Abstract
In the title compound, C10H6ClNO2, the dihedral angle between the benzene and maleimide rings is 47.54 (9)°. Molecules form centrosymmetric dimers through C—H⋯O hydrogen bonds, resulting in rings of graph-set motif R 2 2(8) and chains in the [100] direction. Molecules are also linked by C—H⋯Cl hydrogen bonds along [001]. In this same direction, molecules are connected to other neighbouring molecules by C—H⋯O hydrogen bonds, forming edge-fused R 4 4(24) rings.
Related literature
For general background, see: Etter (1990 ▶); Howell & Zhang (2006 ▶); Miller et al. (2000 ▶, 2001 ▶); Moreno-Fuquen, Valencia, Abonia, Kennedy & Graham (2003 ▶); Nardelli (1995 ▶); Sarma & Desiraju (1986 ▶). 
Experimental
Crystal data
C10H6ClNO2
M r = 207.61
Monoclinic,

a = 10.6504 (7) Å
b = 3.8589 (2) Å
c = 22.0308 (14) Å
β = 100.741 (3)°
V = 889.57 (9) Å3
Z = 4
Mo Kα radiation
μ = 0.40 mm−1
T = 150 K
0.18 × 0.04 × 0.03 mm
Data collection
Bruker–Nonius KappaCCD diffractometer
Absorption correction: multi-scan (DENZO; Otwinowski & Minor, 1997 ▶) T min = 0.951, T max = 0.982
11729 measured reflections
1646 independent reflections
1231 reflections with I > 2σ(I)
R int = 0.089
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.116
S = 1.07
1646 reflections
128 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.31 e Å−3
Data collection: DENZO (Otwinowski & Minor, 1997 ▶) and COLLECT (Nonius, 2000 ▶); cell refinement: DENZO; data reduction: DENZO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: PARST95 (Nardelli, 1995 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680803016X/fj2150sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803016X/fj2150Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C8—H8⋯O1i | 0.93 | 2.58 | 3.493 (3) | 169 |
| C2—H2⋯O1ii | 0.93 | 2.77 | 3.659 (3) | 161 |
| C5—H5⋯O2iii | 0.93 | 2.58 | 3.319 (3) | 137 |
| C9—H9⋯O2iv | 0.93 | 2.64 | 3.326 (3) | 131 |
| C9—H9⋯Cl1v | 0.93 | 2.89 | 3.551 (3) | 129 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
RMF dedicates this work to the memory of Professor J. Valderrama. RMF is grateful to the Instituto de Química Física Rocasolano, CSIC, Spain, for the use of the license of Cambridge Structural Database System (Allen, 2002 ▶). This work was partially supported by the Universidad del Valle, Colombia.
supplementary crystallographic information
Comment
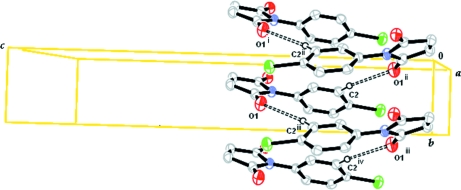
It is known that cyclic unsaturated dicarbonyl compounds such as N-substituted maleimides can be used in free-radical-initiated polymerization processes upon exposure to light (Howell & Zhang, 2006). In order to study the possible application of N-(p-chlorophenylmaleimide) (I) in polymerization processes, and to explain its hydrogen bonding patterns, the synthesis and the study of the crystal structure are reported in this work. N-(p-nitrophenylmaleimide) (4NPMI) (Moreno-Fuquen et al., 2003), N-(o-chlorophenylmaleimide) (2ClPMI) systems (Miller et al., 2001) show a close analogy to the title compounds and are thus employed as a basic reference for comparison. Perspective view of (I), showing the atomic numbering scheme, can be seen in Fig.1. In the arylmaleimide systems the value of the dihedral angle between the benzene and imidic rings influences on the polimerization process, and the presence of different substituents in the benzene ring change the value of this angle (Miller et al., 2000). The photochemical properties