Abstract
Much effort has been spent recently in identifying host factors required for HIV-1 to effectively replicate in cultured human cells. However, much less is known about the genetic factors in vivo that impact viral replication in lymphatic tissue, the primary anatomical site of virus-host interactions where the bulk of viral replication and pathogenesis occur. In order to identify genetic determinants in lymphatic tissue that critically affect HIV-1 replication, we used microarrays to transcriptionally profile and identify host genes expressed in inguinal lymph nodes that were associated determinants of viral load. Strikingly, ~95% of the transcripts (558) in this data set (592 transcripts total) were negatively associated with HIV-1 replication. Genes in this subset (1) inhibit cellular activation/proliferation (ex.: TCFL5, SOCS5 and SCOS7, KLF10), (2) promote heterochromatin formation (ex.: HIC2, CREBZF, ZNF148/ZBP-89), (3) increase collagen synthesis (ex.: PLOD2, POSTN, CRTAP), and (4) reduce cellular transcription and translation. Potential anti-HIV-1 restriction factors were also identified (ex.: NR3C1, HNRNPU, PACT). Only ~5% of the transcripts (34) were positively associated with HIV-1 replication. Paradoxically, nearly all these genes function in innate and adaptive immunity, particularly highlighting a heightened interferon system. We conclude that this conventional host response cannot contain HIV-1 replication and, in fact, could well contribute to increased replication through immune activation. More importantly, genes that have a negative association with virus replication point to target cell availability and potentially new viral restriction factors as principal determinants of viral load.
Introduction
Over the last decade, systems biology has taken on an increasingly important role in investigating microbial diseases, delineating salient features of the host-pathogen relationship, and identifying potential host genes that are critical determinants of microbial replication and pathogenesis. In the case of HIV-1, which like any obligate intracellular pathogen relies on the transcriptional and translational machinery of the host cell to complete its life cycle (1–3), these studies have revealed components of host gene expression that establish a favorable intracellular environment for efficient virus replication. For example, genomics-based approaches have thus far documented changes in gene expression in cultured cells during HIV-1 infection (4), and more recently, siRNA technology has identified hundreds of host genes seemingly indispensable for HIV-1 replication in vitro (5–8).
In contrast, much less is known about host genes that play important roles in viral replication in vivo where HIV-1 replicates in the complex environment of lymphatic tissue (LT)3 in the context of a host responding to infection. In previous microarray studies of HIV-1 infection in LT, we have shown that infection massively perturbs host gene expression and that this transcriptional profile is highly dependent on stage of disease (9). Here we report studies that go beyond this initial identification of stage-specific features of the host response in LT to now identify genes that play important roles in viral replication in vivo. We now show that: (1) there is little overlap between genes in vivo compared to genes in vitro that correlate with viral replication; (2) paradoxically, host immune responses correlate with high viral loads; and (3) ~95% of the correlations are inverse correlations that point to the importance of target cell availability, cellular activation, transcriptional factors, and new inhibitors as determinants of viral load in vivo.
Materials & Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of the University of Minnesota. All patients provided written informed consent for the collection of samples and subsequent analysis.
Lymph node biopsy specimens
Inguinal lymph node biopsies from 22 untreated HIV-1-infected individuals at different clinical stages were obtained for this University of Minnesota Institutional Review Board-approved microarray study. Viral load measurements were obtained the same day as biopsies. Each lymph node biopsy was placed into a Falcon tube and snap frozen by dropping it into liquid nitrogen.
RNA extraction, synthesis of biotin-labeled cRNA probes, and microarray hybridization
Frozen lymph nodes were homogenized with a power homogenizer (Heat Systems Ultrasonic) in TRIzol (Invitrogen) without thawing. Total RNA was isolated according to the manufacturer’s protocol and further purified with an RNeasy Mini Kit (Qiagen). Double stranded cDNA and biotin-labeled cRNA probes were synthesized from 5 µg of total RNA with a MessageAmp II aRNA kit (Ambion). The cRNA probes were column purified and fragmented with a Fragmentation kit (Ambion).
Fifteen micrograms of fragmented cRNA was hybridized to an Affymetrix Human Genome U133 Plus 2.0 array. After hybridization, chips were washed, stained with streptavidin-PE, and scanned with GeneChip Operating Software at the Biomedical Genomics Center at the University of Minnesota. The experiments from each RNA sample were duplicated in the preparation of each cRNA probe and microarray hybridization.
Microarray data analysis
The .cel files produced by the Affymetrix data analysis platform were uploaded into the Expressionist program (Genedata, Pro version 4.5) and the expression level for each of the ~56,000 probe sets on the arrays was quantified using the RMA algorithm. The mean expression level from duplicate chips from the same individual’s RNA was computed and used in the subsequent analysis. The RMA algorithm produces a summary of gene expression that is of log scale; we utilized this log scale for all analyses.
To examine the relationship between gene expression and viral load, a linear regression model was fit with gene expression as the response variable and disease stage and the log of viral load as predictor variables. We controlled for disease stage when examining the relationship between viral load and gene expression because we previously found an association between gene expression and disease stage for HIV-1 infection (9) (additionally, there is an association between disease stage and viral load for HIV-1-infected subjects naïve to HAART). The p-values for the null hypothesis (testing no association between viral load and gene expression) were then transformed to q-values using the q-value package in the statistical software program R. By varying the threshold for the q-values, one can obtain a set of genes that are found to be associated with viral load at a given false discovery rate. As the number of genes found to be associated with viral load increases dramatically as this threshold rises above ~7%, we used the conservative cut-off value of 7% for the false discovery rate. To summarize the association between log of viral load and gene expression, we computed the partial correlation between these two quantities for all genes while controlling for the effect of disease stage.
Immunofluorescence
Immunofluorescence was performed using a biotin-free detection system on 5-µm tissue sections mounted on glass slides. Tissues were deparaffinized and rehydrated in deionized water. Heat-induced epitope retrieval was performed using a water-bath (95–98°C for 10–20 min) in DiVA Decloaker (Biocare Medical) retrieval buffer, followed by cooling to room temperature. Tissues sections were then treated with Proteinase K (4 µg/ml) for 20 min at 37°C followed by blocking with SNIPER Blocking Reagent (Biocare Medical) for 15 min at room temperature. A primary antibody specific for collagen type 1 (Sigma, clone Col-1, catalog # C2456) was diluted 1/100 in TNB (0.1M Tris-HCl, pH 7.5; 0.15M NaCl; 0.05% Tween 20 with Dupont blocking buffer) and incubated overnight at 4°C. After the primary antibody incubation, sections were washed with PBS and then incubated with fluorophore-cojugated secondary antibodies (Alexa Fluor dyes, Invitrogen) in 5% non-fat milk for 2 hr at room temperature. These sections were washed and mounted using Aqua Poly/Mount (Polysciences Inc.). Immunofluorescent micrographs were taken using an Olympus BX61 Fluoview confocal microscope with the following objectives: x20 (0.75 NA), x40 (0.75 NA), and x60 (1.42 NA); images were acquired using Olympus Fluoview software (version 1.7a). Isotype-matched IgG or IgM negative control antibodies in all instances yielded negative staining results.
