Abstract
The title compound, C10H15NO4, also known as N-tert-butyloxycarbonyl-allohydroxy-l-proline lactone, is quite similar to N-acetyl-allohydroxy-l-proline lactone [Lenstra, Petit & Geise (1979 ▶). Cryst. Struct. Commun. 8, 1023–1029], whereby both carbonyl groups point roughly in the same direction because of the trans conformation of the peptide bond.
Related literature
For general background, see: Allen (2002 ▶). For related structures, see: Didier et al. (2004 ▶); Lenstra et al. (1979 ▶); Papaioannou et al. (1989 ▶). For related synthesis, see: Gómez-Vidal & Silverman (2001 ▶). For related literature, see: Flack & Schwarzenbach (1988 ▶).
Experimental
Crystal data
C10H15NO4
M r = 213.23
Monoclinic,
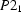
a = 6.0710 (7) Å
b = 9.3703 (11) Å
c = 9.3002 (10) Å
β = 100.013 (5)°
V = 521.00 (10) Å3
Z = 2
Mo Kα radiation
μ = 0.10 mm−1
T = 100 (2) K
0.3 × 0.2 × 0.2 mm
Data collection
Nonius KappaCCD area-detector diffractometer
Absorption correction: none
5951 measured reflections
1143 independent reflections
1054 reflections with I > 2σ(I)
R int = 0.066
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.102
S = 1.20
1143 reflections
139 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.23 e Å−3
Δρmin = −0.22 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SIR92 (Altomare et al., 1994 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808030651/ww2125sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808030651/ww2125Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the Service Commun de Diffraction X (Nancy Université) for providing access to crystallographic experimental facilities.
supplementary crystallographic information
Comment
N-tert-Butyloxycarbonyl-allohydroxy-L-proline lactone is prepared in one step under Mitsunobu conditions starting from corresponding trans-4-hydroxyproline. As previously described (Gómez-Vidal & Silverman, 2001), this lactone is a useful derivative that can be readily transformed to N-Boc-cis-4-hydroxyl-L-prolinemethyl ester by quantitative trans-esterification with methanol in the presence of sodium azide. Further transformation of the hydroxyl group to an azido group in the presence of diphenylphosphorylazide (DPPA) under Mistunobu conditions affords N-Boc-trans-4-azido-L-proline methyl ester, a useful building block for the preparation of 4-aminoproline containing molecules.
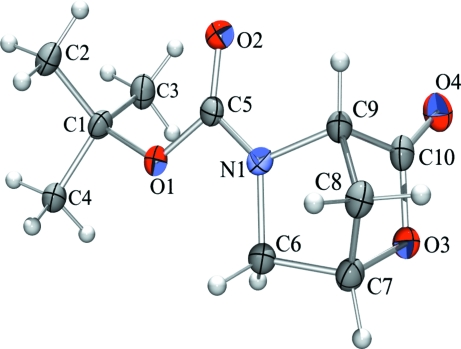
A search of the Cambridge Structural Database (CSD, Version 5.29; Allen, 2002) for allohydroxy-L-proline lacton gave rise to 3 hits: (1S,4S)-N-acetyl-3-oxo-5-aza-2-oxabicyclo[2.2.1]heptane (Lenstra et al., 1979); N-triphenylmethyl-2-oxa-5-azabicyclo[2.2.1]heptan-3-one (Papaioannou et al., 1989); tert-butyl 7-chloro-6-methyl-2,3-dihydro-2-oxo-6H-3,10b-methano-1,4- dioxazino[3,2-c](2,1)benzoxazine-4(4aH)-carboxylate (Didier et al., 2004). In the four structures the pyrrolidine ring (N1/C6/C7/C8/C9 numbering in the title compound) adopts the same envelope conformation with C8 out of the mean plane defined by N1, C6, C7 and C9. Three structures consists of an amide bond: the title compound, the N-acetyl-allohydroxy-L-proline lacton and the tert-butyl- 7-chloro-6-methyl-2,3-dihydro-2-oxo-6H-3,10b-methano-1,4- dioxazino(3,2-c)(2,1)benzoxazine-4(4aH)-carboxylate. The two first structures exhibit a quite similar structure with a nearly planar trans- amide bond. In the last one, the peptide bond is cis- and the nitrogen atom of the pyrrolidine ring exhibits an observable pyramidalization. Indeed, the sum of bond angles around the nitrogen atom is of 347.9° whereas of 357.9° and 357.4° in the two first structures.
Experimental
The title compound was prepared in 80% from N-Boc-trans-4-hydroxyproline following the a described procedure (Gómez-Vidal & Silverman, 2001) and was crystallized by slow evaporation of a cyclohexane/ethyl acetate (3:2, v/v) solution.