of arylmaleimide systems depend on the value of this angle. The dihedral angle between benzene and maleimide planes is 42.98 (5)° for 4NPMI, 66.10 (4) ° for 2ClPMI and 47.54 (9)° for (I). The chlorine atoms, which are pending on the aromatic nucleus, tend to steer the crystal structure to a state characterized by a short axis (Sarma & Desiraju, 1986). For (I), the b axis has a small value [3.8589 (2) Å] and a Cl···Cl nonbonded contact is observed at the same distance. The crystal structure of (I) is stabilized by weak intermolecular C—H···O and C—H···Cl hydrogen-bonds (Nardelli, 1995) (Table 1). The molecules of (I) are linked into a three-dimensional framework by a combination of C—H···O and C—H···Cl hydrogen bonds. The formation of the framework can be explained in terms of three-one substructures. In the first substructure, atom C8 in the molecule at (x,y,z) acts as a hydrogen-bond donor to maleimidic atom O1 in the molecule at (-x,1 - y,1 - z) and atom C9 in the molecule at (x,y,z) acts as a hydrogen-bond donor to maleimidic atom O2 in the molecule at (1 - x,1 - y,1 - z). Both interactions generate dimers containing centrosymmetric rings with graph motif R22(8) (Etter, 1990) (Fig. 2, supp. material). These dimers are linked by C(5) chains which are running parallel to [100] direction. In the second substructure, atom C9 in the molecule at (x,1/2 - y,-1/2 + z) acts as a hydrogen-bond donor to atom Cl1 in the molecule at (x,y,z), similarly, atom C5 in the molecule at (x,y,z) acts as a hydrogen-bond donor to maleimidic atom O2 in the molecule at (x,1 + y,z) so generating a chain of edge-fused R44(24) rings along [001] (Fig. 3, supp. material). The third one-dimensional substructure is built by C—H···O hydrogen bonds. Atom C2 in the molecule at (x,y,z) acts as hydrogen bond donor to maleimidic O1 in the molecule at (-x,-1/2 + y,1/2 - z) so generating a C(7) chains in the [010] direction (Fig.4, supp. material). The low value of the dihedral angle between benzene and maleimide planes, allows to conclude that (I) is not a good candidate to use in a photopolymerization process.
Experimental
All reagents (purchased from Aldrich) and solvents were used as received. Column chromatography was performed using silica gel H60 to purify the intermediates and final products. Thin layer chromatography (TLC) was used to confirm the structure of the individual compounds.
Refinement
The space group P 21/c for (I) was uniquely assigned from the systematic absences. All H-atoms were located from difference maps and then treated as riding atoms [C—H= 0.93Å and Uiso(H)= 1.2Ueq(C)].
Figures
Fig. 1.
An ORTEP-3 (Farrugia, 1997) plot of the (I) compound, with the atomic labelling scheme. The shapes of the ellipsoids correspond to 50% probability contours of atomic displacement and, for the sake of clarity, H atoms are shown as spheres of arbitrary radius.
Fig. 2.
Part of the crystal structure of (I) showing the formation of centrosymmetric R22(8) dimmers rings and C(4) chains which are running parallel to the [100] direction. [Symmetry codes: (i) 1 + x,y + z; (ii)1 - x,1 - y,1 - z; (iii) -x,1 - y,1 - z].
Fig. 3.
Part of the crystal structure of (I) showing the formation of C(9) chains and edge-fused R44(24) rings along [001]. [Symmetry codes: (i) x,1/2 - y,-1/2 + z; (ii) x,1 + y,z; (iii) x,y,-1 + z; (iv) x,1 + y,-1 + z; (v) x,3/2 - y,-1/2 + z; (vi) x,1/2 - y,1/2 + z; (vii) x,1/2 - y,1/2 + z].
Fig. 4.