Microarray data accession number
All microarray results have been deposited in the Gene Expression Omnibus database: http://www.ncbi.nlm.nih.gov/geo/ (accession number: GSE21589).
Results
Predominance of negative correlations with viral replication
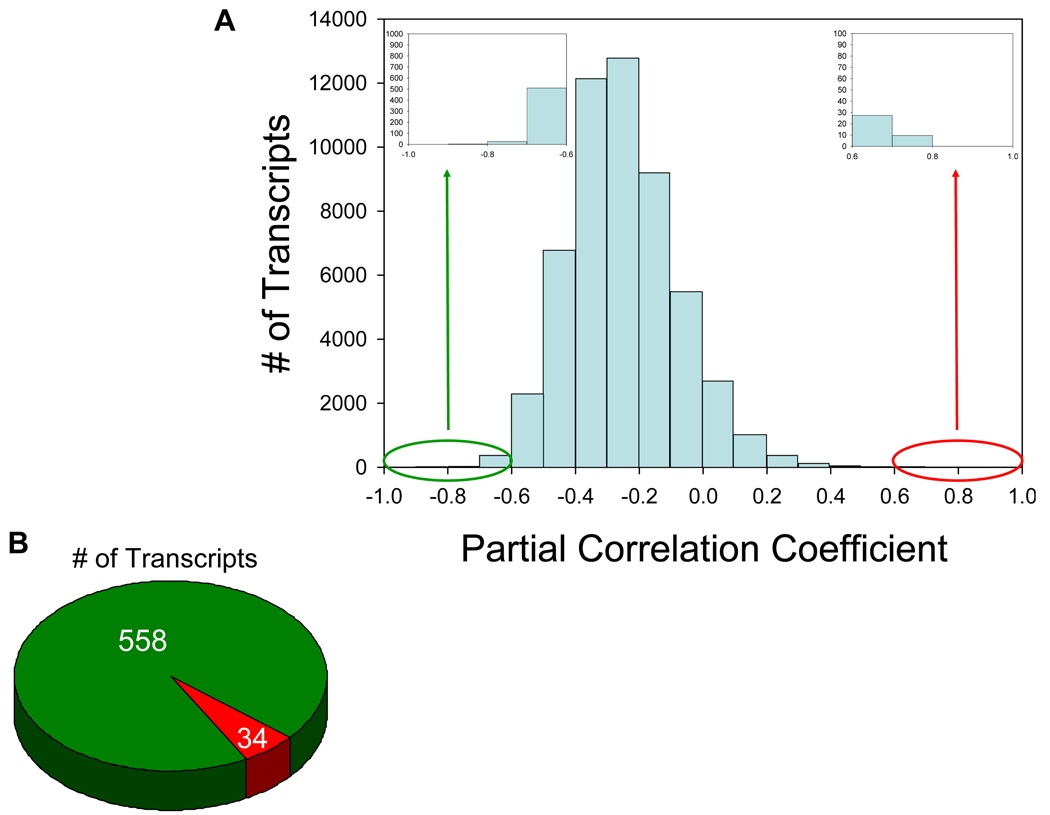
To identify genes within LT related to viral replication, we examined host gene expression and its association with viral load in inguinal lymph nodes from 22 HIV-1-infected individuals (Table 1). We identified 592 transcripts significantly associated with viral load (–0.6 > partial correlation coefficient > 0.6; 7% false discovery rate) (Figure 1A). Strikingly, ~95% of the transcripts (558) in this data set are negatively associated with HIV-1 replication while only ~5% (34 transcripts) have a positive association (Figure 1B). Based on gene ontology/annotations from the NetAffx Analysis Center, Ingenuity Pathways Analysis, and extensive examination of published literature, we classified ~60% of all altered transcripts into functional categories, resulting in a list of 345 genes (the remaining transcripts have as of yet no identification or functional information available) (Figures 2, 4, & Supplementary Table 1).
Table 1.
Clinical Characteristics of Study Subjects
| Patient I.D. |
Disease Stage |
Gender | Age | Race | Peripheral Blood CD4+ T Cell Count (Cells/µl) |
Plasma HIV-1 RNA Levels (Copies/ml) |
|---|---|---|---|---|---|---|
| MH58 | Acute | Male | 51 | Caucasian | 400 | 439,000 |
| JS30 | Acute | Male | 26 | Caucasian | 683 | 3,610 |
| DP24 | Acute | Male | 41 | Caucasian | 494 | 14,696 |
| WB55 | Acute | Male | 23 | African American |
209 | 19,400 |
| PH29 | Acute | Male | 59 | Caucasian | 370 | 484,694 |
| WB91 | Acute | Male | 37 | African American |
234 | 24,718 |
| PR89 | Acute | Male | 32 | Caucasian | 824 | 32,173 |
| TH49 | Acute | Male | 30 | Caucasian | 333 | 100,000 |
| TS35 | Acute | Male | 42 | Caucasian | 663 | 100,000 |
| JR31 | Asymptomatic | Male | 34 | Caucasian | 333 | 71,200 |
| WU59 | Asymptomatic | Male | 36 | Caucasian | 286 | 100,000 |
| KS64 | Asymptomatic | Male | 34 | Caucasian | 202 | 122,000 |
| RJ56 | Asymptomatic | Male | 49 | Caucasian | 478 | 174,000 |
| DS35 | Asymptomatic | Male | 32 | Caucasian | 400 | 15,284 |
| RM08 | Asymptomatic | Male | 39 | Caucasian | 685 | 20,014 |
| CF63 | Asymptomatic | Male | 23 | African American |
259 | 27,200 |
| TK07 | Asymptomatic | Male | 35 | Caucasian | 372 | 31,922 |
| SU36 | Asymptomatic | Male | 63 | Caucasian | 248 | 46,400 |
| GL38 | AIDS | Male | 49 | Caucasian | 147 | 4,960 |
| TK62 | AIDS | Male | 43 | Caucasian | 81 | 35,000 |
| VM27 | AIDS | Female | 40 | African American |
112 | 12,046 |
| OB06 | AIDS | Male | 45 | African American |
188 | 10,684 |
Figure 1. The majority of transcripts in lymphatic tissue are negatively associated with viral load.
