Abstract
As part of an antipsychotic drug discovery program, we report the crystal structure of the title compound, C24H23ClN4O. The molecule has a tricyclic framework with a characteristic buckled V-shaped pyridobenzoxazepine unit, with the central seven-membered heterocycle in a boat configuration. The piperazine ring displays a chair conformation with the 2-phenyl-ethyl substituent assuming an equatorial orientation. There are two crystallographically independent, but virtually identical, molecules in the asymmetric unit.
Related literature
For related literature see: Andreasen et al. (1994 ▶, 2000 ▶); Dupont & Liégeois (2003 ▶); Petcher & Weber (1976 ▶); Capuano et al. (1999 ▶, 2002 ▶, 2003 ▶, 2006 ▶); Gerlach (1991 ▶); Gerson & Meltzer (1992 ▶); Liégeois et al. (1994 ▶, 1997 ▶, 2000 ▶); Mouithys-Mickalad et al. (2001 ▶); Vom (2006 ▶).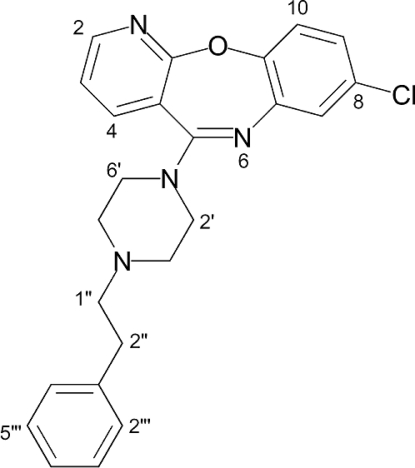
Experimental
Crystal data
C24H23ClN4O
M r = 418.91
Triclinic,
a = 9.9253 (2) Å
b = 15.0549 (3) Å
c = 15.9996 (3) Å
α = 107.774 (2)°
β = 95.487 (1)°
γ = 108.783 (1)°
V = 2104.93 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 123 (2) K
0.30 × 0.25 × 0.20 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: none
25726 measured reflections
9532 independent reflections
6353 reflections with I > 2σ(I)
R int = 0.084
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.158
S = 1.00
9532 reflections
541 parameters
H-atom parameters constrained
Δρmax = 0.48 e Å−3
Δρmin = −0.32 e Å−3
Data collection: COLLECT (Bruker, 2000 ▶); cell refinement: DENZO-SMN (Otwinowski & Minor, 1997 ▶); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: CIFTAB (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808027062/fj2144sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808027062/fj2144Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
We acknowledge support from Monash University and the Australian Research Council
supplementary crystallographic information
Comment
Schizophrenia (Gr., "split mind") is a debilitating mental illness that severely impairs an individual's perception of reality, and damages a variety of emotional, behavioural and cognitive functions that we regard as specifically human (Andreasen et al., 2000). This devastating disease afflicts approximately 1% of the world population thereby making it the most common form of psychosis. The symptoms of schizophrenia are divided into two distinct classifications; positive (delusions and hallucinations) and negative (social and emotional withdrawal) (Andreasen et al., 1994). Antipsychotic drugs are divided into two clinical classes; typical (treat the positive symptoms and have a propensity to induce movement disorders) and atypical (treat both positive and negative symptoms, as well as associated cognitive deficits, and are virtually devoid of movement disorders) (Gerlach, 1991).
Clozapine is an atypical antipsychotic drug far superior in efficacy against treatment-resistant schizophrenia compared to other clinically available therapeutics. However, clozapine has been found to induce the blood disorder agranulocytosis that can, in some cases, be fatal. The synthesis component of this drug discovery programme is a continuation of previous work done in our research group (Capuano et al., 2002, 2003) and is based on the structural hybridisation of two common antipsychotics, namely clozapine and haloperidol. The resulting structural series contains a tricyclic motif attached to piperazine, with an additional π-system anchored to the distal nitrogen atom of the piperazine ring system by a suitable spacer (Capuano et al., 2003). The NH of the central seven-membered ring of clozapine has been isosterically replaced with oxygen, and the adjacent benzene ring replaced with a pyridine ring. This structural class, known as 'pyridobenzoxazepine', has been previously investigated by Liegeois et al., with particular interest to the compound 8-chloro-5-(4-methylpiperazin-1-yl)-11H-pyrido[2,3-b] [1,5]benzoxazepine (also known as JL13), which is currently being clinically evaluated. This alternative tricyclic nucleus is predicted to enhance aqueous solubility and bioavailability compared to previously published compounds. Additionally, isosteric replacement of NH for O in structurally related compounds has been shown to significantly reduce their oxidative sensitivity towards neutrophils, and therefore lessen their hematotoxic potential (Mouithys-Mickalad et al., 2001; Liegeois et al., 2000; Liegeois et al., 1997).
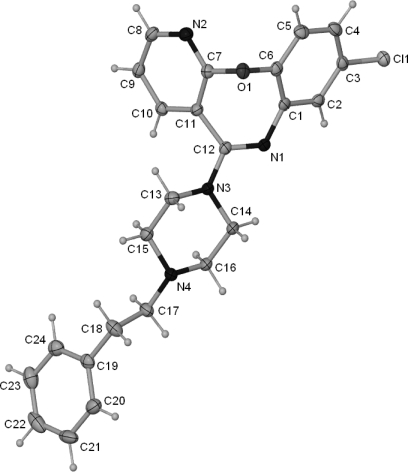
Our interest in the crystal structure of (I) was to examine the geometries of the piperazine ring and the tricyclic nucleus relative to clozapine. The crystal contains two crystallographically independent, but virtually identical molecules. The title compound (I) exhibits the characteristic buckled conformation of the pyridobenzoxazepine nucleus with the central seven-membered heterocycle in a classical boat conformation. The dihedral angle between the planes of the aromatic rings (defined as the obtuse angle subtended by the plane normals) are 112.40 (5)° (molecule 1) and 109.06 (6)° (molecule 2), which are comparable to 114° observed for JL13 (Dupont & Liégeois, 2003) and 115° observed for the prototypical atypical antipsychotic, clozapine (Petcher & Weber, 1976). The dihedral angles between the plane of the four C atoms in the piperazine ring and the chloro-substituted and pyridyl rings are 25.5 (1)° and 43.43 (6)° (molecule 1) and 26.39 (6)° and 45.42 (7)°, respectively (for clozapine, the angles are 40.5° and 31.8°, respectively (Petcher & Weber, 1976)); a consequence of the planarity of the piperazine nitrogen in the amidine moiety and the partial double bond character of N1—C12 (1.297 (3) Å and N5—C36 1.301 (2) Å. The piperazine ring adopts an almost perfect chair conformation with the phenethyl substituent assuming an equatorial orientation, by virtue of the sp2-like nature of the piperazine nitrogen atoms. No significant interactions between molecules of the title compound were observed.
