Abstract
Sirtuin-1 (SIRT1) is an NAD+-dependent protein deacetylase that is sensitive to oxidative signals. Our purpose was to determine whether SIRT1 activity is sensitive to the low molecular weight nitrosothiol, S-nitrosoglutathione (GSNO), which can transduce oxidative signals into physiological responses. SIRT1 formed mixed disulfides with GSNO-Sepharose, and mass spectrometry identified several cysteines that are modified by GSNO, including Cys-67 which was S-glutathiolated. GSNO had no effect on basal SIRT1deacetylase activity, but inhibited stimulation of activity by resveratrol (RSV) with an IC50 of 69 μM. These observations indicate that S-glutathiolation of SIRT1 by low concentrations of reactive glutathione can modulate its enzymatic activity.
Antioxid. Redox Signal. 13, 1023–1032.
Introduction
Sirtuin-1 (SIRT1) is a highly conserved NAD+-dependent deacetylase that plays an instrumental role in cell cycle progression, energy metabolism, and aging. In the vascular endothelium, SIRT1 promotes cellular homeostasis through deacetylating a wide variety of targets, including eNOS, FOXO1, and p53 (20). SIRT1 activity depends upon the availability of NAD+ such that increases in the [NAD+]/[NADH] ratio during caloric restriction activate the enzyme (13) and by doing so can regulate gene expression (6) and apoptosis (1). Recent evidence suggests that SIRT1 may be sensitive to other cellular redox couples. In neural progenitor cells, GSH depletion caused by buthionine sulfoximine shifted terminal cell differentiation in a SIRT1-dependent manner, indicating that SIRT1 may be sensitive to intracellular GSH levels (21).
SIRT1 activity is stimulated by the polyphenol, resveratrol (RSV), a component of red wine, which has a variety of cardioprotective effects, including suppression of NF-κB-dependent endothelial inflammation caused by TNFα (5) or cigarette smoke extract (4). Furthermore, RSV increases eNOS expression and activity, and in turn, improves bioavailability of nitric oxide (•NO) (12, 27). At present, potent small molecule activators of SIRT1 are highly sought after for their therapeutic potential (15).
SIRT1 activity is regulated by post-translational modifications including sumoylation at lysine-734 that increases its activity. UV radiation and hydrogen peroxide decreases SIRT1 sumoylation and deacetylase activity, and therefore increases apoptosis (29). Likewise, SIRT1 activity is stimulated by cyclinB/Cdk1-dependent phosphorylation at threonine-530 and serine-540 that regulates cell cycle progression (23). Direct post-translational modification of SIRT1 by oxidants has not yet been reported.
•NO is an important signaling molecule that mediates a wide variety of cellular effects by either stimulating guanylyl cyclase to increase cyclic GMP or by inducing reversible adducts on cysteine thiols that include S-sulfenation, S-nitrosation, and S-glutathiolation (25). S-nitrosothiols play a prominent role as intermediates in •NO signaling. •NO reacts with GSH, the prevalent low molecular weight thiol in the cell, to generate S-nitrosoglutathione (GSNO) which can then transnitrosate proteinyl thiols, eliciting a change in protein function (14). S-nitrosated thiols can undergo further oxidation to form S-thiolated proteins that are covalently modified by glutathione, cysteine, or homocysteine (7). An increasing body of evidence implicates GSNO as mediating S-glutathiolation of protein thiols in addition to its role in S-transnitrosation. Exogenous and endogenous application of •NO results in S-glutathiolation of hsp70, actin, and vimentin in rat vascular smooth muscle cells (28). In vitro GSNO can S-glutathiolate aldose reductase, glyceraldehyde-3-phosphate dehydrogenase, caspase-3, c-jun and cathepsin-K on specific cysteine thiols (3, 9, 17, 19, 30).
The goal of this work was to determine whether SIRT1 is subject to direct modification and regulation by a cellular oxidant. Our findings clearly demonstrate that GSNO attenuates stimulation of enzymatic activity of purified SIRT1 by RSV without affecting its basal activity. In addition, reduction of GSNO-treated protein with dithiothreitol (DTT) restored SIRT1 responsiveness to RSV stimulation, indicating that GSNO induces a reversible thiol modification. Mass spectrometry analysis conclusively identified several SIRT1 thiols that are sensitive to GSNO-induced oxidation, and S-glutathiolation of Cys-67 was definitively demonstrated by LC-MS/MS. This work presents novel findings that indicate that SIRT1 is directly modified by GSNO, providing a novel redox regulatory mechanism of SIRT1 activity.
Determination of SIRT1 Function after GSNO Treatment
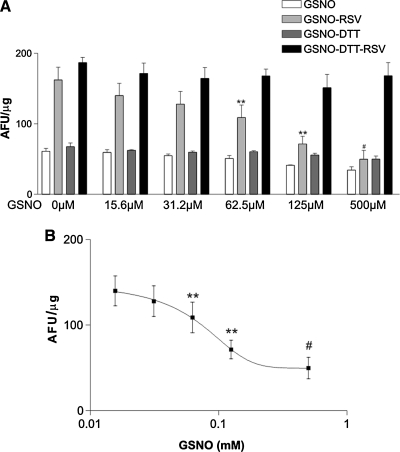
The SIRT1-Flag deacetylation activity assay was tested for specificity in the presence and absence of a specific SIRT1 inhibitor or the activator, RSV. In the presence of 5 μM EX-243 ((S)-6-chloro-2,3,4,9-tetrahydro-1-H-carbazole-1-carboxamide), but not its inactive (R) stereoisomer, EX-242, basal SIRT1 activity was blocked (Supplemental Fig. 1; see www.liebertonline.com/ars) indicating that SIRT1 deacetylase activity measurement was specific. In the presence of RSV (50 μM), SIRT1 activity was stimulated approximately 2-fold, as reported previously (18). Prior to characterizing the effects of GSNO, SIRT1-Flag isolated on Flag Sepharose beads was treated with DTT (2 mmol/l) to ensure global cysteine reduction. After removing DTT by buffer exchange, SIRT1 was incubated with GSNO at concentrations between 156 nM and 500 μM. After an additional buffer exchange, activity was assayed in the presence or absence of RSV (50 μM). Under these conditions, there was no statistically significant effect of GSNO on basal SIRT1 activity. However, GSNO caused a concentration-dependent inhibition of SIRT1 stimulation by RSV (Fig. 1A, white bars). The concentration of GSNO required for half-maximal inhibition of RSV stimulation was 69 μM, with complete inhibition at 500 μM (Fig. 1B). Following GSNO, an additional reduction step with DTT restored RSV stimulation to levels equivalent to that of SIRT1 that was not treated with GSNO (Fig. 1A, dark gray bars). These data demonstrate one or more SIRT1 cysteine residues are susceptible to reversible oxidation by GSNO which inhibits stimulation of enzyme activity by RSV.