(A) Histogram of the partial correlation coefficients for each transcript in the Affymetrix Human Genome U133 Plus 2.0 array. The partial correlation coefficient represents the strength of association between viral load and gene expression while controlling for stage of disease. Transcripts with an absolute value exceeding 0.6 were deemed differentially expressed while controlling for a false discovery rate of 0.7. (B) Pie chart depicting the number of transcripts remaining after applying the selection criteria (−0.6 > partial correlation coefficient > 0.6; 7% false discovery rate). The green and red sectors identify transcripts with a negative and positive association, respectively, between gene expression and viral load.
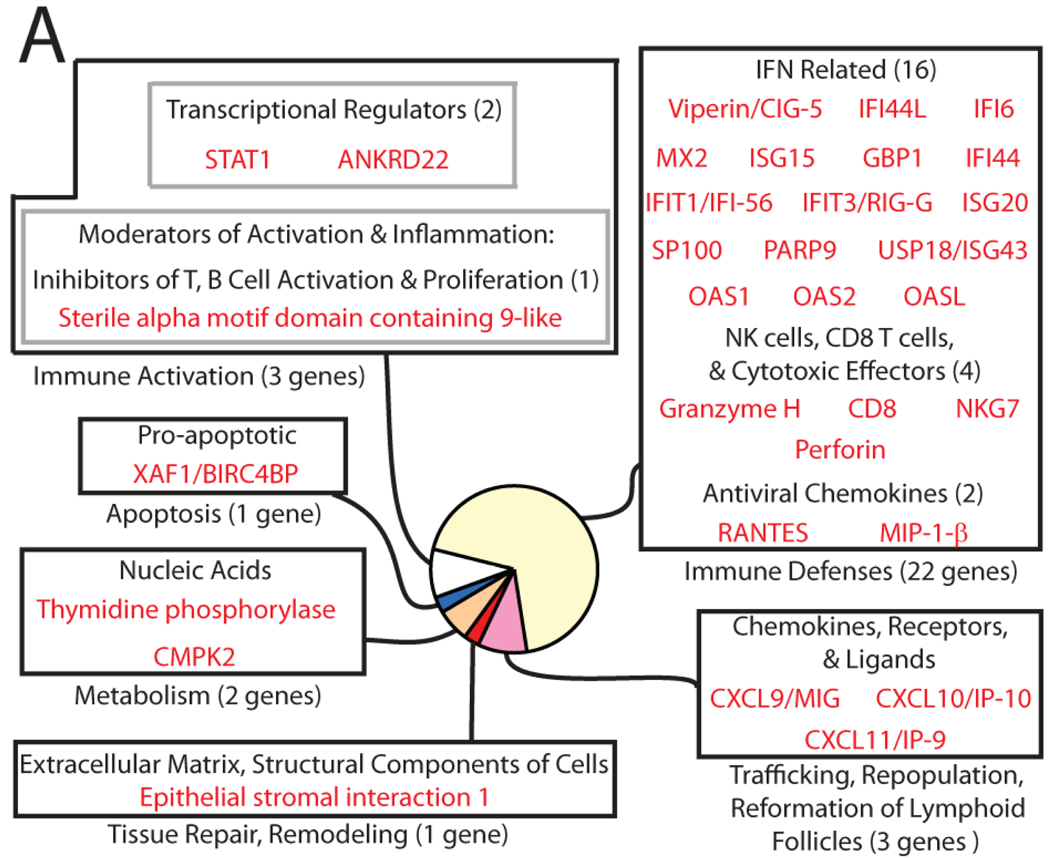
Figure 2. Differentially-expressed genes in HIV-1-infected lymphatic tissue can be classified into functional categories.
Genes positively associated with viral load reveal a heightened IFN response. The size of each sector in a pie diagram is proportional to the number of genes in its category (in parentheses). All genes and their names derived from abbreviations can be found in Supplementary Table 1.
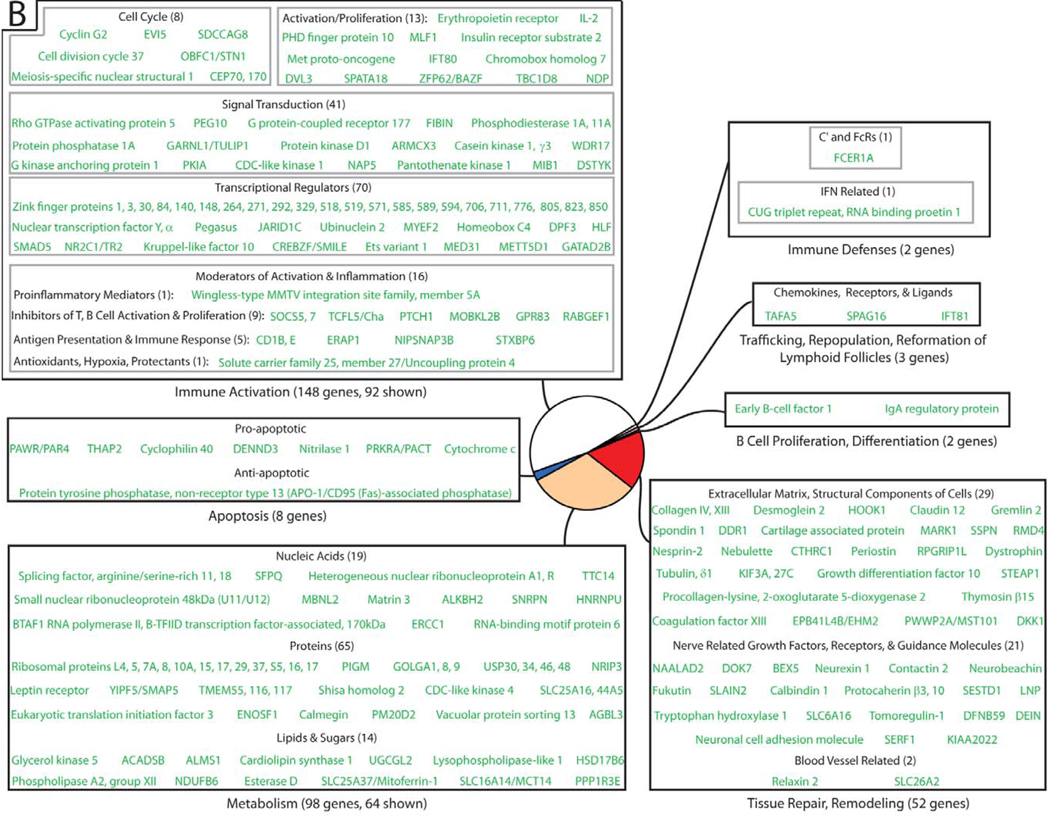
Figure 4. Differentially-expressed genes in HIV-1-infected lymphatic tissue can be classified into functional categories.