Experimental
The title compound (I) was synthesized (Vom, 2006) from the tricyclic lactam, 8-chlorobenzo[b]pyrido[3,2-f][1,4]oxazepin- 5(6H)-one (Liegeois et al., 1994), and the monosubstituted piperazine, 1-phenethylpiperazine (Capuano et al., 2003), in the presence of a Lewis acid, (TiCl4). A stirred solution of 1-phenethylpiperazine (1.00 g, 5.28 mmol) in anhydrous 1,4-dioxane (5 ml) under nitrogen was treated with a solution of titanium tetrachloride in dry toluene (1M, 1.1 ml, 1.1 mmol). The mixture was warmed to 50–55 °C to which a hot solution of the tricyclic lactam (250 mg, 1.01 mmol) in anhydrous dioxane (20 ml) was added. The reaction mixture was heated at reflux for 4 h after which time it was then cooled and evaporated to dryness in vacuo. The resulting yellow/brown residue was partitioned between sodium hydroxide solution (1M, 50 mL) and ethyl acetate (50 ml). The organic layer was removed and the aqueous phase was further extracted with ethyl acetate (3x50 ml). The combined organic fractions were washed with water (50 ml), dried over anhydrous sodium sulfate, and then evaporated to dryness. The resulting residue was purified by flash chromatography (ethyl acetate:methanol 97.5:2.5) and the major product evaporated to dryness. The product was recrystallized from dichloromethane/hexane as pale yellow prisms (216 mg, 51%), which were suitable for X-ray diffraction studies (mp 428 K (softens) 433 K (melts)). IR νmax 1599, 1583, 1549 cm-1. 1H NMR (CDCl3) δ 2.70–2.75 (6H, m, H3', H5', H2''), 2.86–2.91 (2H, m, H1''), 3.64 (4H, br s, H2',H6'), 6.97 (1H, dd, J = 8.5, 2.5 Hz,H9), 7.14–7.32 (8H, m, H3, H7, H10, H2''', H3''', H4''', H5''', H6'''), 7.73 (1H, dd, J 7.5, 2.0 Hz, H4), 8.43 (1H, dd, J 5.0, 2.0 Hz, H2). 13C NMR (CDCl3) δ 33.0 (CH2), 46.9 (CH2), 52.7 (CH2), 60.1 (CH2), 117.6 (Cq), 121.7 (CH), 122.3 (CH), 124.5 (CH), 126.4 (CH), 126.7 (CH), 128.6 (2 x CH), 131.0 (Cq), 139.5 (Cq), 140.7 (Cq), 148.4 (Cq), 151.6 (CH), 157.9 (Cq), 164.2 (Cq). ESI MS (20 V) m/z 421 (M[37Cl]H+, 36%), 419 (M[35Cl]H+, 100%). ESI high resolution MS: Found m/z 419.1630. Calcd. for C24H23ClN4O: m/z 419.1633.
Refinement
All H atoms for the primary molecules were initially located in the difference Fourier map but were placed in geometrically idealized positions and constrained to ride on their parent atoms with C—H distances in the range 0.95–1.00 Å and Uiso(H) = 1.2–1.5 Ueq(C).
Figures
Fig. 1.
Molecular diagram of one of the crystallographically independent molecules of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C24H23ClN4O | Z = 4 |
| Mr = 418.91 | F(000) = 880 |
| Triclinic, P1 | Dx = 1.322 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.9253 (2) Å | Cell parameters from 25726 reflections |
| b = 15.0549 (3) Å | θ = 1.5–27.5° |
| c = 15.9996 (3) Å | µ = 0.21 mm−1 |
| α = 107.774 (2)° | T = 123 K |
| β = 95.487 (1)° | Prismatic, pale yellow |
| γ = 108.783 (1)° | 0.30 × 0.25 × 0.20 mm |
| V = 2104.93 (8) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 6353 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.084 |
| graphite | θmax = 27.5°, θmin = 1.5° |
| 1 deg frames in φ and ω scans | h = −12→12 |
| 25726 measured reflections | k = −19→19 |
| 9532 independent reflections | l = −16→20 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.158 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.086P)2] where P = (Fo2 + 2Fc2)/3 |
| 9532 reflections | (Δ/σ)max = 0.002 |
| 541 parameters | Δρmax = 0.48 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.43755 (6) | 1.38055 (4) | 1.08930 (3) | 0.03010 (15) | |