FIG. 1.
SIRT1 treated with concentrations of up to 0.5 mM GSNO displays attenuated stimulation by RSV (50 μM). SIRT1 was treated with DTT to ensure global cysteine reduction. Increasing concentrations of GSNO had no effect on basal SIRT1 (white bars) activity but did inhibit RSV stimulation (light gray bars). Reversal of SIRT1 oxidation by an additional reduction step reverted SIRT1 activity (dark gray bars) and RSV response (black bars) to control conditions indicating that GSNO induces a reversible OPTM that inhibits SIRT1 stimulation by RSV (A). GSNO inhibited RSV-stimulated SIRT1 activity with an IC50 of 69 μM (B). (n = 3; **p < 0.01 vs. GSNO-RSV; #p < 0.001 vs. GSNO-RSV).
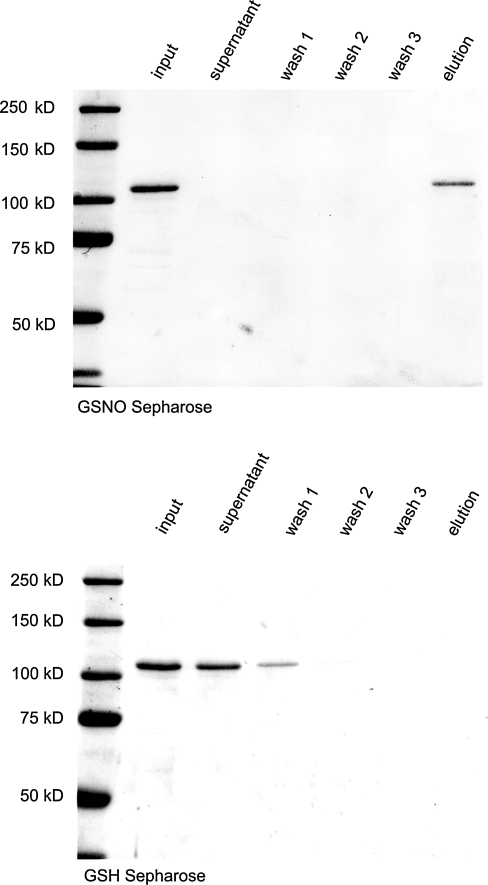
In order to determine the reactivity of SIRT1 cysteines with low molecular weight thiols, GSH-Sepharose was used in either unmodified form or as GSNO-Sepharose after nitrosation with acidified nitrite. As evidenced by the absence of SIRT1 in the supernatant and wash fractions, SIRT1 bound covalently to GSNO-Sepharose, but not to GSH-Sepharose (Fig. 2). BSA, as a negative control, did not bind to either column material (results not shown). The formation of a covalent disulfide adduct with GSNO was demonstrated by elution of the protein from GSNO-Sepharose with DTT. These results indicate that SIRT1 efficiently forms mixed disulfides in vitro with GSNO, but not GSH.
FIG. 2.
SIRT1 (120 kDa) binds to GSNO but not GSH beads. 50 μg recombinant protein was incubated with 100 μl GSNO or GSH beads for 30 min in binding buffer (50 mM Tris pH 8.0, 250 mM NaCl, 1 mM EDTA, 0.01% NP40) at room temperature. The beads were then washed three times, and bound proteins were eluted with 10 mM DTT. The samples were subjected to SDS-PAGE, followed by transfer to nitrocellulose (NC) membranes. NC membranes were stained with Memcode reversible protein stain and proteins were visualized. Lane 1: Protein input. Lane 2: supernatant following SIRT1 incubation with beads. Lane 3–5: Wash fractions. Lane 6: Elution of bound proteins.
Differential Alkylation and Mass Spectrometry Identifies Surface Cysteines Modified by GSNO
In order to identify the specific cysteines modified by GSNO, SIRT1-Flag was subjected to differential cysteine alkylation by NEM and IAM, followed by MALDI-TOF MS and LC-MS/MS. Differential alkylation labels cysteines based on whether the thiols are reduced or oxidized (24). After initial reduction with DTT, SIRT1 was first exposed to GSNO and alkylated with IAM to alkylate remaining free cysteine thiols. These cysteines were recognized during analysis of the mass spectra by the increased mass conferred by IAM (+57 Da). Cysteines that were reversibly oxidized by GSNO were then reduced with DTT and alkylated with NEM, and were recognized by an increase in mass of 125 Da. In all, SIRT1-Flag was subjected to four conditions: GSNO-IAM-DTT-NEM, IAM-DTT-NEM, GSNO-NEM-DTT-IAM, and NEM-DTT-IAM. Analysis of the spectra and comparison between each condition allowed us to identify GSNO-reactive cysteines on SIRT1.