Genes negatively associated with viral load point to immune activation, target cell availability, and transcriptional/translational factors as important determinants for HIV-1 replication. The size of each sector in a pie diagram is proportional to the number of genes in its category (in parentheses). All genes and their names derived from abbreviations can be found in Supplementary Table 1.
Antiviral host response positively correlates with virus replication
Surprisingly, only a small subset of genes was positively associated with viral replication (32 genes + 2 transcripts of unknown function), with most of these genes (~70%) paradoxically coding for proteins involved in innate and adaptive defenses, which might have been expected to decrease viral load (Figure 2). Interferon-stimulated genes were a prominent component of this response (16 of the 22 genes ascribed to immune defenses), but there were also genes encoding proteins involved in cell-mediated cytotoxicity, (granzyme H, perforin, CD8, and NKG7), antiviral chemokines (RANTES and MIP-1β), and ligands for the chemokine receptor CXCR3 (CXCL9, CXCL10, and CXCL11). Finally, STAT1, a master regulator of transcription of numerous immune defense genes, was also positively associated with HIV-1 replication. In sum, this analysis reflects a robust antiviral host response that parallels virus replication but is apparently associated with higher viral loads rather than containment of HIV-1 replication.
In a previous analysis of LT comparing uninfected to HIV-1-infected individuals (9), we found signatures of gene expression dependent on stage of disease, with the most striking signature in early infection where expression of genes that control immune activation and innate and adaptive immune defenses was highly upregulated. Since immune activation is now widely believed to be a factor in overall immune dysfunction and negative prognosticator of disease progression (10, 11), we used a chronic immune activation (CIA) index based on gene expression, developed by Rempel et al. (12), to characterize cellular immune dysfunction during HIV-1 infection. This index is comprised of genes related to immune activation that are elevated during infection and can be computed for each infected individual by taking the mean fluorescence intensity of specified genes for each sample and calculated as follows:
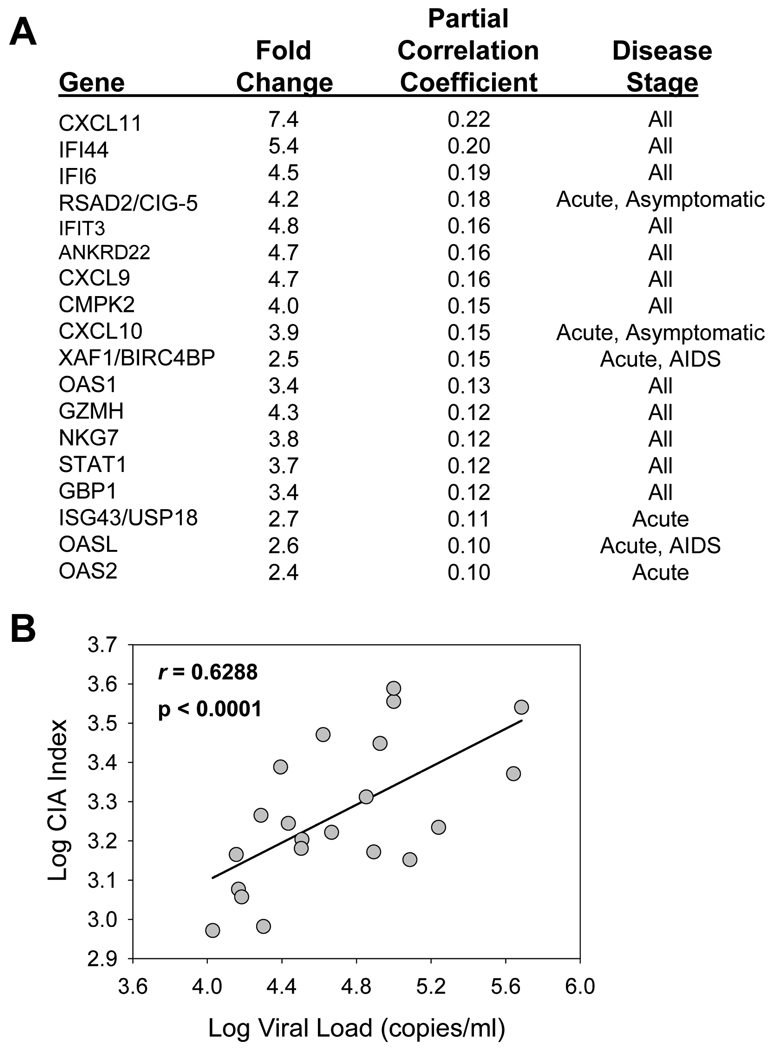
where n equals the number of selected genes. Here, we explored the relationship between CIA and viral replication in our study cohort. We used 18 genes for the CIA index, genes which had the highest increase in expression in LT during HIV-1 infection (9) (Figure 3A). The CIA values and viral load measurements were plotted for each individual, revealing a significant association between CIA and viral load (r = 0.6288, p < 0.0001) (Figure 3B). Additionally, these LT genes with the highest upregulation upon infection also have the strongest positive correlation to viral load (Figure 3A). Overall, this analysis provides a glimpse into the intricate relationship between immune defenses, immune activation, and viral replication, whereby individuals with a higher CIA index are more likely to have higher viral loads.
Figure 3. Chronic Immune activation correlates with HIV-1 viral load.
(A) The top 18 genes in lymphatic tissue during HIV-1 infection with the highest increase in expression (9) and strongest correlation to viral load were chosen for the CIA index computation. The stage of disease at which the gene’s expression is increased is also listed. (B) The CIA index for HIV-1-infected individuals is significantly correlated with peripheral blood viral load.
Negative correlations: immune activation and transcriptional accessibility important for HIV-1 replication
Strikingly, the vast majority of host genes expressed in LT during HIV-1 infection were negatively associated with virus replication (313 genes + 245 transcripts of unknown function). A large proportion of genes in this data set encode for proteins involved in immune activation, target cell availability, and transcriptional/translational metabolism (Figure 4), highlighting the importance of activation status and cellular transcriptional/translational machinery in virus replication. In this list of genes negatively correlated with viral load (Supplementary Table 2), there are genes involved in inhibiting cellular activation and proliferation, such as the suppressors of cytokine signaling, SOCS5 and SCOS7; TCFL5/Cha, a transcription factor implicated in the maintenance of a resting state in T cells; CCNG2, an unconventional cyclin which blocks cell cycle entry; KLF10, a transcription factor involved in inhibiting cellular proliferation through the TGF-β signaling pathway; and GPR83, a G protein-coupled receptor involved in the generation of Foxp3+ TReg cells.
We also identified genes that regulate the intracellular environment for virus replication, such as genes involved in suppressing cellular transcription, particularly through the modification of DNA histones (Supplementary Table 3). Examples include genes whose products recruit histone deacetylases, such as CREBZF and ZNF148/ZBP-89; genes whose products interact/recruit histone methyltransferases, such as HP1BP3, HIC2, and DPF3. This analysis suggests that these gene products adversely affect virus replication by modifying DNA histones, promoting heterochromatin formation, and suppressing cellular and viral transcription.