| Cl2 | 0.12302 (6) | 0.14811 (4) | 0.23248 (3) | 0.03132 (15) | |
| O1 | 0.82246 (16) | 1.34506 (11) | 0.83292 (9) | 0.0255 (3) | |
| O2 | −0.04349 (15) | 0.18117 (10) | 0.58032 (8) | 0.0248 (3) | |
| N1 | 0.55743 (18) | 1.17733 (13) | 0.80785 (10) | 0.0223 (4) | |
| N2 | 1.02695 (19) | 1.32173 (13) | 0.88624 (11) | 0.0265 (4) | |
| N3 | 0.59265 (18) | 1.05244 (13) | 0.69781 (10) | 0.0245 (4) | |
| N4 | 0.47574 (18) | 0.86689 (13) | 0.54748 (10) | 0.0252 (4) | |
| N5 | 0.05941 (18) | 0.35809 (13) | 0.53319 (10) | 0.0224 (4) | |
| N6 | 0.13408 (19) | 0.17182 (13) | 0.67722 (10) | 0.0245 (4) | |
| N7 | 0.10986 (18) | 0.48198 (12) | 0.67195 (10) | 0.0221 (4) | |
| N8 | 0.11770 (18) | 0.67465 (12) | 0.77966 (10) | 0.0219 (4) | |
| C1 | 0.6055 (2) | 1.26737 (16) | 0.88188 (12) | 0.0217 (4) | |
| C2 | 0.5157 (2) | 1.27768 (16) | 0.94409 (12) | 0.0230 (4) | |
| H2 | 0.4327 | 1.2217 | 0.9400 | 0.028* | |
| C3 | 0.5486 (2) | 1.36993 (16) | 1.01146 (12) | 0.0238 (5) | |
| C4 | 0.6674 (2) | 1.45349 (16) | 1.01968 (13) | 0.0279 (5) | |
| H4 | 0.6862 | 1.5163 | 1.0654 | 0.034* | |
| C5 | 0.7592 (2) | 1.44418 (16) | 0.95978 (13) | 0.0276 (5) | |
| H5 | 0.8423 | 1.5005 | 0.9646 | 0.033* | |
| C6 | 0.7283 (2) | 1.35212 (16) | 0.89302 (12) | 0.0230 (4) | |
| C7 | 0.8879 (2) | 1.28001 (16) | 0.84410 (12) | 0.0228 (4) | |
| C8 | 1.0898 (2) | 1.25901 (17) | 0.89859 (13) | 0.0280 (5) | |
| H8 | 1.1890 | 1.2871 | 0.9293 | 0.034* | |
| C9 | 1.0194 (2) | 1.15579 (17) | 0.86940 (12) | 0.0260 (5) | |
| H9 | 1.0689 | 1.1145 | 0.8800 | 0.031* | |
| C10 | 0.8740 (2) | 1.11413 (16) | 0.82406 (12) | 0.0226 (4) | |
| H10 | 0.8224 | 1.0436 | 0.8029 | 0.027* | |
| C11 | 0.8060 (2) | 1.17713 (15) | 0.81026 (12) | 0.0209 (4) | |
| C12 | 0.6466 (2) | 1.13941 (15) | 0.77075 (12) | 0.0208 (4) | |
| C13 | 0.6737 (2) | 1.03001 (16) | 0.62703 (12) | 0.0262 (5) | |
| H13A | 0.7797 | 1.0629 | 0.6530 | 0.031* | |
| H13B | 0.6506 | 1.0567 | 0.5804 | 0.031* | |
| C14 | 0.4348 (2) | 1.00197 (16) | 0.66368 (14) | 0.0295 (5) | |
| H14A | 0.4021 | 1.0285 | 0.6193 | 0.035* | |
| H14B | 0.3838 | 1.0148 | 0.7137 | 0.035* | |
| C15 | 0.6332 (2) | 0.91819 (16) | 0.58523 (13) | 0.0266 (5) | |
| H15A | 0.6870 | 0.9036 | 0.5370 | 0.032* | |
| H15B | 0.6617 | 0.8924 | 0.6313 | 0.032* | |
| C16 | 0.3981 (2) | 0.88978 (16) | 0.61953 (13) | 0.0277 (5) | |
| H16A | 0.4238 | 0.8630 | 0.6655 | 0.033* | |
| H16B | 0.2919 | 0.8558 | 0.5946 | 0.033* | |
| C17 | 0.4348 (2) | 0.75783 (16) | 0.50652 (13) | 0.0283 (5) | |
| H17A | 0.3273 | 0.7252 | 0.4932 | 0.034* | |
| H17B | 0.4753 | 0.7336 | 0.5502 | 0.034* | |
| C18 | 0.4887 (3) | 0.72652 (17) | 0.42041 (14) | 0.0345 (6) | |
| H18A | 0.4413 | 0.7450 | 0.3746 | 0.041* | |
| H18B | 0.5951 | 0.7634 | 0.4323 | 0.041* | |
| C19 | 0.4567 (2) | 0.61571 (17) | 0.38432 (13) | 0.0270 (5) | |
| C20 | 0.5436 (2) | 0.57522 (18) | 0.42136 (13) | 0.0321 (5) | |
| H20 | 0.6227 | 0.6183 | 0.4708 | 0.039* | |
| C21 | 0.5179 (3) | 0.47376 (19) | 0.38809 (15) | 0.0398 (6) | |
| H21 | 0.5809 | 0.4482 | 0.4133 | 0.048* | |
| C22 | 0.4013 (3) | 0.40982 (19) | 0.31860 (17) | 0.0458 (7) | |
| H22 | 0.3828 | 0.3399 | 0.2961 | 0.055* | |
| C23 | 0.3108 (3) | 0.4477 (2) | 0.28147 (15) | 0.0447 (7) | |
| H23 | 0.2294 | 0.4037 | 0.2338 | 0.054* | |
| C24 | 0.3391 (2) | 0.55034 (19) | 0.31401 (14) | 0.0367 (6) | |
| H24 | 0.2773 | 0.5760 | 0.2878 | 0.044* | |
| C25 | 0.0493 (2) | 0.26283 (15) | 0.47717 (12) | 0.0209 (4) | |