Part of the crystal structure of (I) showing the formation of C(7) chains along [010]. [Symmetry codes: (i) x,-1 + y,z; (ii) -x,-1/2 + y,1/2 + z; (iii) -x,1/2 + y,1/2 - z; (iv) x,1 + y,z].
Fig. 5.
The formation of the title compound.
Crystal data
| C10H6ClNO2 | F(000) = 424 |
| Mr = 207.61 | Dx = 1.550 Mg m−3 |
| Monoclinic, P21/c | Melting point: 384(1) K |
| Hall symbol: -P 2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.6504 (7) Å | Cell parameters from 11729 reflections |
| b = 3.8589 (2) Å | θ = 2.9–25.4° |
| c = 22.0308 (14) Å | µ = 0.40 mm−1 |
| β = 100.741 (3)° | T = 150 K |
| V = 889.57 (9) Å3 | Needle, pale-yellow |
| Z = 4 | 0.18 × 0.04 × 0.03 mm |
Data collection
| Bruker–Nonius KappaCCD diffractometer | 1646 independent reflections |
| Radiation source: fine-focus sealed tube | 1231 reflections with I > 2σ(I) |
| graphite | Rint = 0.089 |
| φ and ω scans | θmax = 25.4°, θmin = 3.0° |
| Absorption correction: multi-scan (DENZO; Otwinowski & Minor, 1997) | h = −12→12 |
| Tmin = 0.951, Tmax = 0.982 | k = −4→4 |
| 11729 measured reflections | l = −24→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.116 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0501P)2 + 0.491P] where P = (Fo2 + 2Fc2)/3 |
| 1646 reflections | (Δ/σ)max < 0.001 |
| 128 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.26337 (8) | 0.6828 (2) | 0.13972 (3) | 0.0486 (3) | |
| O1 | 0.04581 (16) | 0.6461 (5) | 0.40894 (8) | 0.0419 (5) | |
| O2 | 0.45773 (15) | 0.2532 (5) | 0.43624 (8) | 0.0342 (5) | |
| N1 | 0.25175 (18) | 0.4677 (5) | 0.40446 (9) | 0.0278 (5) | |
| C1 | 0.2590 (2) | 0.6168 (6) | 0.21732 (11) | 0.0303 (6) | |
| C2 | 0.1564 (2) | 0.4474 (7) | 0.23409 (11) | 0.0314 (6) | |
| H2 | 0.0899 | 0.3657 | 0.2040 | 0.038* | |
| C3 | 0.1537 (2) | 0.4004 (6) | 0.29608 (11) | 0.0282 (6) | |
| H3 | 0.0852 | 0.2870 | 0.3081 | 0.034* | |
| C4 | 0.2542 (2) | 0.5240 (6) | 0.34041 (11) | 0.0269 (6) | |
| C5 | 0.3567 (2) | 0.6933 (6) | 0.32329 (12) | 0.0283 (6) | |
| H5 | 0.4237 | 0.7748 | 0.3532 | 0.034* | |
| C6 | 0.3585 (2) | 0.7399 (6) | 0.26132 (12) | 0.0303 (6) | |
| H6 | 0.4266 | 0.8541 | 0.2492 | 0.036* | |
| C7 | 0.1462 (2) | 0.5170 (7) | 0.43329 (12) | 0.0323 (6) | |
| C8 | 0.1852 (2) | 0.3901 (7) | 0.49762 (11) | 0.0340 (6) | |
| H8 | 0.1336 | 0.3862 | 0.5273 | 0.041* | |
| C9 | 0.3057 (2) | 0.2833 (7) | 0.50601 (12) | 0.0317 (6) | |