Negative correlations: collagen deposition and virus replication
One pathological hallmark of HIV-1 infection is the aberrant deposition of collagen and consequent fibrotic damage in both gut and LT (13), a process which may have adverse consequences for the maintenance and preservation of CD4+ T cells and overall immune function in these anatomical niches. Here we show that genes related to collagen synthesis/deposition are negatively associated with virus replication (Supplementary Table 4). Examples include PLOD2, an enzyme responsible for the irreversible cross-linking of collagen fibers; POSTN, a regulator of collagen fibrillogenesis; CRTAP, a scaffolding protein linking collagen-synthesizing enzymes to their precursor substrates; CTHRC1, a regulator of TGF-β responsiveness and their target genes such as collagens; COL4 and COL13, collagen members important for the structure of the basement membrane. The products of these genes likely increase overall collagen deposition and fibrosis in LT and negatively affect virus replication by potentially decreasing the viability/availability of target immune cells for HIV-1.
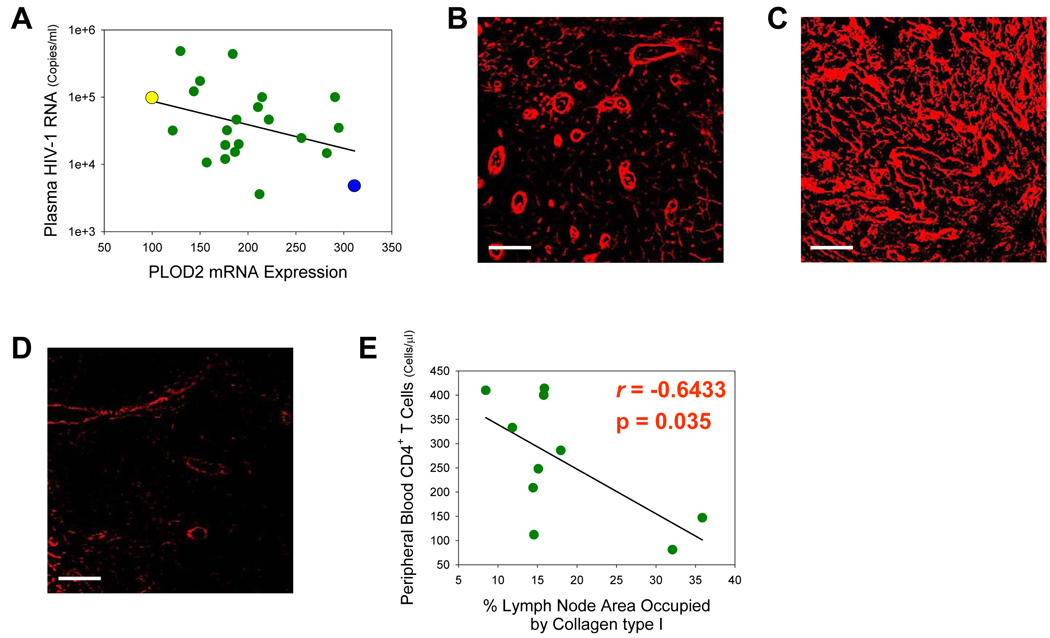
To examine this in more detail, we explored the relationship between mRNA expression of a collagen-synthesizing gene and the resulting end product of this process, protein levels of collagen deposited in LT. One gene we looked at was PLOD2, a telopeptide lysyl hydroxylase primarily responsible for increasing pyridinoline cross-links within collagen molecules during fibrillogenesis, a process which leads to the irreversible accumulation of collagen in fibrotic tissues (14). We used immunofluorescence to visualize collagen type I deposition in inguinal lymph nodes from HIV-1-infected individuals in our study cohort. We initially compared subject WU59 with GL38 because of their divergent PLOD2 mRNA expression (Figure 5A) and found a relationship between PLOD2 mRNA expression and collagen type I deposition, whereby greater levels of fibrosis were detected in the lymph node of subject GL38 coincident with higher levels of PLOD2 mRNA (compare Figures 5B & 5C). Previously reported to be increased during HIV-1 infection (15), much less collagen deposition was seen in the lymph node of an uninfected individual (Figure 5D). There was also a negative correlation between collagen deposition in LT and numbers of peripheral blood CD4+ T cells in a group of our study subjects (Figure 5E), in agreement with previous reports (15, 16), suggesting that increased fibrosis during HIV-1 infection may negatively impact viral replication by adversely affecting CD4+ T cell viability and reducing access to and availability of target cells for viral replication.
Figure 5. Increases in PLOD2 mRNA levels in lymphatic tissue correspond to increased collagen type I synthesis/deposition and decreased CD4+ T cells.
(A) PLOD2 mRNA expression in inguinal lymph node is negatively associated with viral load. Yellow, blue, and green points represent subject #’s WU59, GL38, and remaining cohort individuals, respectively. (B, C, & D) Representative immunofluorescent micrographs reveal collagen type I deposition (red) in the paracortical T cell zone of inguinal lymph nodes from subject #’s WU59 (B), GL38 (C), and an uninfected individual (D). (E) Collagen type I deposition in inguinal lymph nodes is inversely correlated with numbers of peripheral blood CD4+ T cells (Pearson’s correlation coefficient). Original magnifications: X400; scale bars: 50 µm.
Candidates for new host restriction factors
In our list of genes negatively associated with viral load, we identified genes potentially encoding anti-HIV-1 restriction factors (Supplementary Table 5). One such gene is an intracellular glucocorticoid receptor known as nuclear receptor subfamily 3, group C, member 1 (NR3C1). This receptor has been shown to impede proviral integration in peripheral blood mononuclear cells in the absence of steroidal ligands (17); this block is alleviated in the presence of NR3C1’s ligand and requires the HIV-1 protein, Vpr. Another candidate restriction factor is CUG triplet repeat, RNA binding protein 1 (CUGBP1), a previously unrecognized downstream effector of IFN-β signaling in primary macrophages that induces a transcriptional inhibitory protein that works to suppress HIV/SIV replication (18). Heterogeneous nuclear ribonucleoprotein U (HNRNPU) is another candidate identified in our analysis, a ribonucleoprotein that has been shown to specifically target the 3' long terminal repeat in the viral mRNA and block the cytoplasmic accumulation of HIV-1 mRNAs (19).