| C26 | 0.0878 (2) | 0.25284 (16) | 0.39394 (12) | 0.0233 (4) | |
| H26 | 0.1293 | 0.3107 | 0.3789 | 0.028* | |
| C27 | 0.0653 (2) | 0.15853 (16) | 0.33345 (12) | 0.0235 (5) | |
| C28 | 0.0012 (2) | 0.07163 (16) | 0.35102 (13) | 0.0251 (5) | |
| H28 | −0.0174 | 0.0076 | 0.3073 | 0.030* | |
| C29 | −0.0354 (2) | 0.08040 (16) | 0.43400 (12) | 0.0238 (5) | |
| H29 | −0.0781 | 0.0221 | 0.4483 | 0.029* | |
| C30 | −0.0094 (2) | 0.17435 (15) | 0.49555 (12) | 0.0226 (4) | |
| C31 | 0.0867 (2) | 0.22962 (16) | 0.64466 (12) | 0.0225 (4) | |
| C32 | 0.2615 (2) | 0.21837 (17) | 0.73820 (13) | 0.0273 (5) | |
| H32 | 0.2987 | 0.1789 | 0.7629 | 0.033* | |
| C33 | 0.3407 (2) | 0.32015 (16) | 0.76676 (12) | 0.0244 (5) | |
| H33 | 0.4299 | 0.3497 | 0.8100 | 0.029* | |
| C34 | 0.2875 (2) | 0.37851 (16) | 0.73110 (12) | 0.0222 (4) | |
| H34 | 0.3399 | 0.4488 | 0.7495 | 0.027* | |
| C35 | 0.1563 (2) | 0.33270 (15) | 0.66807 (12) | 0.0205 (4) | |
| C36 | 0.0998 (2) | 0.38808 (15) | 0.62012 (12) | 0.0211 (4) | |
| C37 | 0.0757 (2) | 0.50015 (15) | 0.76178 (12) | 0.0235 (5) | |
| H37A | 0.1046 | 0.4567 | 0.7895 | 0.028* | |
| H37B | −0.0305 | 0.4830 | 0.7564 | 0.028* | |
| C38 | 0.0800 (2) | 0.54901 (15) | 0.62959 (12) | 0.0246 (5) | |
| H38A | −0.0260 | 0.5346 | 0.6170 | 0.029* | |
| H38B | 0.1126 | 0.5384 | 0.5720 | 0.029* | |
| C39 | 0.1557 (2) | 0.60878 (15) | 0.82070 (12) | 0.0238 (5) | |
| H39A | 0.1299 | 0.6206 | 0.8803 | 0.029* | |
| H39B | 0.2621 | 0.6246 | 0.8295 | 0.029* | |
| C40 | 0.1610 (2) | 0.65674 (15) | 0.69272 (12) | 0.0230 (4) | |
| H40A | 0.2672 | 0.6715 | 0.7023 | 0.028* | |
| H40B | 0.1406 | 0.7027 | 0.6649 | 0.028* | |
| C41 | 0.1924 (2) | 0.78003 (15) | 0.83789 (12) | 0.0251 (5) | |
| H41A | 0.1826 | 0.8231 | 0.8035 | 0.030* | |
| H41B | 0.2974 | 0.7935 | 0.8554 | 0.030* | |
| C42 | 0.1336 (2) | 0.80871 (16) | 0.92268 (13) | 0.0273 (5) | |
| H42A | 0.0302 | 0.7997 | 0.9056 | 0.033* | |
| H42B | 0.1379 | 0.7633 | 0.9556 | 0.033* | |
| C43 | 0.2189 (2) | 0.91604 (16) | 0.98384 (12) | 0.0245 (5) | |
| C44 | 0.3466 (2) | 0.93923 (17) | 1.04455 (13) | 0.0263 (5) | |
| H44 | 0.3809 | 0.8875 | 1.0460 | 0.032* | |
| C45 | 0.4243 (2) | 1.03739 (17) | 1.10308 (13) | 0.0286 (5) | |
| H45 | 0.5102 | 1.0520 | 1.1449 | 0.034* | |
| C46 | 0.3771 (2) | 1.11341 (17) | 1.10059 (13) | 0.0313 (5) | |
| H46 | 0.4300 | 1.1803 | 1.1407 | 0.038* | |
| C47 | 0.2520 (2) | 1.09162 (17) | 1.03929 (13) | 0.0302 (5) | |
| H47 | 0.2199 | 1.1439 | 1.0366 | 0.036* | |
| C48 | 0.1736 (2) | 0.99370 (16) | 0.98195 (13) | 0.0279 (5) | |
| H48 | 0.0873 | 0.9794 | 0.9406 | 0.033* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0300 (3) | 0.0328 (3) | 0.0256 (3) | 0.0151 (2) | 0.0060 (2) | 0.0043 (2) |
| Cl2 | 0.0398 (3) | 0.0298 (3) | 0.0227 (3) | 0.0145 (3) | 0.0059 (2) | 0.0056 (2) |
| O1 | 0.0273 (8) | 0.0225 (8) | 0.0304 (8) | 0.0101 (7) | 0.0088 (6) | 0.0127 (6) |
| O2 | 0.0203 (7) | 0.0238 (8) | 0.0245 (7) | 0.0041 (6) | 0.0025 (5) | 0.0060 (6) |
| N1 | 0.0191 (9) | 0.0228 (10) | 0.0223 (8) | 0.0083 (8) | 0.0006 (6) | 0.0050 (7) |
| N2 | 0.0218 (9) | 0.0254 (10) | 0.0269 (9) | 0.0048 (8) | 0.0048 (7) | 0.0062 (7) |
| N3 | 0.0176 (9) | 0.0247 (10) | 0.0246 (9) | 0.0065 (8) | 0.0023 (7) | 0.0020 (7) |
| N4 | 0.0220 (9) | 0.0218 (10) | 0.0260 (9) | 0.0075 (8) | 0.0031 (7) | 0.0020 (7) |
| N5 | 0.0236 (9) | 0.0216 (10) | 0.0213 (8) | 0.0098 (8) | 0.0010 (7) | 0.0060 (7) |
| N6 | 0.0314 (10) | 0.0224 (10) | 0.0240 (9) | 0.0136 (8) | 0.0074 (7) | 0.0100 (7) |