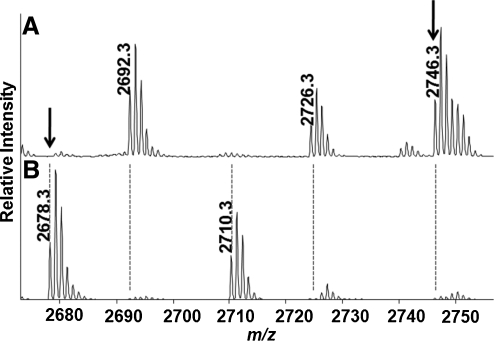
By MALDI-TOF MS, sequence coverage of differentially alkylated mouse SIRT1 samples varied between 56% and 65%. Peaks were assigned to known cysteine-containing peptides of SIRT1 (Table I). In a representative mass spectrum from SIRT1 not previously treated with GSNO (IAM-DTT-NEM), a peak was observed at m/z 2678.277 that corresponds to the SIRT1 peptide containing amino acids 46–74 in which Cys-61 (human Cys-67) is alkylated with IAM (+57 Da) (Fig. 3B). A peak corresponding to the NEM-alkylated form of the same peptide (+125 Da) at m/z 2746.313 in the GSNO-IAM-DTT-NEM-treated sample confirmed that the site was modified by GSNO. These results indicate that cysteine-61 was reversibly oxidized by GSNO, reduced by DTT, and alkylated with NEM (Fig. 3A). The peak observed in the sample not treated with GSNO (m/z 2678.277) did not appear in the GSNO-IAM-DTT-NEM sample, indicating that the oxidation of Cys-61 was nearly quantitative and confirming that GSNO forms a reducible modification on Cys-61. Cys-61 was confirmed to be alkylated with IAM by LC-MS/MS in the IAM-DTT-NEM sample, but not in the GSNO-IAM-DTT-NEM treated sample (Supplemental Fig. 2; see www.liebertonline.com/ars). Analysis of all spectra yielded a list of putative GSNO-reactive cysteines on SIRT1 that followed a similar pattern of differential alkylation as Cys-61 that included Cys-C245, Cys-C260, Cys-C318, and Cys-C613 (Table I). These five cysteines identified on mouse SIRT1 are potential sites at which reversible oxidation of SIRT1 by GSNO may regulate SIRT1 function.
Table I.
Identification of SIRT1 Reactive Cysteines
| Residue | GSNO-IAM-NEM | IAM-DTT-NEM | GSNO-NEM-DTT-IAM | NEM-DTT-IAM |
|---|---|---|---|---|
| C61 | NEM | IAM | both | |
| C245 | IAM | IAM | ||
| C260 | NEM | IAM | IAM | both |
| C318 | NEM | IAM | IAM | both |
| C613 | NEM | both | IAM | both |
Differential alkylation of SIRT1-Flag reveals 5 out of a total of 19 cysteines on SIRT1 that are reactive with GSNO. In GSNO-treated samples, the second alkylating agent is expected to label the cysteine residue because that cysteine thiol is reduced by DTT after the reversible glutathione adduct blocked alkylation with the first agent. In the control protein, not treated with GSNO, the first alkylating agent is expected to label the same thiol. In each box of the table, the alkylating agent, either IAM (+57) or NEM (+125), found on each cysteine-containing peptide is indicated. Boxes labeled “both” indicate that both peptides were identified in the spectra, but that the height of the peak corresponding to the appropriate alkylated peptide indicated that the cysteine thiol was reversibly modified by GSNO. Note that numbers of cysteine residues in the table refer to those of mouse SIRT1.
FIG. 3.
Differential alkylation of SIRT1-Flag. Purified mouse SIRT1 was incubated in the presence of 2.0 mM GSNO for 15 min at 37°C. Unreacted thiols were blocked with NEM. Initial alkylation is followed by addition of DTT to remove GSH adducts, and then a second alkylation step with IAM. (A) Thiols that were reactive with GSNO were expected to be labeled by the second alkylating agent following reduction with DTT. A major peak with m/z 2746.313 was identified that corresponds to the peptide R46SPGEPSAAVAPAA AGCEAASAAAPAALWR74 with an additional +125 Da, indicating that the cysteine is alkylated with NEM. (B) Control protein that was not exposed to GSNO was also differentially alkylated and both spectra were compared. In this spectrum a peak was identified with m/z 2678.277 corresponding to the same peptide bearing a carbamidomethylation (+57 Da) rather than modification by NEM. Samples were subjected to SDS-PAGE, in-gel trypsin digestion, and analysis by MALDI-TOF. The converse experiment in which IAM was used initially, followed by NEM labeling of cysteines reduced by DTT, was used to control for varying reactivities of alkylating reagents, and as indicated in Table I showed similar results.
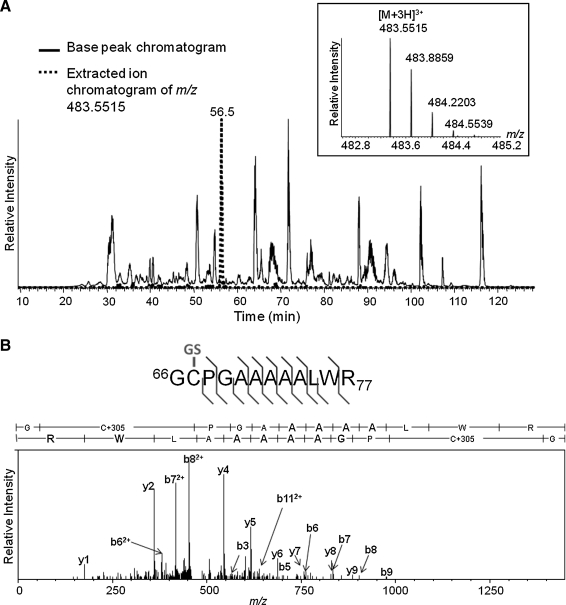
In order to confirm the specific modification induced by GSNO that was detected by differential alkylation, human SIRT1 was treated with GSNO, followed by alkylation of remaining free thiols with NEM. SIRT1 then was subjected to in-solution digest and analyzed by LC-MS/MS. An [M + 3H]3+ ion corresponding to m/z 483.55 (Fig. 4A) was identified as the SIRT1 peptide containing amino acids 66–77 with an additional mass of 305 Da, consistent with the addition of a single glutathione adduct. The fragmentation ion spectrum clearly demonstrated that human SIRT1 is S-glutathiolated on cysteine 67 (Fig. 4B).
FIG. 4.