Discussion
The major and unexpected finding of this study is the unique transcriptome profile in LT during HIV-1 infection related to viral replication, whereby the vast majority of changes in mRNA expression are negatively associated with HIV-1 replication while only a small subset of genes (~5%) is positively associated with virus levels. Surprisingly, most of the genes in this small subset mediate innate and adaptive defenses mounted by the host to contain HIV-1, a host response that would have been expected to negatively correlate with viral load. While counter-intuitive, this LT analysis is in agreement with a transcriptome analysis of primary, peripheral blood CD4+ T cells from HIV-1-infected individuals (20), where expression of a large proportion of interferon-related genes also correlated with increasing levels of virus (Supplementary Table 6).
In addition to a heightened IFN response, key genes important for cell-mediated immunity (e.g., perforin, granzyme H, CD8, NKG7) (Figure 2) and recruitment of immune cells to sites of infection (e.g., CXCL9, CXCL10, CXCL11) (21) were also positively associated with HIV-1 replication. Interestingly, granzyme H, in addition to its pro-apoptotic function as a serine protease highly expressed in natural killer cells and closely related to granzyme B (22, 23), has recently been shown to mediate antiviral activity through direct cleavage of intracellular viral substrates (24, 25). Genes related to chemotaxis may provide some clues regarding the surprising association between host defense genes and high viral loads—the chemotactic response may actually be part of a double-edged sword, on the one hand, serving to recruit HIV-1-specific cytotoxic CD8+ T cells to eliminate virus-infected cells (26) but, on the other hand, serving to fuel the infection further by recruiting susceptible, activated CD4+ T cells to the virus, ultimately aiding in viral dissemination (27).
Surprisingly, we did not identify classical activation markers such as Ki-67, HLA-DR, CD69, CD38 in our data set, genes predicted to be positively correlated with virus replication due, in part, to their role in cellular activation as a means to provide the virus with additional target cells for infection. One interpretation would be that these gene products actually represent general activation of the immune system as a whole and are not specifically indicative of activation in the subsets of cells that are specific targets of the virus. In other words, these classical activation markers may not be the primary determinants of a suitable intracellular environment for permissiveness in target cells. The genes we did detect with a positive correlation to viral load may actually be more representative of cellular activation and permissiveness in individual target cells. The significant association between the CIA index and viral load suggests that this may indeed be the case (Figure 3).
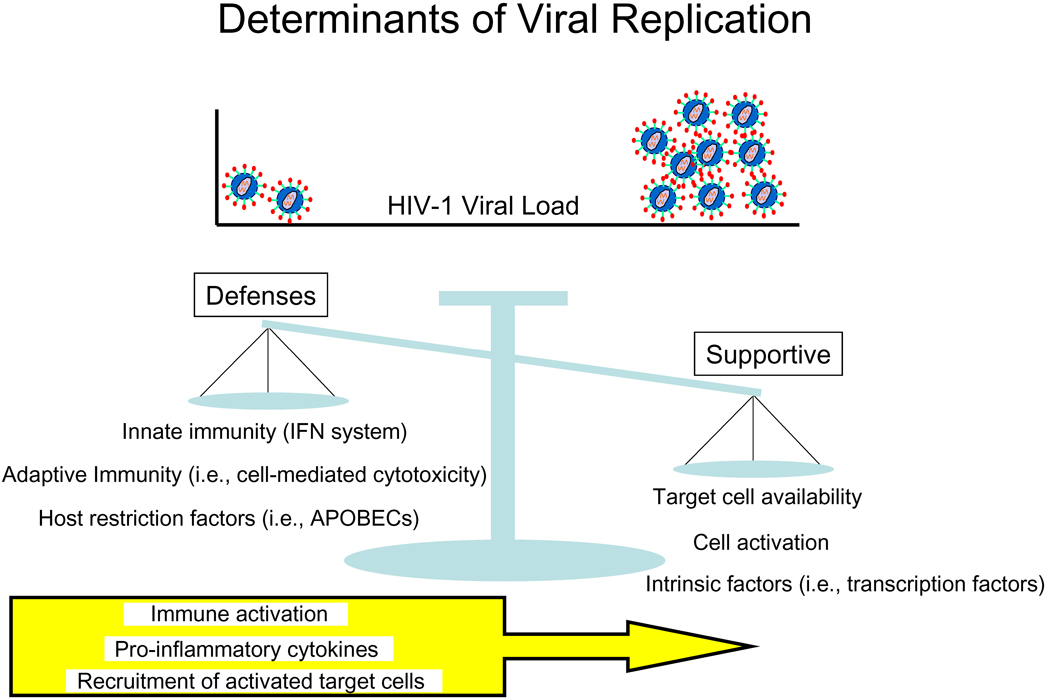
How then to explain the paradoxical, positive association between host defenses and viral load, and even more strikingly, the negative correlations that dominate the dataset? One explanation could be that HIV-1 itself is driving host gene expression; for example, as virus replication increases, the host responds comparably by increasing expression of genes controlling immune defenses. However, due to the complex relationship between immune defenses, immune activation, and virus replication, another explanation could be a model in which the level of viral replication is determined by a balance between both factors that provide virus access to the largest number of susceptible host cells and increase replication and factors that decrease target cell availability/permissiveness and inhibit viral replication (Figure 6). This schematic is consistent with a previous model proposed from transcriptional profiling of the response to HAART (28), aimed to explain the slow progress of HIV-1 infection. On one side of the balance are the supportive determinants of viral replication, such as target cell availability, the activation state of CD4+ T cells and other susceptible cells, and intrinsic intracellular factors that support replication. On the other side of the balance are innate and adaptive defenses and host restriction factors that suppress HIV-1 replication. In this model, host defenses likely inhibit viral replication to some extent, yet, because they are inseparable from immune activation, related pro-inflammatory cytokines, and recruitment of activated target cells, on balance host defenses actually may contribute to increased viral load.
Figure 6. HIV-1 infection in lymphatic tissue is determined by a balance of factors that increase or decrease virus replication.
In this hypothetical model, HIV-1 replication within lymphatic tissue is largely determined by a balance of factors that support or inhibit virus replication. Supportive determinants include target cell availability, cell activation, and intracellular intrinsic factors that promote a favorable transcriptional/translational environment. These supportive determinants are counterbalanced by inhibitory determinants such as innate and adaptive immune responses as well as intracellular host restriction factors that suppress virus replication. This delicate balance, though, is altered by inflammation, a chemotactic response that recruits susceptible target cells to sites of infection, and chronic immune activation required to maintain immune defenses, ultimately tipping the scale towards continued HIV-1 replication and pathology.
In this model, many of the negatively associated genes that dominate the data set reflect the importance of target cell availability and the ability of host intracellular machinery to support viral replication. Collagen deposition in this model negatively impacts viral load through fibrotic damage of the lymph node niche and its documented effects of decreasing CD4+ T cells (Figure 5E) (15, 16), essentially decreasing access and opportunities for virus to interact with target cells.