| N7 | 0.0289 (10) | 0.0198 (9) | 0.0201 (8) | 0.0131 (8) | 0.0030 (7) | 0.0071 (7) |
| N8 | 0.0252 (9) | 0.0193 (9) | 0.0227 (8) | 0.0101 (8) | 0.0032 (7) | 0.0083 (7) |
| C1 | 0.0212 (10) | 0.0249 (11) | 0.0203 (10) | 0.0126 (9) | −0.0013 (7) | 0.0073 (8) |
| C2 | 0.0211 (11) | 0.0236 (11) | 0.0233 (10) | 0.0090 (9) | −0.0012 (8) | 0.0083 (8) |
| C3 | 0.0265 (11) | 0.0286 (12) | 0.0199 (10) | 0.0161 (10) | 0.0038 (8) | 0.0076 (8) |
| C4 | 0.0350 (13) | 0.0218 (12) | 0.0249 (11) | 0.0137 (10) | −0.0007 (9) | 0.0042 (8) |
| C5 | 0.0301 (12) | 0.0199 (11) | 0.0304 (11) | 0.0074 (9) | 0.0017 (9) | 0.0090 (9) |
| C6 | 0.0237 (11) | 0.0255 (12) | 0.0234 (10) | 0.0125 (9) | 0.0030 (8) | 0.0107 (8) |
| C7 | 0.0230 (11) | 0.0232 (11) | 0.0226 (10) | 0.0087 (9) | 0.0055 (8) | 0.0086 (8) |
| C8 | 0.0207 (11) | 0.0389 (14) | 0.0228 (10) | 0.0115 (10) | 0.0030 (8) | 0.0088 (9) |
| C9 | 0.0236 (11) | 0.0329 (13) | 0.0245 (10) | 0.0143 (10) | 0.0051 (8) | 0.0101 (9) |
| C10 | 0.0239 (11) | 0.0225 (11) | 0.0213 (10) | 0.0087 (9) | 0.0077 (8) | 0.0068 (8) |
| C11 | 0.0182 (10) | 0.0226 (11) | 0.0196 (9) | 0.0062 (9) | 0.0036 (7) | 0.0061 (8) |
| C12 | 0.0187 (10) | 0.0229 (11) | 0.0216 (10) | 0.0078 (9) | 0.0025 (7) | 0.0097 (8) |
| C13 | 0.0233 (11) | 0.0273 (12) | 0.0226 (10) | 0.0048 (9) | 0.0039 (8) | 0.0067 (9) |
| C14 | 0.0189 (11) | 0.0278 (13) | 0.0330 (11) | 0.0094 (9) | −0.0001 (8) | 0.0000 (9) |
| C15 | 0.0211 (11) | 0.0295 (13) | 0.0248 (10) | 0.0084 (9) | 0.0046 (8) | 0.0048 (9) |
| C16 | 0.0174 (10) | 0.0268 (12) | 0.0308 (11) | 0.0070 (9) | 0.0025 (8) | 0.0017 (9) |
| C17 | 0.0268 (12) | 0.0231 (12) | 0.0284 (11) | 0.0069 (10) | 0.0066 (9) | 0.0026 (9) |
| C18 | 0.0446 (15) | 0.0251 (13) | 0.0303 (12) | 0.0113 (11) | 0.0112 (10) | 0.0061 (9) |
| C19 | 0.0288 (12) | 0.0280 (12) | 0.0220 (10) | 0.0088 (10) | 0.0110 (8) | 0.0061 (9) |
| C20 | 0.0304 (13) | 0.0361 (14) | 0.0253 (11) | 0.0086 (11) | 0.0085 (9) | 0.0079 (9) |
| C21 | 0.0511 (16) | 0.0411 (16) | 0.0432 (14) | 0.0258 (13) | 0.0256 (12) | 0.0227 (12) |
| C22 | 0.0585 (18) | 0.0264 (14) | 0.0490 (15) | 0.0109 (13) | 0.0326 (13) | 0.0077 (11) |
| C23 | 0.0328 (14) | 0.0386 (16) | 0.0348 (13) | −0.0027 (12) | 0.0079 (10) | −0.0077 (11) |
| C24 | 0.0261 (12) | 0.0428 (15) | 0.0320 (12) | 0.0101 (11) | 0.0045 (9) | 0.0044 (10) |
| C25 | 0.0196 (10) | 0.0216 (11) | 0.0201 (10) | 0.0096 (9) | −0.0023 (7) | 0.0053 (8) |
| C26 | 0.0236 (11) | 0.0228 (11) | 0.0232 (10) | 0.0093 (9) | −0.0008 (8) | 0.0088 (8) |
| C27 | 0.0254 (11) | 0.0282 (12) | 0.0165 (9) | 0.0128 (9) | −0.0002 (8) | 0.0057 (8) |
| C28 | 0.0262 (11) | 0.0209 (11) | 0.0237 (10) | 0.0108 (9) | −0.0023 (8) | 0.0021 (8) |
| C29 | 0.0189 (10) | 0.0201 (11) | 0.0268 (11) | 0.0039 (9) | −0.0033 (8) | 0.0068 (8) |
| C30 | 0.0193 (10) | 0.0248 (12) | 0.0208 (10) | 0.0080 (9) | −0.0007 (7) | 0.0057 (8) |
| C31 | 0.0227 (11) | 0.0250 (12) | 0.0197 (10) | 0.0094 (9) | 0.0061 (8) | 0.0070 (8) |
| C32 | 0.0337 (12) | 0.0309 (13) | 0.0257 (11) | 0.0193 (10) | 0.0081 (9) | 0.0132 (9) |
| C33 | 0.0222 (11) | 0.0309 (12) | 0.0226 (10) | 0.0119 (9) | 0.0037 (8) | 0.0110 (9) |
| C34 | 0.0223 (11) | 0.0237 (11) | 0.0203 (10) | 0.0086 (9) | 0.0056 (8) | 0.0073 (8) |
| C35 | 0.0211 (10) | 0.0215 (11) | 0.0209 (10) | 0.0097 (9) | 0.0057 (7) | 0.0080 (8) |
| C36 | 0.0171 (10) | 0.0214 (11) | 0.0240 (10) | 0.0067 (9) | 0.0029 (7) | 0.0079 (8) |
| C37 | 0.0273 (11) | 0.0219 (11) | 0.0251 (10) | 0.0120 (9) | 0.0087 (8) | 0.0097 (8) |
| C38 | 0.0321 (12) | 0.0222 (11) | 0.0207 (10) | 0.0137 (10) | 0.0005 (8) | 0.0072 (8) |