MS detection and sequencing of S-glutathiolated human SIRT1 peptide 66–77. Purified recombinant human SIRT1 was treated in vitro with GSNO, followed by NEM labeling of unreacted cysteines, and in-solution tryptic digestion. Peptides were subjected to nano-flow reversed-phase chromatography and MS/MS on an LTQ-Orbitrap mass spectrometer. (A) Base peak chromatogram (black trace) over the portion of the LC-MS run over which peptide elution occurs. Overlaid, the extracted ion chromatogram of the [M + 3H]3+ ion at m/z 483.5514 (red trace), which demonstrates that this species elutes distinctly during the 4 h total LC run at a retention time of ca. 56 min. Chromatographic peaks are labeled with the m/z value of their base peak. Inset (above) into the chromatogram is the mass spectrum of the [M + 3H]3+ ion species at m/z 483.5514 at its elution peak. (B) Fragment ion tandem mass spectrum resulting from the collisional dissociation of the [M + 3H]3+m/z 483.5514 precursor ion. Prominent fragment ions are marked with their assignments to b-type ions (right) and y-type ions (left) of the SIRT1 peptide 66GCPGAAAAALWR77 bearing a glutathione moiety on its cysteine. Above the spectrum, a summary of the fragment ion data, including less abundant fragment ions detected, is indicated on the peptide sequence by flags indicating b (right) and y (left) ions. The site of S-glutathiolation (indicated in green on the sequence) is identified unambiguously to the first three residues, therefore to Cys-67, by numerous prominent diagnostic b and y ions.
Concluding Remarks and Future Directions
The lack of a definitive crystal structure of SIRT1 and its large size makes it otherwise difficult to determine which of its 19 total cysteines might be reactive surface cysteines. Therefore, identifying a small subset of SIRT1 cysteines that are reactive to GSNO is an important advance in determining how intracellular SIRT1 might be redox regulated.
Using MALDI-TOF MS and LC-MS/MS in combination with differential alkylation, five cysteines were identified on mouse SIRT1 that were S-glutathiolated after exposure to GSNO, and S-glutathiolation of Cys-67 was confirmed by LC-MS/MS on human SIRT1. All five SIRT1 reactive cysteines identified through MALDI-TOF MS are candidates for OPTMs that regulate enzymatic activity. Autiero et al. recently utilized modeling to predict the structure of SIRT1 (2). Via personal correspondence, Autiero confirmed that human SIRT1 Cys-67, Cys-268, and Cys-623 were solvent-exposed in their computer-generated model, and may therefore be susceptible to thiol modification. SIRT1 is regulated by post-translational modifications, including phosphorylation and sumoylation, which are localized to the N- and C-terminal regions of the enzyme (2). S-glutathiolation by GSNO at Cys-67 and Cys-623 are consistent with post-translational regulation at these terminal regions of SIRT1. Cys-268 is of interest because it lies in the NAD+ binding region identified by modeling of SIRT1 and other members of the SIRT family, in which the catalytic core is highly conserved (2, 16). Binding of NAD+ results in changes in SIRT1 conformation that allow catalysis to proceed. It is possible that S-glutathiolation of this cysteine might reduce binding affinity of NAD+.
S-glutathiolation of SIRT1 has no effect on basal activity, but does inhibit SIRT1 activation by RSV with an IC50 of 69 μM. This effect was reversed following reduction with DTT, demonstrating that GSNO induces a reversible modification on SIRT1. This was confirmed by demonstrating that SIRT1 formed mixed disulfides with GSNO-Sepharose. Considering the normal millimolar intracellular GSH concentrations and the low IC50 of 69 μM for GSNO to inhibit SIRT1, our findings strongly suggest that SIRT1 stimulation by RSV may be regulated in vivo by direct redox modification(s) of SIRT1.
By demonstrating that SIRT1 can be regulated by S-glutathiolation of specific Cys residues, our results have uncovered the potential for SIRT1 to be directly regulated by oxidative modifications that may link its activity to GSH redox and production of cellular reactive oxygen and nitrogen species. These novel findings also indicate that the GSH redox state may affect the response of SIRT1 to polyphenols. These results imply that antioxidants might significantly potentiate the response to small molecule activators of SIRT1. Our findings open an exciting new avenue of SIRT1 regulation that will be important in understanding SIRT1 as a therapeutic target for the treatment of several chronic diseases.
Notes
Adenoviral transfer and clones
pAd-track SIRT1-Flag containing full-length mouse SIRT1 was obtained from Addgene, Cambridge MA (plasmid 8438) (22). pAd-track SIRT1-Flag was linearized and recombined with pAdeasy in BJ5183 cells to generate cosmids. Suitable cosmids were digested with PAC-1 and transfected into HEK293 cells utilizing calcium phosphate. Adenoviral colonies were isolated and propagated in HEK293A cells. Adenovirus was isolated using double cesium chloride gradient centrifugation. Titers were determined by OD260 assay and expressed as OPU/ml (optical particle units per milliliter).
Protein purification
Plasmids containing human GST-SIRT1 fusion protein were supplied by Dr. Tony Kouzarides at The Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge (11). SIRT1 protein was cloned into pcDNA cmv-6x His and prepared in transformed BL21 bacteria by standard methods. SIRT1-His E. Coli Rosetta-2 DE3 cells (Novagen, Darmstadt, Germany) were transformed with a plasmid containing a construct for 6x His-tagged SIRT1 (human) protein. Transformed bacterial cells were then grown up to an OD600 of 0.6–1.0, induced with 1 mmol/L IPTG (AppliChem, Darmstadt, Germany) and protein expression proceeded at 30°C for 4 h. Bacterial cells were then spun down, and the bacterial pellet was incubated in lysis buffer (50 mmol/L Tris pH 8.0, 10 mmol/L NaCl, 1 mmol/L DTT, 1 mmol/L PMSF) containing 1 mg/ml lysozyme. Lysed bacteria were sonicated, 25 Units benzonase (BD Biosciences, San Jose, CA) were added, and the mixture was incubated on ice for 15 min. The lysed bacteria were spun down at 17000 g for 10 min at 4°C. The supernatant was collected and subjected to ammonium sulfate (Acros Organics, Geel, Belgium) precipitation at a concentration of 40% saturation. The mixture was then spun down at 17000 g for 5 min at 4°C and the supernatant was discarded. The pellet was resuspended in 50 mmol/L Tris pH 8.0, 500 mmol/L NaCl, 0.1% Triton, 300 mmol/L sucrose, 1 mmol/L PMSF, 1 mmol/L DTT, and 100 μM zinc sulfate, and dialyzed overnight at 4°C in lysis buffer. The dialyzed sample was then purified via the 6X His tag using a His60 Nickel column (ClonTech, Mountain View, CA).
Cell culture
HEK293T cells (ATCC, Manassas, VA) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. Bovine aortic endothelial cells (BAEC, Cell Systems Corporation, Kirkland, WA) were cultured in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin, 0.1% DMSO, and 1 mmol/L ascorbic acid in dishes pre-coated with 0.1% gelatin. Media was refreshed every 48 h.