There may also be a complex relationship between collagen deposition, regulatory T cells (TReg cells), and HIV-1 replication, suggested by findings in the rhesus macaque model of SIV infection that TGF-β1-producing TReg cells are primarily responsible for inducing collagen synthesis/deposition in LT (29). In HIV-1 infection, TReg cells might dampen cellular activation in bystander immune cells, a classical feature of TReg cells (30), thus decreasing overall viral output in HIV-infected cells. This would explain why GPR83, a G protein-coupled receptor reported to be involved in the peripheral generation of TReg cells in vivo (31, 32), along with mediators of the TGF-β signaling pathway (e.g., ITGB8, SMAD5, PEG10, GDF10, KLF10), are all negatively associated with HIV-1 replication.
Beyond the principal hypothesis of target cell availability and permissiveness as the key determinant of viral load, there may be new, host restriction factors that also play an important role. By identifying genes that are both negatively associated with virus replication and code for proteins that display antiviral properties, we found several candidate genes that fit into this category (Supplementary Table 5). One gene in this list, PACT, warrants additional comment. PACT encodes a protein kinase that acts upstream of the important antiviral, sentinel-like molecule, dsRNA-dependent protein kinase (PKR) (33). PACT has been shown to serve as a cellular activator of PKR in the absence of viral RNA (34) but has also recently been demonstrated to possess a role in type I IFN production during viral infection, specifically bypassing PKR activation during amplification of the IFN response (35). Thus, we have a gene that acts upstream of the IFN-response pathway and is negatively associated with viral replication in a data set in which all other IFN-responsive genes are positively associated with HIV-1 replication. This may indicate that PACT is acting outside the IFN-response pathway and adversely affecting HIV-1 replication through its other functions, such as inhibiting cellular translation and inducing apoptosis (36) or amplifying levels of micro RNAs (37) which may serve to inhibit HIV-1 replication (38, 39). We are currently investigating PACT and other candidate genes for their potential role as novel anti-HIV-1 restriction factors.
We have previously documented changes in LT gene expression during HIV-1 infection and found that infection substantially alters the transcriptional profile compared to uninfected individuals (9) and that these transcriptional changes are dependent on stage of disease. A comparison between the present analysis and our previous microarray findings is challenging because the previous study stratified changes in gene expression based on disease stage while our current analysis stratifies gene expression based strictly on virus levels, variables which do not share high concordance (virus levels vs. disease stage). Nevertheless, when comparing the present analysis with our previous microarray findings, we find that the majority of genes positively associated with virus levels are significantly upregulated early in infection (acute stage of disease), overlapping data illustrating a shared relationship between immune defense genes and viral levels in early disease. In contrast, most of the genes negatively associated with virus levels do not overlap with our previous dataset, illustrating the overall discordance between disease stage and virus levels. One would expect to observe greater concordance between datasets if viral levels were indeed correlated with disease stage, but even in our limited study cohort, viral levels vary appreciably across stage. Thus, this comparative analysis highlights two sets of genes—one set of genes may be directly influencing virus replication (current study) and another set of genes that are stage dependent and, on a global scale, may impact virus replication through direct or indirect means (9).
In this microarray study, we utilized whole lymph nodes for processing and generation of the template RNA for microarray chip hybridization as a means to capture the sum of all interactions in an important microenvironment (lymph node) where the bulk of HIV-1 replication and pathogenesis occur. As such, cells were not first separated into individual subpopulations for RNA preparation (i.e., CD4+ T cells). Thus, one complexity is that the cellular composition within the lymph node is not defined at the outset of the experiment. However, this type of tissue microarray study can be most advantageous in that RNA from all the essential cell populations residing within the lymph node microenvironment are present during chip hybridization and subsequent analysis. Genes identified in this study are a good starting point for further investigations of their roles in HIV-1 replication and pathogenesis using in situ technologies to identify the types of cells in which gene expression has been altered and immunohistochemistry/immunofluorescence to identify potential changes in protein expression. For example, we used immunofluorescence to identify the cellular expression of SP100, an IFN-responsive gene that codes for one of the major components of a nuclear transcriptional complex known as the Promyelocytic Leukemia Nuclear Body (40) and has been reported to play an important role in innate antiviral defense against a number of different DNA and RNA viruses (41, 42). We found the size and number of SP100-containing nuclear bodies increased in LT CD3+ T cells and CD163+ macrophages during HIV-1 infection compared to uninfected individuals (Supplementary Figure 1). In sum, we believe this global, tissue microarray methodology is an important first step in a systems-biology approach to understand HIV-1 infection that provides a future framework for focused investigations examining the roles of specific genes impacting HIV-1 replication and pathogenesis.
This microarray study revealed very little overlap (< 3%) with recent small-interfering RNA-knockdown screens (5–8) designed to identify host cell factors required for productive HIV-1 infection in vitro (Supplementary Table 7). This could be due to differences between in vitro systems using laboratory-adapted strains of HIV-1 and cell lines vs. biopsied lymph nodes from HIV-1-infected persons.
In summary, this work yields key insights into systems biology of HIV-1 infection in LT, generates fruitful starting points for additional investigations into candidate genes that may play an important role in aiding or inhibiting HIV-1 infection, and may identify adjunctive approaches to improve therapies and immune reconstitution.
Supplementary Material
Acknowledgements
We thank all of the donor participants in this study.
Footnotes
This work was supported by National Institute of Health (NIH) grant R01 AI056997 (A. T. H.).
Author Contributions
A.J.S. and Q. L. designed and performed experiments; A.J.S., Q. L., S.W.W., and A.T.H. analyzed the data; T.W.S. recruited subjects and procured biopsy samples; C.S.R. performed statistical analyses; A.J.S. and A.T.H. wrote the manuscript.
Abbreviations used in this paper: LT, lymphatic tissue; CIA, chronic immune activation; PKR, dsRNA-dependent protein kinase.