| C39 | 0.0276 (11) | 0.0241 (12) | 0.0210 (10) | 0.0120 (9) | 0.0048 (8) | 0.0073 (8) |
| C40 | 0.0272 (11) | 0.0234 (11) | 0.0211 (10) | 0.0117 (9) | 0.0047 (8) | 0.0093 (8) |
| C41 | 0.0273 (12) | 0.0214 (11) | 0.0257 (10) | 0.0091 (9) | 0.0040 (8) | 0.0075 (8) |
| C42 | 0.0294 (12) | 0.0228 (12) | 0.0272 (11) | 0.0085 (10) | 0.0074 (8) | 0.0062 (9) |
| C43 | 0.0273 (12) | 0.0263 (12) | 0.0206 (10) | 0.0111 (10) | 0.0078 (8) | 0.0074 (8) |
| C44 | 0.0285 (12) | 0.0297 (13) | 0.0278 (11) | 0.0144 (10) | 0.0100 (8) | 0.0146 (9) |
| C45 | 0.0247 (12) | 0.0361 (14) | 0.0237 (10) | 0.0091 (10) | 0.0051 (8) | 0.0112 (9) |
| C46 | 0.0299 (13) | 0.0275 (13) | 0.0290 (11) | 0.0078 (10) | 0.0084 (9) | 0.0026 (9) |
| C47 | 0.0346 (13) | 0.0259 (12) | 0.0301 (11) | 0.0152 (10) | 0.0064 (9) | 0.0059 (9) |
| C48 | 0.0292 (12) | 0.0265 (12) | 0.0273 (11) | 0.0122 (10) | 0.0030 (8) | 0.0077 (9) |
Geometric parameters (Å, °)
| Cl1—C3 | 1.745 (2) | C18—H18A | 0.9900 |
| Cl2—C27 | 1.7442 (19) | C18—H18B | 0.9900 |
| O1—C7 | 1.381 (2) | C19—C20 | 1.388 (3) |
| O1—C6 | 1.408 (2) | C19—C24 | 1.389 (3) |
| O2—C31 | 1.393 (2) | C20—C21 | 1.381 (3) |
| O2—C30 | 1.410 (2) | C20—H20 | 0.9500 |
| N1—C12 | 1.297 (3) | C21—C22 | 1.373 (4) |
| N1—C1 | 1.401 (2) | C21—H21 | 0.9500 |
| N2—C7 | 1.326 (3) | C22—C23 | 1.386 (4) |
| N2—C8 | 1.338 (3) | C22—H22 | 0.9500 |
| N3—C12 | 1.363 (2) | C23—C24 | 1.392 (3) |
| N3—C14 | 1.466 (3) | C23—H23 | 0.9500 |
| N3—C13 | 1.469 (2) | C24—H24 | 0.9500 |
| N4—C16 | 1.463 (2) | C25—C26 | 1.398 (3) |
| N4—C17 | 1.467 (3) | C25—C30 | 1.405 (3) |
| N4—C15 | 1.470 (3) | C26—C27 | 1.385 (3) |
| N5—C36 | 1.301 (2) | C26—H26 | 0.9500 |
| N5—C25 | 1.406 (2) | C27—C28 | 1.385 (3) |
| N6—C31 | 1.320 (3) | C28—C29 | 1.389 (3) |
| N6—C32 | 1.348 (3) | C28—H28 | 0.9500 |
| N7—C36 | 1.369 (3) | C29—C30 | 1.379 (3) |
| N7—C38 | 1.462 (3) | C29—H29 | 0.9500 |
| N7—C37 | 1.472 (2) | C31—C35 | 1.390 (3) |
| N8—C41 | 1.464 (2) | C32—C33 | 1.380 (3) |
| N8—C39 | 1.466 (3) | C32—H32 | 0.9500 |
| N8—C40 | 1.467 (2) | C33—C34 | 1.387 (3) |
| C1—C6 | 1.402 (3) | C33—H33 | 0.9500 |
| C1—C2 | 1.405 (3) | C34—C35 | 1.391 (3) |
| C2—C3 | 1.387 (3) | C34—H34 | 0.9500 |
| C2—H2 | 0.9500 | C35—C36 | 1.486 (3) |
| C3—C4 | 1.380 (3) | C37—C39 | 1.508 (3) |
| C4—C5 | 1.391 (3) | C37—H37A | 0.9900 |
| C4—H4 | 0.9500 | C37—H37B | 0.9900 |
| C5—C6 | 1.384 (3) | C38—C40 | 1.517 (3) |
| C5—H5 | 0.9500 | C38—H38A | 0.9900 |
| C7—C11 | 1.395 (3) | C38—H38B | 0.9900 |
| C8—C9 | 1.384 (3) | C39—H39A | 0.9900 |
| C8—H8 | 0.9500 | C39—H39B | 0.9900 |
| C9—C10 | 1.393 (3) | C40—H40A | 0.9900 |
| C9—H9 | 0.9500 | C40—H40B | 0.9900 |
| C10—C11 | 1.384 (3) | C41—C42 | 1.524 (3) |
| C10—H10 | 0.9500 | C41—H41A | 0.9900 |
| C11—C12 | 1.495 (3) | C41—H41B | 0.9900 |
| C13—C15 | 1.506 (3) | C42—C43 | 1.510 (3) |
| C13—H13A | 0.9900 | C42—H42A | 0.9900 |
| C13—H13B | 0.9900 | C42—H42B | 0.9900 |
| C14—C16 | 1.520 (3) | C43—C48 | 1.388 (3) |
| C14—H14A | 0.9900 | C43—C44 | 1.394 (3) |
| C14—H14B | 0.9900 | C44—C45 | 1.392 (3) |
| C15—H15A | 0.9900 | C44—H44 | 0.9500 |
| C15—H15B | 0.9900 | C45—C46 | 1.379 (3) |
| C16—H16A | 0.9900 | C45—H45 | 0.9500 |
| C16—H16B | 0.9900 | C46—C47 | 1.386 (3) |
| C17—C18 | 1.521 (3) | C46—H46 | 0.9500 |
| C17—H17A | 0.9900 | C47—C48 | 1.385 (3) |
| C17—H17B | 0.9900 | C47—H47 | 0.9500 |
| C18—C19 | 1.501 (3) | C48—H48 | 0.9500 |
| C7—O1—C6 | 108.51 (15) | C21—C22—C23 | 119.7 (2) |
| C31—O2—C30 | 107.85 (14) | C21—C22—H22 | 120.1 |