Refinement
Because of the lack of any significant anomalous dispersion effects, the absolute configurations of the title compound could not be determined from the diffraction experiments but was known from the method of synthesis. The origin was fixed by floating-origin restraints (Flack & Schwarzenbach, 1988). All H atoms were located in difference Fourier maps. The C-bonded H atoms were placed at calculated positions and refined using a riding model, with C—H distances of 0.93–0.96 Å. The H-atom Uiso parameters were fixed at 1.2Ueq(C) for methine and methylene C—H and at 1.5Ueq(C) for methyl C—H.
Figures
Fig. 1.
The molecular structure of title compound showing the atom-numbering scheme. All non-H atoms are represented by 50% probability displacement ellipsoids.
Crystal data
| C10H15NO4 | F(000) = 228 |
| Mr = 213.23 | Dx = 1.359 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.7107 Å |
| Hall symbol: P 2yb | Cell parameters from 11845 reflections |
| a = 6.0710 (7) Å | θ = 0.4–26.4° |
| b = 9.3703 (11) Å | µ = 0.11 mm−1 |
| c = 9.3002 (10) Å | T = 100 K |
| β = 100.013 (5)° | Prism, colourless |
| V = 521.00 (10) Å3 | 0.3 × 0.2 × 0.2 mm |
| Z = 2 |
Data collection
| Nonius KappaCCD area-detector diffractometer | 1054 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.066 |
| graphite | θmax = 26.6°, θmin = 3.1° |
| ω and φ scans | h = −7→7 |
| 5951 measured reflections | k = −11→11 |
| 1143 independent reflections | l = −11→11 |
Refinement
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.052 | w = 1/[σ2(Fo2) + (0.0035P)2 + 0.6362P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.102 | (Δ/σ)max < 0.001 |
| S = 1.20 | Δρmax = 0.23 e Å−3 |
| 1143 reflections | Δρmin = −0.22 e Å−3 |
| 139 parameters |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.3991 (4) | 0.3322 (3) | 0.3133 (3) | 0.0243 (6) | |
| O2 | 0.0303 (4) | 0.2997 (3) | 0.2103 (3) | 0.0280 (7) | |
| O3 | 0.3864 (4) | −0.0967 (3) | 0.1079 (3) | 0.0262 (7) | |
| O4 | 0.0299 (5) | −0.1077 (3) | 0.1467 (3) | 0.0319 (7) | |
| N1 | 0.3137 (5) | 0.1896 (4) | 0.1226 (4) | 0.0230 (7) | |
| C1 | 0.3539 (7) | 0.4316 (4) | 0.4288 (4) | 0.0249 (9) | |
| C2 | 0.2354 (7) | 0.5650 (4) | 0.3607 (5) | 0.0306 (10) | |
| H2A | 0.3194 | 0.6052 | 0.2893 | 0.046* | |
| H2B | 0.2265 | 0.6356 | 0.4372 | 0.046* | |
| H2C | 0.0842 | 0.54 | 0.3117 | 0.046* | |
| C3 | 0.2204 (8) | 0.3563 (5) | 0.5297 (5) | 0.0315 (10) | |
| H3A | 0.075 | 0.3274 | 0.4744 | 0.047* | |
| H3B | 0.1977 | 0.4212 | 0.6085 | 0.047* | |
| H3C | 0.3024 | 0.2717 | 0.5715 | 0.047* | |
| C4 | 0.5884 (6) | 0.4670 (4) | 0.5062 (5) | 0.0266 (9) | |
| H4A | 0.6574 | 0.3816 | 0.5556 | 0.04* | |
| H4B | 0.5813 | 0.5423 | 0.5784 | 0.04* | |
| H4C | 0.678 | 0.5001 | 0.4347 | 0.04* | |
| C5 | 0.2283 (6) | 0.2759 (4) | 0.2159 (4) | 0.0227 (8) | |
| C6 | 0.5472 (7) | 0.1381 (4) | 0.1373 (5) | 0.0250 (9) | |