Immunoprecipitation of Flag-SIRT1
Infected HEK293T cells or BAECs were lysed in RIPA buffer (50 mmol/L Tris pH 7.4, 150 mmol/L NaCl, 1% NP40, 0.1% SDS, 0.5% deoxycholine, 1 mmol/L PMSF, 1X protease inhibitor cocktail (Roche)). Lysates were subjected to sonication, then rotated at 4°C for 30 min, followed by centrifugation for 10 min at maximum speed to remove insoluble material. Anti-Flag M2 agarose affinity gel (Sigma-Aldrich, St. Louis, MO) was prepared, 40 μl gel suspension was pipetted into microcentrifuge tubes, and the suspension was washed three times with 1 ml ice-cold wash buffer (PBS, 150 mmol/L NaCl). Processed cell lysates were then added to the affinity gel and rotated at 4°C for at least 2 h. The protein was incubated with 2 mmol/L DTT in wash buffer (PBS, 150 mmol/L NaCl) and incubated at 37°C for 30 min, followed by buffer exchange to remove DTT.
GSNO, GSH-Sepharose, and GSNO-Sepharose preparation
S-nitrosoglutathione, GSH- and GSNO-Sepharose were prepared as previously described (8, 10, 14). To make GSNO, 0.75 g reduced L-glutathione was dissolved in 5.25 ml ice-cold 0.5 N HCl. 170 mg of sodium nitrite was then added to generate nitrous acid (HNO2). The resulting red mixture was stirred at 4°C for 40 min. 2 ml chilled acetone was then added and the mixture stirred for an additional 10 min at 4°C to allow precipitation of the GSNO. The mixture was then centrifuged at 2000 g for 5 min. The supernatant was carefully decanted, and the product was washed three times with acetone and three washes with ether. The GSNO was then dried via SpeedVac and stored in the dark at −20°C. GSNO was prepared freshly for each experiment and stored on ice. GSNO was diluted to indicated concentrations and samples were treated for 15 min at 37°C.
GSH- and GSNO-Sepharose were prepared and used as previously described (10). All solutions were degassed with nitrogen prior to use. One gram of activated Sepharose 4B (Amersham Biosciences, Piscataway, NJ) was reconstituted in 10 ml PBS at room temperature for 15 min. The beads were then washed twice with 10 ml PBS. Thiol reduction was performed by incubating the beads in 50 mmol/L Tris pH 8.5, 0.3 M NaHCO3, 1 mmol/L EDTA, and 1% (w/v) DTT for 45 min on a shaker. The beads were then rinsed with several column volumes of wash buffer (50 mmol/L Tris pH 7.4, 250 mmol/L NaCl, 1 mmol/L EDTA). The activated thiol Sepharose 4B (GSH-Sepharose) was stored at 4°C in wash buffer. Free thiols were determined by Ellman's reagent. Half of the bead volume was reserved to generate GSNO-Sepharose. These beads were rinsed in several bead volumes of 10 mmol/L HCl, followed by resuspension in 2 ml 10 mmol/L HCl. 2 ml 10 mmol/L NaNO2 was added to form nitrous acid. The mixture was incubated on a shaker for 15 min in buffer A (50 mmol/L Tris pH 8.0, 250 mmol/L NaCl, 1 mmol/L EDTA, 0.01% NP40). The resulting GSNO-Sepharose was rinsed in several bead volumes of buffer A and stored in the dark at 4°C. For experiments, 100 μl beads were rinsed three times in buffer A and then incubated with 100 μg recombinant SIRT1 on a shaker for 30 min at room temperature. The beads were spun down for 1 min at 8000 g and the beads containing bound SIRT1 were washed three times with Buffer A. All fractions were reserved for analysis by SDS-PAGE, followed by transfer to nitrocellulose membranes and Memcode reversible protein stain (Pierce, Rockford, IL).
Measurement of SIRT1 activity
SIRT1 activity was measured according to modified published methods (26). The assay is a two-step process in which the purified SIRT1 is first incubated with a peptide substrate corresponding to the p53 acetylation target site spanning amino acids 379–382 (Arg-His-Lys-Lys(Ac)-AMC). This step involves a 30 min incubation at 37°C with 1 mmol/L NAD+ and 50 μM substrate peptide to allow SIRT1 to freely deacetylate the peptide in activity assay buffer (50 mmol/L Tris-Cl, pH 8.0, 137 mmol/L NaCl, 2.7 mmol/L KCl, 1 mmol/L MgCl2). Deacetylation of the peptide exposes the lysine to tryptic cleavage. In the second step, 1 mmol/L nicotinamide (NAM) is added to block further SIRT1 activity, and trypsin is then added to cleave any deacetylated substrate. Lysine cleavage by trypsin releases the fluorescent AMC fluorophore allowing the quantification of the amount of substrate deacetylated by SIRT1 during the first step. The amount of fluorescence is directly proportional to the level of SIRT1 activity in the sample. The trypsin reaction was allowed to proceed for 60 min. The cleaved fluorescent tag was excited at 375 nm, and fluorescence emission was recorded at 465 nm in a Fluoroskan Ascent microplate fluorometer (Thermo Scientific, Waltham, MA). Total protein content was determined by SDS-PAGE, followed by GelCode blue staining. Densitometry of the SIRT1 bands was performed using ImageJ (NIH) and quantified according to a BSA standard curve run on the same gel.