| C12—N1—C1 | 122.45 (17) | C23—C22—H22 | 120.1 |
| C7—N2—C8 | 116.26 (19) | C22—C23—C24 | 120.0 (2) |
| C12—N3—C14 | 119.65 (17) | C22—C23—H23 | 120.0 |
| C12—N3—C13 | 122.36 (17) | C24—C23—H23 | 120.0 |
| C14—N3—C13 | 111.97 (15) | C19—C24—C23 | 120.8 (2) |
| C16—N4—C17 | 109.81 (16) | C19—C24—H24 | 119.6 |
| C16—N4—C15 | 108.62 (15) | C23—C24—H24 | 119.6 |
| C17—N4—C15 | 111.58 (17) | C26—C25—C30 | 117.01 (18) |
| C36—N5—C25 | 121.46 (18) | C26—C25—N5 | 118.53 (18) |
| C31—N6—C32 | 116.07 (18) | C30—C25—N5 | 124.21 (17) |
| C36—N7—C38 | 120.12 (15) | C27—C26—C25 | 119.9 (2) |
| C36—N7—C37 | 120.21 (17) | C27—C26—H26 | 120.0 |
| C38—N7—C37 | 112.65 (16) | C25—C26—H26 | 120.0 |
| C41—N8—C39 | 110.58 (15) | C26—C27—C28 | 122.30 (19) |
| C41—N8—C40 | 110.43 (16) | C26—C27—Cl2 | 119.27 (17) |
| C39—N8—C40 | 107.81 (16) | C28—C27—Cl2 | 118.42 (15) |
| N1—C1—C6 | 124.50 (18) | C27—C28—C29 | 118.42 (18) |
| N1—C1—C2 | 117.77 (18) | C27—C28—H28 | 120.8 |
| C6—C1—C2 | 117.31 (18) | C29—C28—H28 | 120.8 |
| C3—C2—C1 | 119.8 (2) | C30—C29—C28 | 119.5 (2) |
| C3—C2—H2 | 120.1 | C30—C29—H29 | 120.3 |
| C1—C2—H2 | 120.1 | C28—C29—H29 | 120.3 |
| C4—C3—C2 | 122.16 (19) | C29—C30—C25 | 122.73 (18) |
| C4—C3—Cl1 | 118.81 (15) | C29—C30—O2 | 118.25 (18) |
| C2—C3—Cl1 | 119.03 (17) | C25—C30—O2 | 119.02 (17) |
| C3—C4—C5 | 118.86 (19) | N6—C31—C35 | 125.67 (19) |
| C3—C4—H4 | 120.6 | N6—C31—O2 | 116.04 (18) |
| C5—C4—H4 | 120.6 | C35—C31—O2 | 118.28 (18) |
| C6—C5—C4 | 119.4 (2) | N6—C32—C33 | 123.6 (2) |
| C6—C5—H5 | 120.3 | N6—C32—H32 | 118.2 |
| C4—C5—H5 | 120.3 | C33—C32—H32 | 118.2 |
| C5—C6—C1 | 122.44 (19) | C32—C33—C34 | 118.69 (19) |
| C5—C6—O1 | 118.19 (19) | C32—C33—H33 | 120.7 |
| C1—C6—O1 | 119.34 (17) | C34—C33—H33 | 120.7 |
| N2—C7—O1 | 116.16 (19) | C33—C34—C35 | 119.05 (19) |
| N2—C7—C11 | 125.0 (2) | C33—C34—H34 | 120.5 |
| O1—C7—C11 | 118.86 (18) | C35—C34—H34 | 120.5 |
| N2—C8—C9 | 124.1 (2) | C31—C35—C34 | 116.89 (19) |
| N2—C8—H8 | 117.9 | C31—C35—C36 | 120.65 (17) |
| C9—C8—H8 | 117.9 | C34—C35—C36 | 122.14 (19) |
| C8—C9—C10 | 118.2 (2) | N5—C36—N7 | 119.44 (19) |
| C8—C9—H9 | 120.9 | N5—C36—C35 | 124.18 (18) |
| C10—C9—H9 | 120.9 | N7—C36—C35 | 115.85 (16) |
| C11—C10—C9 | 119.0 (2) | N7—C37—C39 | 110.22 (16) |
| C11—C10—H10 | 120.5 | N7—C37—H37A | 109.6 |
| C9—C10—H10 | 120.5 | C39—C37—H37A | 109.6 |
| C10—C11—C7 | 117.41 (18) | N7—C37—H37B | 109.6 |
| C10—C11—C12 | 122.08 (19) | C39—C37—H37B | 109.6 |
| C7—C11—C12 | 120.12 (19) | H37A—C37—H37B | 108.1 |
| N1—C12—N3 | 119.41 (18) | N7—C38—C40 | 108.73 (15) |
| N1—C12—C11 | 123.82 (17) | N7—C38—H38A | 109.9 |
| N3—C12—C11 | 116.22 (17) | C40—C38—H38A | 109.9 |
| N3—C13—C15 | 109.85 (17) | N7—C38—H38B | 109.9 |
| N3—C13—H13A | 109.7 | C40—C38—H38B | 109.9 |
| C15—C13—H13A | 109.7 | H38A—C38—H38B | 108.3 |
| N3—C13—H13B | 109.7 | N8—C39—C37 | 110.54 (16) |
| C15—C13—H13B | 109.7 | N8—C39—H39A | 109.5 |
| H13A—C13—H13B | 108.2 | C37—C39—H39A | 109.5 |
| N3—C14—C16 | 109.06 (17) | N8—C39—H39B | 109.5 |
| N3—C14—H14A | 109.9 | C37—C39—H39B | 109.5 |
| C16—C14—H14A | 109.9 | H39A—C39—H39B | 108.1 |
| N3—C14—H14B | 109.9 | N8—C40—C38 | 111.45 (16) |
| C16—C14—H14B | 109.9 | N8—C40—H40A | 109.3 |
| H14A—C14—H14B | 108.3 | C38—C40—H40A | 109.3 |
| N4—C15—C13 | 111.03 (18) | N8—C40—H40B | 109.3 |
| N4—C15—H15A | 109.4 | C38—C40—H40B | 109.3 |
| C13—C15—H15A | 109.4 | H40A—C40—H40B | 108.0 |
| N4—C15—H15B | 109.4 | N8—C41—C42 | 113.44 (17) |
| C13—C15—H15B | 109.4 | N8—C41—H41A | 108.9 |
| H15A—C15—H15B | 108.0 | C42—C41—H41A | 108.9 |
