwen B. F., Hsieh C. E., Mattheyses A. L., Rieder C. L. (1998). A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107, 366-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., Volkov V. A., Cheeseman I. M., Desai A., Mastronarde D. N., Ataullakhanov F. I. (2008). Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell 135, 322-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C. (2005). The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12, 138-143 [DOI] [PubMed] [Google Scholar]
- Miranda J. J., King D. S., Harrison S. C. (2007). Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol. Biol. Cell 18, 2503-2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Kirschner M. W. (1985). Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J. Cell Biol. 101, 755-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. (1992). Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119, 569-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov M. I., Grishchuk E. L., Efremov A. K., McIntosh J. R., Ataullakhanov F. I. (2005a). Force production by depolymerizing microtubules: a theoretical study. Proc. Natl. Acad. Sci. USA 102, 4353-4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov M. I., Ermakova E. A., Shnol E. E., Grishchuk E. L., McIntosh J. R., Ataullakhanov F. I. (2005b). A molecular-mechanical model of the microtubule. Biophys. J. 88, 3167-3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T., Chretien D., Severin F., Hyman A. A. (1998). Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate. Proc. Natl. Acad. Sci. USA 95, 3661-3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B. (1997). How cells get the right chromosomes. Science 275, 632-637 [DOI] [PubMed] [Google Scholar]
- Nogales E., Downing K. H., Amos L. A., Lowe J. (1998). Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5, 451-458 [DOI] [PubMed] [Google Scholar]
- Oestergren G. (1949). Luzula and the mechanism for chromosome movement. Hereditas 35, 445-468 [Google Scholar]
- Osawa M., Anderson D. E., Erickson H. P. (2008). Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D. E., Erickson H. P. (2009). Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28, 3476-3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M., McIntosh J. R. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017-2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E. T., McDonald K. L., Mantler J., McIntosh J. R., Hyman A. A., Muller-Reichert T. (2003). Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163, 451-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin C. S., Oster G. F. (1995). Force production by depolymerizing microtubules: load-velocity curves and run-pause statistics. Biophys. J. 69, 2268-2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453-465 [DOI] [PubMed] [Google Scholar]
- Powers A. F., Franck A. D., Gestaut D. R., Cooper J., Gracyzk B., Wei R. R., Wordeman L., Davis T. N., Asbury C. L. (2009). The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136, 865-875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redick S. D., Stricker J., Briscoe G., Erickson H. P. (2005). Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187, 2727-2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. M., Montabana E. A., Agard D. A. (2008). The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA 105, 5378-5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel R. E., Salmon E. D., Briehl R. W. (1990). Nucleation and growth of fibres and gel formation in sickle cell haemoglobin. Nature 345, 833-835 [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T., Millar J. B. (2005). The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24, 2931-2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R. B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., Lehner C. F. (2007). Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. (2000). Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2, 922-930 [DOI] [PubMed] [Google Scholar]
- Stewart R. J., Thaler J. P., Goldstein L. S. (1993). Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophila ncd protein. Proc. Natl. Acad. Sci. USA 90, 5209-5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T. U. (2007). Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178, 269-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G. (1968). Studies on the microtubules in heliozoa. IV. The effect of colchicine on the formation and maintenance of the axopodia and the redevelopment of pattern in Actinosphaerium nucleofilum (Barrett). J. Cell Sci. 3, 549-562 [DOI] [PubMed] [Google Scholar]
- Wan X., O'Quinn R. P., Pierce H. L., Joglekar A. P., Gall W. E., DeLuca J. G., Carroll C. W., Liu S. T., Yen T. J., McEwen B. F., et al. (2009). Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. W., Ramey V. H., Westermann S., Leschziner A. E., Welburn J. P., Nakajima Y., Drubin D. G., Barnes G., Nogales E. (2007). Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol. 14, 721-726 [DOI] [PubMed] [Google Scholar]
- Wei R. R., Al-Bassam J., Harrison S. C. (2007). The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14, 54-59 [DOI] [PubMed] [Google Scholar]
- Welburn J. P., Grishchuk E. L., Backer C. B., Wilson-Kubalek E. M., Yates J. R., 3rd, Cheeseman I. M. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 16, 374-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., Drubin D. G., Nogales E., Barnes G. (2005). Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17, 277-290 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H. W., Avila-Sakar A., Drubin D. G., Nogales E., Barnes G. (2006). The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440, 565-569 [DOI] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., von Dassow G. (2007). MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179, 869-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U. S., Wadsworth P., Rieder C. L. (2007). Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17, 973-980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.