| H6A | 0.6091 | 0.1101 | 0.239 | 0.03* | |
| H6B | 0.646 | 0.21 | 0.1032 | 0.03* | |
| C7 | 0.5098 (7) | 0.0093 (4) | 0.0356 (4) | 0.0265 (9) | |
| H7 | 0.648 | −0.0281 | 0.004 | 0.032* | |
| C8 | 0.3299 (7) | 0.0625 (5) | −0.0887 (4) | 0.0259 (9) | |
| H8A | 0.3762 | 0.1478 | −0.1388 | 0.031* | |
| H8B | 0.273 | −0.0127 | −0.1603 | 0.031* | |
| C9 | 0.1701 (6) | 0.0969 (4) | 0.0167 (4) | 0.0233 (9) | |
| H9 | 0.0204 | 0.1361 | −0.0275 | 0.028* | |
| C10 | 0.1710 (7) | −0.0459 (4) | 0.0960 (4) | 0.0256 (9) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0249 (14) | 0.0180 (14) | 0.0299 (14) | −0.0012 (12) | 0.0044 (12) | −0.0053 (12) |
| O2 | 0.0234 (14) | 0.0253 (16) | 0.0352 (16) | 0.0018 (13) | 0.0050 (12) | −0.0054 (13) |
| O3 | 0.0253 (15) | 0.0189 (14) | 0.0349 (16) | −0.0018 (13) | 0.0067 (12) | 0.0007 (13) |
| O4 | 0.0344 (17) | 0.0263 (16) | 0.0359 (16) | −0.0089 (14) | 0.0091 (13) | 0.0022 (14) |
| N1 | 0.0215 (17) | 0.0192 (17) | 0.0275 (18) | 0.0024 (14) | 0.0017 (14) | −0.0023 (15) |
| C1 | 0.027 (2) | 0.018 (2) | 0.031 (2) | 0.0018 (17) | 0.0092 (17) | −0.0050 (17) |
| C2 | 0.034 (2) | 0.018 (2) | 0.039 (2) | 0.0029 (18) | 0.0049 (19) | −0.0017 (19) |
| C3 | 0.037 (2) | 0.023 (2) | 0.035 (2) | −0.0045 (18) | 0.0101 (19) | −0.0047 (19) |
| C4 | 0.026 (2) | 0.020 (2) | 0.034 (2) | −0.0009 (17) | 0.0064 (17) | −0.0039 (18) |
| C5 | 0.023 (2) | 0.0157 (19) | 0.029 (2) | −0.0012 (16) | 0.0028 (16) | 0.0015 (16) |
| C6 | 0.024 (2) | 0.021 (2) | 0.030 (2) | −0.0009 (17) | 0.0057 (16) | −0.0024 (17) |
| C7 | 0.028 (2) | 0.022 (2) | 0.030 (2) | −0.0006 (18) | 0.0099 (19) | −0.0045 (18) |
| C8 | 0.031 (2) | 0.019 (2) | 0.028 (2) | −0.0005 (17) | 0.0056 (17) | −0.0007 (18) |
| C9 | 0.0238 (19) | 0.0187 (19) | 0.027 (2) | −0.0022 (17) | 0.0049 (17) | −0.0039 (16) |
| C10 | 0.031 (2) | 0.020 (2) | 0.025 (2) | −0.0038 (18) | 0.0024 (17) | −0.0047 (17) |
Geometric parameters (Å, °)
| O1—C5 | 1.359 (5) | C3—H3B | 0.98 |
| O1—C1 | 1.484 (5) | C3—H3C | 0.98 |
| O2—C5 | 1.215 (5) | C4—H4A | 0.98 |
| O3—C10 | 1.377 (5) | C4—H4B | 0.98 |
| O3—C7 | 1.475 (5) | C4—H4C | 0.98 |
| O4—C10 | 1.196 (5) | C6—C7 | 1.526 (6) |
| N1—C5 | 1.353 (5) | C6—H6A | 0.99 |
| N1—C9 | 1.478 (5) | C6—H6B | 0.99 |
| N1—C6 | 1.481 (5) | C7—C8 | 1.529 (6) |
| C1—C4 | 1.516 (5) | C7—H7 | 1 |
| C1—C3 | 1.516 (6) | C8—C9 | 1.528 (6) |
| C1—C2 | 1.524 (6) | C8—H8A | 0.99 |
| C2—H2A | 0.98 | C8—H8B | 0.99 |
| C2—H2B | 0.98 | C9—C10 | 1.528 (6) |
| C2—H2C | 0.98 | C9—H9 | 1 |
| C3—H3A | 0.98 | ||
| C5—O1—C1 | 120.7 (3) | O2—C5—O1 | 126.3 (4) |
| C10—O3—C7 | 106.3 (3) | N1—C5—O1 | 109.0 (3) |
| C5—N1—C9 | 122.1 (3) | N1—C6—C7 | 99.4 (3) |
| C5—N1—C6 | 127.1 (3) | N1—C6—H6A | 111.9 |
| C9—N1—C6 | 108.3 (3) | C7—C6—H6A | 111.9 |
| O1—C1—C4 | 101.8 (3) | N1—C6—H6B | 111.9 |
| O1—C1—C3 | 110.0 (3) | C7—C6—H6B | 111.9 |