| N4—C16—C14 | 112.14 (18) | N8—C41—H41B | 108.9 |
| N4—C16—H16A | 109.2 | C42—C41—H41B | 108.9 |
| C14—C16—H16A | 109.2 | H41A—C41—H41B | 107.7 |
| N4—C16—H16B | 109.2 | C43—C42—C41 | 112.03 (17) |
| C14—C16—H16B | 109.2 | C43—C42—H42A | 109.2 |
| H16A—C16—H16B | 107.9 | C41—C42—H42A | 109.2 |
| N4—C17—C18 | 113.11 (18) | C43—C42—H42B | 109.2 |
| N4—C17—H17A | 109.0 | C41—C42—H42B | 109.2 |
| C18—C17—H17A | 109.0 | H42A—C42—H42B | 107.9 |
| N4—C17—H17B | 109.0 | C48—C43—C44 | 118.2 (2) |
| C18—C17—H17B | 109.0 | C48—C43—C42 | 121.78 (19) |
| H17A—C17—H17B | 107.8 | C44—C43—C42 | 120.0 (2) |
| C19—C18—C17 | 111.98 (18) | C45—C44—C43 | 120.7 (2) |
| C19—C18—H18A | 109.2 | C45—C44—H44 | 119.7 |
| C17—C18—H18A | 109.2 | C43—C44—H44 | 119.7 |
| C19—C18—H18B | 109.2 | C46—C45—C44 | 120.2 (2) |
| C17—C18—H18B | 109.2 | C46—C45—H45 | 119.9 |
| H18A—C18—H18B | 107.9 | C44—C45—H45 | 119.9 |
| C20—C19—C24 | 117.9 (2) | C45—C46—C47 | 119.6 (2) |
| C20—C19—C18 | 120.6 (2) | C45—C46—H46 | 120.2 |
| C24—C19—C18 | 121.5 (2) | C47—C46—H46 | 120.2 |
| C21—C20—C19 | 121.6 (2) | C48—C47—C46 | 120.1 (2) |
| C21—C20—H20 | 119.2 | C48—C47—H47 | 119.9 |
| C19—C20—H20 | 119.2 | C46—C47—H47 | 119.9 |
| C22—C21—C20 | 120.0 (3) | C47—C48—C43 | 121.1 (2) |
| C22—C21—H21 | 120.0 | C47—C48—H48 | 119.4 |
| C20—C21—H21 | 120.0 | C43—C48—H48 | 119.4 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2144).
References
- Andreasen, N. C. (2000). Brain Res. Brain Res. Rev.31, 106–112. [DOI] [PubMed]
- Andreasen, N. C., Nopoulos, P., Schultz, S., Miller, D., Gupta, S., Swayze, V. & Flaum, M. (1994). Acta Psychiatr. Scand. Suppl.384, 51–59. [DOI] [PubMed]
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2000). COLLECT. Bruker AXS BV, Delft, The Netherlands.
- Capuano, B. (1999). Molecules, 4, 329-332.
- Capuano, B., Crosby, I. T., Forsyth, C. M., Lloyd, E. J., Vom, A. & Yuriev, E. (2006). Acta Cryst. E62, o5434–o5436. [DOI] [PMC free article] [PubMed]
- Capuano, B., Crosby, I. T., Lloyd, E. J. & Taylor, D. A. (2002). Aust. J. Chem.55, 565–576.
- Capuano, B., Crosby, I. T., Lloyd, E. J. & Taylor, D. A. (2003). Aust. J. Chem.56, 875–886.
- Dupont, L. & Liégeois, J.-F. (2003). Acta Cryst. E59, o1962–o1963.
- Gerlach, J. (1991). Schizophr. Bull.17, 289–309. [DOI] [PubMed]
- Gerson, S. L. & Meltzer, H. (1992). Drug Safety, 7 (Suppl. 1), 17–25. [DOI] [PubMed]
- Liégeois, J.-F., Mouithys-Mickalad, A., Bruhwyler, J., Petit, C., Kauffmann, J. M. & Lamy, M. (1997). Biochem. Biophys. Res. Commun.238, 252–255. [DOI] [PubMed]
- Liégeois, J.-F., Rogistert, F. A., Bruhwyler, J., Damas, J., Nguyen, T. P., Inarejos, M.-O., Chleide, E. M. G., Mercier, M. G. A. & Delarget, J. E. (1994). J. Med. Chem.37, 519–525. [DOI] [PubMed]
- Liégeois, J.-F., Zahid, N., Bruhwyler, J. & Uetrecht, J. (2000). Arch. Pharm. Wienheim, Ger.333, 63–67. [DOI] [PubMed]
- Mouithys-Mickalad, A., Kauffmann, J. M., Petit, C., Bruhwyler, J., Liao, Y., Wikstrom, H., Damas, J., Delarge, J., Deby-Dupont, G., Geczy, J. & Liegeois, J.-F. (2001). J. Med. Chem.44, 769–776. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Petcher, T. J. & Weber, H.-P. (1976). J. Chem. Soc. Perkin Trans. 2, pp. 1415–1420.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Vom, A. (2006). Honours Thesis, Monash University (Parkville), Victoria, Australia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808027062/fj2144sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808027062/fj2144Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report