References
- 1.Tasara T, Hottiger MO, Hübscher U. Functional genomics in HIV-1 virus replication: protein-protein interactions as a basis for recruiting the host cell machinery for viral propagation. Biol Chem. 2001;382:993–999. doi: 10.1515/BC.2001.125. [DOI] [PubMed] [Google Scholar]
- 2.Trkola A. HIV-host interactions: vital to the virus and key to its inhibition. Curr Opin Microbiol. 2004;7:555–559. doi: 10.1016/j.mib.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Sorin M, Kalpana GV. Dynamics of virus-host interplay in HIV-1 replication. Curr HIV Res. 2006;4:117–130. doi: 10.2174/157016206776055048. [DOI] [PubMed] [Google Scholar]
- 4.Giri MS, Nebozhyn M, Showe S, Montaner LJ. Microarray data on gene modulation by HIV-1 in immune cells: 2000–2006. J Leu Biol. 2006;80:1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 5.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 6.König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Micro. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Smith AJ, Schacker TW, Carlis JV, Duan L, Reilly CS, Haase AT. Microarray Analysis of Lymphatic Tissue Reveals Stage-Specific, Gene-Expression Signatures in HIV-1 Infection. J Immunol. 2009;183:1975–1982. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, Hamann D, Prins M, Miedema F. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 11.Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010 doi: 10.1186/1742-4690-7-54. PMID: 20569472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS. 2010;24:1415–1423. doi: 10.1097/QAD.0b013e32833ac623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes JD, Haase AT, Schacker TW. The role of collagen deposition in depleting CD4+ T cells and limiting reconstitution in HIV-1 and SIV infections through damage to the secondary lymphoid organ niche. Semin Immunol. 2008;20:181–186. doi: 10.1016/j.smim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Slot AJ, Zuurmond AM, Bardoel AF, Wijmenga C, Pruijs HE, Sillence DO, Brinckmann J, Abraham DJ, Black CM, Verzijl N, DeGroot J, Hanemaaijer R, TeKoppele JM, Huizinga TW, Bank RA. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 15.Schacker TW, Nguyen PL, Beilman GJ, Wolinsky S, Larson M, Reilly C, Haase AT. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J Clin Invest. 2002;110:1133–1139. doi: 10.1172/JCI16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schacker TW, Brenchley JM, Beilman GJ, Reilly C, Pambuccian SE, Taylor J, Skarda D, Larson M, Douek DC, Haase AT. Lymphatic tissue fibrosis is associated with reduced numbers of naive CD4+ T cells in human immunodeficiency virus type 1 infection. Clin Vaccine Immunol. 2006;13:556–560. doi: 10.1128/CVI.13.5.556-560.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiegers K, Schwarck D, Reimer R, Bohn W. Activation of the glucocorticoid receptor releases unstimulated PBMCs from an early block in HIV-1 replication. Virology. 2008;375:73–84. doi: 10.1016/j.virol.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 18.Dudaronek JM, Barber SA, Clements JE. CUGBP1 is required for IFNbeta-mediated induction of dominant-negative CEBPbeta and suppression of SIV replication in macrophages. J Immunol. 2007;179:7262–7269. doi: 10.4049/jimmunol.179.11.7262. [DOI] [PubMed] [Google Scholar]
- 19.Valente ST, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol Cell. 2006;23:597–605. doi: 10.1016/j.molcel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Günthard HF, Goldstein DB, Telenti A. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farber JM. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol. 1997;61:246–257. [PubMed] [Google Scholar]
- 22.Fellows E, Gil-Parrado S, Jenne DE, Kurschus FC. Natural killer cell-derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood. 2007;110:544–552. doi: 10.1182/blood-2006-10-051649. [DOI] [PubMed] [Google Scholar]
- 23.Hou Q, Zhao T, Zhang H, Lu H, Zhang Q, Sun L, Fan Z. Granzyme H induces apoptosis of target tumor cells characterized by DNA fragmentation and Bid-dependent mitochondrial damage. Mol Immunol. 2008;45:1044–1055. doi: 10.1016/j.molimm.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 24.Andrade F, Fellows E, Jenne DE, Rosen A, Young CS. Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. EMBO J. 2007;26:2148–2157. doi: 10.1038/sj.emboj.7601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero V, Fellows E, Jenne DE, Andrade F. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ. 2009;16:340–348. doi: 10.1038/cdd.2008.165. [DOI] [PubMed] [Google Scholar]
- 26.Brainard DM, Tager AM, Misdraji J, Frahm N, Lichterfeld M, Draenert R, Brander C, Walker BD, Luster AD. Decreased CXCR3+ CD8 T cells in advanced human immunodeficiency virus infection suggest that a homing defect contributes to cytotoxic T-lymphocyte dysfunction. J Virol. 2007;81:8439–8450. doi: 10.1128/JVI.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foley JF, Yu CR, Solow R, Yacobucci M, Peden KW, Farber JM. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J Immunol. 2005;174:4892–4900. doi: 10.4049/jimmunol.174.8.4892. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Schacker T, Carlis J, Beilman G, Nguyen P, Haase AT. Functional genomic analysis of the response of HIV-1-infected lymphatic tissue to antiretroviral therapy. J Infect Dis. 2004;189:572–582. doi: 10.1086/381396. [DOI] [PubMed] [Google Scholar]
- 29.Estes JD, Wietgrefe S, Schacker T, Southern P, Beilman G, Reilly C, Milush JM, Lifson JD, Sodora DL, Carlis JV, Haase AT. Simian immunodeficiency virus-induced lymphatic tissue fibrosis is mediated by transforming growth factor beta 1-positive regulatory T cells and begins in early infection. J Infect Dis. 2007;195:551–561. doi: 10.1086/510852. [DOI] [PubMed] [Google Scholar]
- 30.Allan SE, Broady R, Gregori S, Himmel ME, Locke N, Roncarolo MG, Bacchetta R, Levings MK. CD4+ T-regulatory cells: toward therapy for human diseases. Immunol Rev. 2008;223:391–421. doi: 10.1111/j.1600-065X.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, Sakaguchi S. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 32.Hansen W, Loser K, Westendorf AM, Bruder D, Pfoertner S, Siewert C, Huehn J, Beissert S, Buer J. G protein-coupled receptor 83 overexpression in naive CD4+CD25- T cells leads to the induction of Foxp3+ regulatory T cells in vivo. J Immunol. 2006;177:209–215. doi: 10.4049/jimmunol.177.1.209. [DOI] [PubMed] [Google Scholar]
- 33.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Acquisto F, Ghosh S. PACT and PKR: turning on NF-kappa B in the absence of virus. Sci STKE. 2001;89 doi: 10.1126/stke.2001.89.re1. re1. [DOI] [PubMed] [Google Scholar]
- 35.Iwamura T, Yoneyama M, Koizumi N, Okabe Y, Namiki H, Samuel CE, Fujita T. PACT, a double-stranded RNA binding protein acts as a positive regulator for type I interferon gene induced by Newcastle disease virus. Biochem Biophys Res Commun. 2001;282:515–523. doi: 10.1006/bbrc.2001.4606. [DOI] [PubMed] [Google Scholar]
- 36.Peters GA, Li S, Sen GC. Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J Biol Chem. 2006;281:35129–35136. doi: 10.1074/jbc.M607714200. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, Jacquet JM, Reynes J, Levy Y, Saib A, Bennasser Y, Benkirane M. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batty E, Jensen K, Freemont P. PML nuclear bodies and their spatial relationships in the mammalian cell nucleus. Front Biosci. 2009;14:1182–1196. doi: 10.2741/3302. [DOI] [PubMed] [Google Scholar]
- 41.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 42.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defense. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.