| H9 | 0.3524 | 0.1958 | 0.5427 | 0.038* | |
| C10 | 0.3538 (2) | 0.3260 (6) | 0.44722 (11) | 0.0291 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0730 (6) | 0.0459 (5) | 0.0305 (4) | 0.0071 (4) | 0.0186 (4) | 0.0053 (3) |
| O1 | 0.0297 (10) | 0.0600 (13) | 0.0376 (11) | 0.0126 (9) | 0.0101 (8) | 0.0056 (9) |
| O2 | 0.0285 (10) | 0.0420 (11) | 0.0320 (10) | 0.0049 (8) | 0.0055 (8) | −0.0004 (8) |
| N1 | 0.0251 (11) | 0.0339 (12) | 0.0249 (11) | 0.0031 (9) | 0.0061 (9) | 0.0013 (9) |
| C1 | 0.0401 (15) | 0.0260 (14) | 0.0262 (14) | 0.0073 (11) | 0.0102 (11) | 0.0018 (11) |
| C2 | 0.0325 (14) | 0.0296 (14) | 0.0304 (15) | 0.0052 (11) | 0.0009 (11) | −0.0018 (11) |
| C3 | 0.0242 (13) | 0.0275 (14) | 0.0337 (14) | 0.0016 (10) | 0.0074 (11) | 0.0025 (11) |
| C4 | 0.0274 (13) | 0.0262 (13) | 0.0277 (13) | 0.0047 (10) | 0.0064 (10) | 0.0008 (10) |
| C5 | 0.0278 (13) | 0.0252 (13) | 0.0316 (14) | 0.0035 (10) | 0.0045 (10) | −0.0025 (11) |
| C6 | 0.0301 (13) | 0.0260 (14) | 0.0382 (16) | 0.0034 (11) | 0.0151 (12) | 0.0020 (11) |
| C7 | 0.0286 (14) | 0.0368 (15) | 0.0329 (15) | 0.0005 (12) | 0.0097 (11) | −0.0052 (12) |
| C8 | 0.0361 (15) | 0.0392 (16) | 0.0288 (14) | −0.0014 (12) | 0.0121 (11) | −0.0005 (12) |
| C9 | 0.0346 (15) | 0.0338 (14) | 0.0260 (14) | −0.0009 (12) | 0.0036 (11) | −0.0013 (11) |
| C10 | 0.0311 (14) | 0.0261 (13) | 0.0292 (14) | 0.0017 (11) | 0.0036 (11) | −0.0025 (11) |
Geometric parameters (Å, °)
| Cl1—C1 | 1.738 (2) | C3—H3 | 0.9300 |
| O1—C7 | 1.210 (3) | C4—C5 | 1.384 (3) |
| O2—C10 | 1.209 (3) | C5—C6 | 1.380 (4) |
| N1—C7 | 1.404 (3) | C5—H5 | 0.9300 |
| N1—C10 | 1.410 (3) | C6—H6 | 0.9300 |
| N1—C4 | 1.433 (3) | C7—C8 | 1.484 (4) |
| C1—C6 | 1.380 (4) | C8—C9 | 1.327 (4) |
| C1—C2 | 1.381 (4) | C8—H8 | 0.9300 |
| C2—C3 | 1.383 (3) | C9—C10 | 1.489 (4) |
| C2—H2 | 0.9300 | C9—H9 | 0.9300 |
| C3—C4 | 1.392 (3) | ||
| C7—N1—C10 | 109.5 (2) | C4—C5—H5 | 120.4 |
| C7—N1—C4 | 126.0 (2) | C1—C6—C5 | 120.0 (2) |
| C10—N1—C4 | 124.3 (2) | C1—C6—H6 | 120.0 |
| C6—C1—C2 | 121.1 (2) | C5—C6—H6 | 120.0 |
| C6—C1—Cl1 | 118.86 (19) | O1—C7—N1 | 124.8 (2) |
| C2—C1—Cl1 | 120.01 (19) | O1—C7—C8 | 128.8 (2) |
| C1—C2—C3 | 119.3 (2) | N1—C7—C8 | 106.4 (2) |
| C1—C2—H2 | 120.4 | C9—C8—C7 | 109.1 (2) |
| C3—C2—H2 | 120.4 | C9—C8—H8 | 125.5 |
| C2—C3—C4 | 119.5 (2) | C7—C8—H8 | 125.5 |
| C2—C3—H3 | 120.2 | C8—C9—C10 | 109.0 (2) |