Differential alkylation for MS analysis
SIRT1-Flag protein was immuno-precipitated with anti-Flag M2 affinity gel (Sigma-Aldrich) and divided into four equal aliquots. Two of the aliquots were treated with 2 mmol/L GSNO at 37°C for 10 min to react with any reactive surface cysteines. The other two samples were left untreated to serve as controls. All four samples were then washed with wash buffer (PBS, 150 mmol/L NaCl). One GSNO-treated sample and one control sample were treated with iodoacetamide (IAM, 40 mmol/L, Sigma) for 1 h on ice in the dark to alkylate free cysteines. The other set of GSNO-treated and control samples were alkylated in parallel under the same conditions, but with N-ethylmaleimide (NEM, 40 mmol/L, Sigma) instead of IAM. All samples were washed 3x with wash buffer and boiled at 95°C for 5 min in reducing Laemmli buffer. Samples were then subjected to SDS-PAGE using PAGEr Gold Precast 4–12% Tris-Glycine Gels (Lonza, Basel, Switzerland). The gel was then washed three times in MilliQ water for 5 min each time, followed by GelCode Blue (Pierce) staining overnight. The gel was again washed three times in MilliQ water for 5 min each time to destain the gel. The protein bands were excised and placed in separate, fresh 1.5 mL SafeLock tubes (Eppendorf, Hamburg, Germany). The gel pieces were washed to remove SDS, salts, and other contaminants from the gel pieces before digestion, and then underwent reduction of reversibly oxidized thiols with DTT and alkylation with either IAM or NEM, whichever one was not used previously on the sample.
In-solution digest of GSNO treated SIRT1-His
An in-solution digestion method was employed without reduction to preserve and identify S-glutathiolated cysteine residue(s) using mass spectrometry. Purified human SIRT1-His (20 μg) at a protein concentration of 0.22 μg/μl, was treated with GSNO (2 mmol/L) and incubated for 15 min at 37°C. The sample was then alkylated with 40 mmol/L NEM dissolved in 100 mmol/L ABC pH 8.0. The samples were alkylated in the dark, at room temperature for 30 min. Untreated SIRT1-His was also alkylated with 40 mM NEM and run in parallel as a negative control.
To remove the alkylating reagent and buffer salts, both samples were subjected to a ReadyPrep 2D Cleanup Kit (BioRad, Hercules, CA). After precipitation, the protein pellet was resuspended in 5 μl of 100 mM ABC pH 8.0. 20 μL of acid-labile RapiGest™ SF Surfactant (Waters, Milford, MA) solution was added to each sample, along with 200 ng of trypsin. The samples were digested overnight at 37°C. TFA (1 μl) was added to degrade the detergent and the samples were then divided in half in preparation for MALDI-TOF MS and Orbitrap LC-MS/MS. All samples were desalted using micro-C18 ZipTip™ (Millipore, Temecula, CA) columns. Approximately 1–5 μg of peptide samples were loaded onto the ZipTip™ column and eluted into 10 μl of 50% ACN, 0.1% TFA.
Mass spectrometry and data analysis
MALDI-TOF MS was performed on Reflex IV™ mass spectrometer (Bruker-Daltonics, Billerica, MA) and LC-MS/MS was acquired on an LTQ-Orbitrap™ hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a TriVersa NanoMate ion source (Advion, Ithaca, NY). A washing regimen consisted of adding 100 μl of 100 mM ammonium bicarbonate (ABC) (Fluka) at pH 8.0, then 100 μl of 100 mM ABC, 50% HPLC-grade acetonitrile (ACN) (Fisher Scientific), then 100 μl of 100% ACN with light vortexing in a Thermomixer (Eppendorf) for 15 min for each step. The washing regimen was repeated a total of three times. After the washing regimen was completed, the gel pieces were dried using the SpeedVac evaporator for 15 min with no heat. Gel pieces were rehydrated in fresh reduction buffer, consisting of 30 μl of 20 mM DTT (Thermo Fisher Scientific, San Jose, CA), 100 mM ABC, 5% CAN, and incubated for 1 h at 55°C. The supernatant was then removed and the gel pieces were washed in 100 μl of 100 mM ABC, followed by dehydration using 100 μl of 100% ACN for 20 min with light vortexing. The supernatant was removed and the samples that were initially alkylated with IAM were now alkylated with 40 mM NEM for 30 min in the dark, at room temperature. The samples that were initially alkylated with NEM were alkylated a second time with 40 mM IAM. After in-gel alkylation, the previously described washing regimen was used again to clean the gel samples in preparation for digestion. MS-grade Trypsin Gold (Promega Corporation, Madison, WI) was prepared in 50 mM ABC, 5% ACN buffer to obtain a 1:10 enzyme to sample weight ratio per 25 μl of trypsin buffer. The dried gel pieces were then rehydrated in 25 μl of trypsin buffer and incubated at 37°C overnight. Excess trypsin was washed off the exterior of the gel pieces by quickly adding then decanting 20 mM ABC (pH 8.0). An extraction regimen was then used to extract and collect the peptides from the gel pieces. ABC (100 μl of 20 mM) was added to the gel pieces, lightly vortexed for 20 min, and the supernatant was collected in a fresh 1.5 mL SafeLock tube, followed by 100 μl of 1% trifluoroacetic acid (TFA) (Thermo Fisher Scientific), 50% ACN, followed by 100 μl of 100% ACN. The extraction regimen was repeated once more, pooling all the supernatants. The supernatants were then dried down in the SpeedVac evaporator. The remaining salt pellet was then resuspended in 500 μl of 50% ACN and dried down in the SpeedVac evaporator to eliminate the remaining volatile salts from the sample. The peptides were resuspended in 20 μl of 0.1% TFA. Ten μl of the sample was used for MALDI-TOF MS while the retaining the remaining sample for LC-MS/MS analysis. 0.5 μl of the desalted peptide sample was then crystallized onto the AnchorChip™ target (Bruker-Daltonics, Billerica, MA) with 2,5-dihydroxybenzoic acid (DHB) as the matrix. Spectra were taken on samples using a Reflex IVTM matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer (Bruker-Daltonics). Spectra were taken over the range m/z 300–4000. Each output spectra was taken from an average of the approximately 300–500 laser shots of each sample.
All MS spectra obtained from MALDI-TOF MS were analyzed using the program “M over Z” (Genomic Solutions, Ann Arbor, MI). Each spectrum was smoothed to improve the isotopic peak resolution and decrease background noise. The spectrum was internally calibrated against three known, unique, and unmodified SIRT1 peptide masses. The peaks in the spectrum were then auto-labeled and a mass list of monoisotopic peak masses was then generated and searched using the MS-Fit program on Protein Prospector (UCSF; http://prospector.ucsf.edu/prospector/mshome.htm, accessed June 6, 2010). Search settings allowed for carbamidomethylation, N-ethylmaleimide alkylation, and methionine oxidation. Two missed cleavages and an error tolerance of 50 ppm were allowed for the database search.