| C4—C1—C3 | 111.5 (4) | H6A—C6—H6B | 109.6 |
| O1—C1—C2 | 110.3 (3) | O3—C7—C6 | 106.4 (3) |
| C4—C1—C2 | 110.7 (3) | O3—C7—C8 | 102.2 (3) |
| C3—C1—C2 | 112.0 (3) | C6—C7—C8 | 102.7 (3) |
| C1—C2—H2A | 109.5 | O3—C7—H7 | 114.7 |
| C1—C2—H2B | 109.5 | C6—C7—H7 | 114.7 |
| H2A—C2—H2B | 109.5 | C8—C7—H7 | 114.7 |
| C1—C2—H2C | 109.5 | C9—C8—C7 | 91.9 (3) |
| H2A—C2—H2C | 109.5 | C9—C8—H8A | 113.3 |
| H2B—C2—H2C | 109.5 | C7—C8—H8A | 113.3 |
| C1—C3—H3A | 109.5 | C9—C8—H8B | 113.3 |
| C1—C3—H3B | 109.5 | C7—C8—H8B | 113.3 |
| H3A—C3—H3B | 109.5 | H8A—C8—H8B | 110.6 |
| C1—C3—H3C | 109.5 | N1—C9—C10 | 103.9 (3) |
| H3A—C3—H3C | 109.5 | N1—C9—C8 | 100.6 (3) |
| H3B—C3—H3C | 109.5 | C10—C9—C8 | 100.1 (3) |
| C1—C4—H4A | 109.5 | N1—C9—H9 | 116.5 |
| C1—C4—H4B | 109.5 | C10—C9—H9 | 116.5 |
| H4A—C4—H4B | 109.5 | C8—C9—H9 | 116.5 |
| C1—C4—H4C | 109.5 | O4—C10—O3 | 122.4 (4) |
| H4A—C4—H4C | 109.5 | O4—C10—C9 | 132.2 (4) |
| H4B—C4—H4C | 109.5 | O3—C10—C9 | 105.4 (3) |
| O2—C5—N1 | 124.7 (4) | ||
| C5—O1—C1—C4 | −178.9 (3) | O3—C7—C8—C9 | 53.1 (3) |
| C5—O1—C1—C3 | 62.7 (5) | C6—C7—C8—C9 | −57.1 (3) |
| C5—O1—C1—C2 | −61.3 (4) | C5—N1—C9—C10 | −93.9 (4) |
| C9—N1—C5—O2 | −9.6 (6) | C6—N1—C9—C10 | 69.1 (4) |
| C6—N1—C5—O2 | −169.3 (4) | C5—N1—C9—C8 | 162.7 (3) |
| C9—N1—C5—O1 | 171.3 (3) | C6—N1—C9—C8 | −34.2 (4) |
| C6—N1—C5—O1 | 11.6 (5) | C7—C8—C9—N1 | 53.9 (3) |
| C1—O1—C5—O2 | 0.9 (6) | C7—C8—C9—C10 | −52.5 (3) |
| C1—O1—C5—N1 | 179.9 (3) | C7—O3—C10—O4 | −178.8 (4) |
| C5—N1—C6—C7 | 159.9 (4) | C7—O3—C10—C9 | −1.3 (4) |
| C9—N1—C6—C7 | −2.1 (4) | N1—C9—C10—O4 | 109.5 (5) |
| C10—O3—C7—C6 | 73.2 (4) | C8—C9—C10—O4 | −146.8 (4) |
| C10—O3—C7—C8 | −34.2 (4) | N1—C9—C10—O3 | −67.7 (3) |
| N1—C6—C7—O3 | −69.0 (4) | C8—C9—C10—O3 | 36.1 (4) |
| N1—C6—C7—C8 | 38.0 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WW2125).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Altomare, A., Cascarano, G., Giacovazzo, C., Guagliardi, A., Burla, M. C., Polidori, G. & Camalli, M. (1994). J. Appl. Cryst.27, 435.
- Didier, C., Crichter, D. J., Walshe, N. D., Kojima, Y., Yamauchi, Y. & Barrett, A. G. M. (2004). J. Org. Chem.69, 7875–7879. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Flack, H. D. & Schwarzenbach, D. (1988). Acta Cryst. A44, 499–506.
- Gómez-Vidal, J. A. & Silverman, R. B. (2001). Org. Lett.3, 2481–2484. [DOI] [PubMed]
- Lenstra, A. T. H., Petit, G. H. & Geise, H. J. (1979). Cryst. Struct. Commun.8, 1023–1029.
- Nonius (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Papaioannou, D., Stavropoulos, G., Nastopoulos, V., Voliotis, S. & Leban, I. (1989). Acta Cryst. C45, 1651–1652.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808030651/ww2125sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808030651/ww2125Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report