| C4—C3—H3 | 120.2 | C8—C9—H9 | 125.5 |
| C5—C4—C3 | 120.9 (2) | C10—C9—H9 | 125.5 |
| C5—C4—N1 | 120.0 (2) | O2—C10—N1 | 125.3 (2) |
| C3—C4—N1 | 119.1 (2) | O2—C10—C9 | 128.7 (2) |
| C6—C5—C4 | 119.2 (2) | N1—C10—C9 | 106.0 (2) |
| C6—C5—H5 | 120.4 | ||
| C6—C1—C2—C3 | 0.0 (4) | C10—N1—C7—O1 | 177.3 (3) |
| Cl1—C1—C2—C3 | −179.27 (18) | C4—N1—C7—O1 | −7.5 (4) |
| C1—C2—C3—C4 | −0.1 (4) | C10—N1—C7—C8 | −1.2 (3) |
| C2—C3—C4—C5 | 0.0 (4) | C4—N1—C7—C8 | 174.0 (2) |
| C2—C3—C4—N1 | −178.7 (2) | O1—C7—C8—C9 | −177.0 (3) |
| C7—N1—C4—C5 | 136.1 (3) | N1—C7—C8—C9 | 1.5 (3) |
| C10—N1—C4—C5 | −49.4 (3) | C7—C8—C9—C10 | −1.1 (3) |
| C7—N1—C4—C3 | −45.1 (3) | C7—N1—C10—O2 | 179.8 (2) |
| C10—N1—C4—C3 | 129.4 (3) | C4—N1—C10—O2 | 4.4 (4) |
| C3—C4—C5—C6 | 0.1 (4) | C7—N1—C10—C9 | 0.6 (3) |
| N1—C4—C5—C6 | 178.9 (2) | C4—N1—C10—C9 | −174.7 (2) |
| C2—C1—C6—C5 | 0.2 (4) | C8—C9—C10—O2 | −178.8 (3) |
| Cl1—C1—C6—C5 | 179.46 (18) | C8—C9—C10—N1 | 0.3 (3) |
| C4—C5—C6—C1 | −0.3 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C8—H8···O1i | 0.93 | 2.58 | 3.493 (3) | 169 |
| C2—H2···O1ii | 0.93 | 2.77 | 3.659 (3) | 161 |
| C5—H5···O2iii | 0.93 | 2.58 | 3.319 (3) | 137 |
| C9—H9···O2iv | 0.93 | 2.64 | 3.326 (3) | 131 |
| C9—H9···Cl1v | 0.93 | 2.89 | 3.551 (3) | 129 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x, y−1/2, −z+1/2; (iii) x, y+1, z; (iv) −x+1, −y, −z+1; (v) x, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2150).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Etter, M. (1990). Acc. Chem. Res.23, 120–126.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Howell, B. & Zhang, J. (2006). J. Therm. Anal. Calorim.83, 83–86.
- Miller, C. W., Hoyle, C. E., Valente, E. J., Zobkowski, J. D. & Jönsson, E. S. (2000). J. Chem. Cryst.30, 9, 563–571.
- Miller, C. W., Jönsson, E. S., Hoyle, C. E., Viswanathan, K. & Valente, E. J. (2001). J. Phys. Chem. B, 105, 2707–2717.
- Moreno-Fuquen, R., Valencia, H., Abonia, R., Kennedy, A. R. & Graham, D. (2003). Acta Cryst. E59, o1717–o1718.
- Nardelli, M. (1995). J. Appl. Cryst.28, 659.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sarma, J. A. R. P. & Desiraju, G. R. (1986). Acc. Chem. Res.19, 222–228.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680803016X/fj2150sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803016X/fj2150Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report