Any cysteine-containing peptides from the search results were compiled in a list and the masses were manually searched through each spectrum. Peaks for cysteine-containing peptides showing differential labeling between the four samples were recorded. This search culminated in the identification of five specific cysteine residues of interest. These cysteines showed reactivity toward the alkylating agents, were labeled differentially following modification by GSNO, and are cysteines that likely accessible on the surface of the native protein and are potential sites for regulation.
For LC-MS/MS, peptides were subjected to purification using ZipTip™ (Millipore) micro reversed-phase extraction. LC-MS was performed on the resulting peptides using a nanoAcquity UPLC™ capillary high-performance liquid chromatography system (Waters Corp., Milford, MA) coupled to an LTQ-Orbitrap™ hybrid mass spectrometer (ThermoFisher Scientific) equipped with a TriVersa NanoMate ion source (Advion, Ithaca, NY). Sample concentration and desalting were performed online using a nanoAcquity UPLC™ trapping column. (180 μm x 20 mm, packed with 5 μm, 100 Å Symmetry C18 material, Waters Corp.) at a flow rate of 15 μl/min for 1 min, while separation was accomplished across a nanoAcquity UPLC™ capillary column (100 μm x 100 mm, packed with 1.7 μm, 130 Å BEH C18 material, Waters Corp.) A linear gradient of A and B buffers. (buffer A: 3% CAN/0.1%FA; buffer B: 97% CAN/0.1% FA) over 240 min was used at a flow rate of 0.5 μl/min to elute peptides into the mass spectrometer. Columns were washed and re-equilibrated between LC-MS experiments. Electrospray ionization was carried out with the NanoMate at 1.75 kV, with the LTQ heated capillary set to 225°C. Mass spectra were acquired in the Orbitrap in the positive ion mode over the range m/z 300–2000 at a resolution of 60,000 (approximately 1 mass spectrum/ s). Mass accuracy after internal calibration (using lock mass on siloxane ions) was typically within +/− 2 ppm. Simultaneously, tandem MS (MS/MS) were acquired using the LTQ in data-dependent, inclusion list-directed mode: Inclusion lists (parent ion lists) were generated for cysteine-containing SIRT1 peptides through in silico tryptic digestion of the human SIRT1 protein sequence, including up to four missed cleavages and glutathiolation, di- or tri-oxidation, NEM or IAM-modification of each cysteine alone and in combination, calculating m/z values for charge states 2+, 3+, and 4+. The five most abundant, multiply-charged species from the list that were found in the mass spectrum with signal intensities >8000 NL were chosen for MS/MS. Alternatively, if the mass spectrum contained no entries from the list, then the most abundant multiply charged detected ions were chosen for MS/MS. MS/MS collision energies were set at 35% using helium as the collision gas, and MS/MS were acquired over a range of m/z values dependent on the precursor ion. Dynamic exclusion was set such that MS/MS for each species was acquired maximally twice. All spectra were recorded in profile mode for further processing and analysis. Xcalibur™ and Proteome Discoverer™ software (ThermoFisher Scientific) were used for MS and MS/MS data analysis, while peptide and protein assignments were conducted with Mascot™ to search against the human and mouse species subsets of the IPI database, employing an error window of 8 ppm on the precursor ions and 1.2 Da on the fragment ions. The error tolerant search function of Mascot™ was employed. Alternatively, variable modifications were set to include methionine oxidation, cysteine carbamidomethylation, N-ethyl maleimide modification, di- and tri-oxidation, and glutathiolation, asparagines and glutamine deamidation, and glutamate and glutamine pyroglutamic acid formation. Potential assignments by Mascot™ were verified and refined by manual interpretation of the original spectra. Scaffold™ (Proteome software) was used for meta-analysis of the Mascot™ search results.
Results
To further confirm the identification of peptides of SIRT1 that was not treated with GSNO, but alkylated in the IAM-DTT-NEM treated samples, we subjected samples to LC-MS/MS. We detected a [M + 2H]2+ ion corresponding to m/z 1339.661 in the control spectrum (Supplemental Fig. 2). The ion was then subjected to collision-induced fragmentation in the mass spectrometer. The fragmentation ion spectrum of this ion identified the peptide fragment as amino acids 46–74 containing a carbamidomethyl group (+57 Da) on Cys-61. This observation indicates that under control conditions in which SIRT1 is not treated with GSNO, the residue is modified by the initial alkylation with IAM. Furthermore, this peptide does not appear in the GSNO-IAM-DTT-NEM sample because GSNO induces a reversible modification that blocks alkylation by IAM. The peptide containing Cys-61 should instead be modified by NEM after DTT reduction, and will therefore have a different mass. This peptide (Cys-61 + 125 Da) was not identified in spectra derived from LTQ-Orbitrap, but was positively identified in the MALDI-TOF spectra (Fig. 3). These results indicate that GSNO treatment of SIRT1 results in a site-specific S-glutathiolation. Although we identified five putative reactive cysteines by analysis with MALDI-TOF MS, only mouse SIRT1 Cys-61 was identified as being modified by GSNO using both MALDI-TOF MS and LC-MS/MS methods.
Supplementary Material
Abbreviations Used
- Cys
cysteine
- DTT
dithiothreitol
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- IAM
iodoacetamide
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- MS
mass spectrometry
- NEM
N-ethylmaleimide
- OPTM
oxidative post-translational modification
- RSV
resveratrol
- SIRT
sirtuin-1
Acknowledgments
This work was presented and published in abstract form at annual meetings of the Society of Free Radical Biology and Medicine, Free Rad Biol Med 45 Suppl. 1 S76, 2008, 47 Suppl. 1 S83, 2009. This work was supported by NIH Grant PO1 HL068758, the National Heart, Lung, and Blood Institute sponsored Boston University Cardiovascular Proteomics Center (Contract No. N01-HV-28178, NIH NCRR P41-RR10888, S10-R15942, S10-RR20946) and NIH pre-doctoral training grant HL007969-06A1. We thank Dr. Mengwei Zang for expert assistance and for providing valuable reagents. The authors would also like to thank Dr. Jessica Fry for providing a critical reading of the manuscript and helpful scientific discussion.
References
- 1.Alcendor RR. Kirshenbaum LA. Imai S. Vatner SF. Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 2.Autiero I. Costantini S. Colonna G. Human sirt-1: Molecular modeling and structure-function relationships of an unordered protein. PLoS ONE. 2008;4:e7350. doi: 10.1371/journal.pone.0007350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandra A. Srivastava S. Petrash JM. Bhatnagar A. Srivastava SK. Modification of aldose reductase by S-nitrosoglutathione. Biochemistry. 1997;36:15801–15809. doi: 10.1021/bi9714722. [DOI] [PubMed] [Google Scholar]
- 4.Csiszar A. Labinskyy N. Podlutsky A. Kaminski PM. Wolin MS. Zhang C. Mukhopadhyay P. Pacher P. Hu F. de CR. Ballabh P. Ungvari Z. Vasoprotective effects of resveratrol and SIRT1: Attenuation of cigarette smoke-induced oxidative stress and proinflammatory phenotypic alterations. Am J Physiol Heart Circ Physiol. 2008;294:H2721–H2735. doi: 10.1152/ajpheart.00235.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csiszar A. Smith K. Labinskyy N. Orosz Z. Rivera A. Ungvari Z. Resveratrol attenuates TNF-alpha-induced activation of coronary arterial endothelial cells: Role of NF-kappaB inhibition. Am J Physiol Heart Circ Physiol. 2006;291:H1694–H1699. doi: 10.1152/ajpheart.00340.2006. [DOI] [PubMed] [Google Scholar]
- 6.Fulco M. Schiltz RL. Iezzi S. King MT. Zhao P. Kashiwaya Y. Hoffman E. Veech RL. Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 7.Giustarini D. Milzani A. Aldini G. Carini M. Rossi R. le–Donne I. S-nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid Redox Signal. 2005;7:930–939. doi: 10.1089/ars.2005.7.930. [DOI] [PubMed] [Google Scholar]
- 8.Hart TW. Some observations concerning the S-nitroso and S-phenylsulphonyl derivatives of L-cysteine and glutathione. Tetrahedron Lett. 1985;26:2013–2016. [Google Scholar]
- 9.Klatt P. Molina EP. Lamas S. Nitric oxide inhibits c-Jun DNA binding by specifically targeted S-glutathionylation. J Biol Chem. 1999;274:15857–15864. doi: 10.1074/jbc.274.22.15857. [DOI] [PubMed] [Google Scholar]
- 10.Klatt P. Pineda ME. Perez–Sala D. Lamas S. Novel application of S-nitrosoglutathione-Sepharose to identify proteins that are potential targets for S-nitrosoglutathione-induced mixed-disulphide formation. Biochem J. 2000;349:567–578. doi: 10.1042/0264-6021:3490567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langley E. Pearson M. Faretta M. Bauer UM. Frye RA. Minucci S. Pelicci PG. Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leikert JF. Rathel TR. Wohlfart P. Cheynier V. Vollmar AM. Dirsch VM. Red wine polyphenols enhance endothelial nitric oxide synthase expression and subsequent nitric oxide release from endothelial cells. Circulation. 2002;106:1614–1617. doi: 10.1161/01.cir.0000034445.31543.43. [DOI] [PubMed] [Google Scholar]
- 13.Lin SJ. Ford E. Haigis M. Liszt G. Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z. Rudd MA. Freedman JE. Loscalzo J. S-Transnitrosation reactions are involved in the metabolic fate and biological actions of nitric oxide. J Pharmacol Exp Ther. 1998;284:526–534. [PubMed] [Google Scholar]
- 15.Milne JC. Lambert PD. Schenk S. Carney DP. Smith JJ. Gagne DJ. Jin L. Boss O. Perni RB. Vu CB. Bemis JE. Xie R. Disch JS. Ng PY. Nunes JJ. Lynch AV. Yang H. Galonek H. Israelian K. Choy W. Iffland A. Lavu S. Medvedik O. Sinclair DA. Olefsky JM. Jirousek MR. Elliott PJ. Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min J. Landry J. Sternglanz R. Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 17.Mohr S. Hallak H. de BA. Lapetina EG. Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 18.Napper AD. Hixon J. McDonagh T. Keavey K. Pons JF. Barker J. Yau WT. Amouzegh P. Flegg A. Hamelin E. Thomas RJ. Kates M. Jones S. Navia MA. Saunders JO. DiStefano PS. Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 19.Percival MD. Ouellet M. Campagnolo C. Claveau D. Li C. Inhibition of cathepsin K by nitric oxide donors: Evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574–13583. doi: 10.1021/bi991028u. [DOI] [PubMed] [Google Scholar]
- 20.Potente M. Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 21.Prozorovski T. Schulze–Topphoff U. Glumm R. Baumgart J. Schroter F. Ninnemann O. Siegert E. Bendix I. Brustle O. Nitsch R. Zipp F. Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 22.Rodgers JT. Lerin C. Haas W. Gygi SP. Spiegelman BM. Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki T. Maier B. Koclega KD. Chruszcz M. Gluba W. Stukenberg PT. Minor W. Scrable H. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schilling B. Yoo CB. Collins CJ. Gibson BW. Determining cysteine oxidation status using differential alkylation. Int J Mass Spectrom. 2004;236:117–127. [Google Scholar]
- 25.Shelton MD. Chock PB. Mieyal JJ. Glutaredoxin: Role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 26.Vaziri H. Dessain SK. Ng EE. Imai SI. Frye RA. Pandita TK. Guarente L. Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 27.Wallerath T. Deckert G. Ternes T. Anderson H. Li H. Witte K. Forstermann U. Resveratrol, a polyphenolic phytoalexin present in red wine, enhances expression and activity of endothelial nitric oxide synthase. Circulation. 2002;106:1652–1658. doi: 10.1161/01.cir.0000029925.18593.5c. [DOI] [PubMed] [Google Scholar]
- 28.West MB. Hill BG. Xuan YT. Bhatnagar A. Protein glutathiolation by nitric oxide: An intracellular mechanism regulating redox protein modification. FASEB J. 2006;20:1715–1717. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y. Fu W. Chen J. Olashaw N. Zhang X. Nicosia SV. Bhalla K. Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zech B. Wilm M. van ER. Brune B. Mass spectrometric analysis of nitric oxide-modified caspase-3. J Biol Chem. 1999;274:20931–20936. doi: 10.1074/jbc.274.30.20931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.