Abstract
In the title coordination polymer, {[Mn(C7H3NO4)(H2O)2]·2H2O}n, the MnII ion is coordinated in a distorted octahedral environment by the O atoms of two water molecules, one N and one O atoms of the chelating pyridine-2,3-dicarboxylate (PDC) dianion, and two axial bridging carboxylate O atoms from two adjacent PDC ligands. The fully deprotonated PDC anion acts a μ3-bridging ligand, establishing a chain structure along the a axis. These polymeric chains are connected into a three-dimensional framework via several intermolecular O—H⋯O hydrogen bonds.
Related literature
For related literature, see: Aghabozorg et al. (2007 ▶); Baruah et al. (2007 ▶); Drew et al. (1971 ▶); Ghoer & Youssef (1993 ▶); Kang et al. (2006 ▶); Li et al. (2006 ▶); Manteghi et al. (2007 ▶); Patrick et al. (2003 ▶); Sun et al. (2006 ▶); Takusagawa & Koetzle (1978 ▶); Turner & Batten (2007 ▶); Zhang & You (2003 ▶); Zhang et al. (2003 ▶).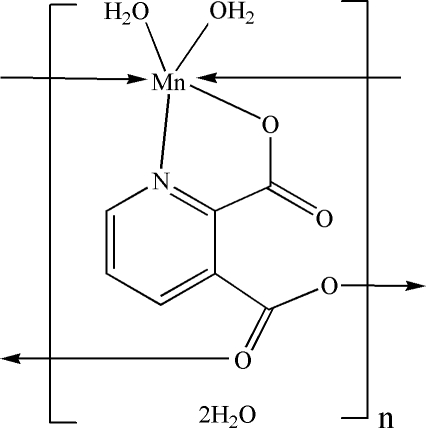
Experimental
Crystal data
[Mn(C7H3NO4)(H2O)2]·2H2O
M r = 292.11
Monoclinic,
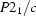
a = 6.5719 (8) Å
b = 7.6703 (9) Å
c = 20.566 (3) Å
β = 93.3540 (10)°
V = 1034.9 (2) Å3
Z = 4
Mo Kα radiation
μ = 1.31 mm−1
T = 291 (2) K
0.38 × 0.15 × 0.11 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.639, T max = 0.865
6428 measured reflections
1921 independent reflections
1774 reflections with I > 2σ(I)
R int = 0.015
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.095
S = 1.07
1921 reflections
154 parameters
3 restraints
H-atom parameters constrained
Δρmax = 0.49 e Å−3
Δρmin = −0.74 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2; data reduction: SAINT (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808029413/sg2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808029413/sg2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Mn1—O3 | 2.098 (3) |
| Mn1—O4 | 2.139 (3) |
| Mn1—O8i | 2.144 (2) |
| Mn1—O5 | 2.160 (3) |
| Mn1—O6ii | 2.242 (3) |
| Mn1—N1i | 2.263 (3) |
| O3—Mn1—O4 | 96.07 (11) |
| O3—Mn1—O8i | 168.80 (10) |
| O3—Mn1—O5 | 87.98 (10) |
| O4—Mn1—O5 | 87.54 (10) |
| O8i—Mn1—O5 | 95.89 (9) |
| O3—Mn1—O6ii | 84.63 (10) |
| O5—Mn1—O6ii | 164.11 (10) |
| O4—Mn1—N1i | 166.71 (11) |
Symmetry codes: (i) 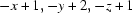 ; (ii)
; (ii) 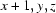 .
.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1W⋯O8 | 0.84 | 1.95 | 2.793 (4) | 177 |
| O1—H2W⋯O2iii | 0.84 | 2.09 | 2.845 (4) | 149 |
| O2—H4W⋯O6iv | 0.84 | 2.07 | 2.884 (4) | 165 |
| O2—H4W⋯O4v | 0.84 | 2.56 | 3.043 (4) | 118 |
| O2—H3W⋯O4vi | 0.85 | 1.94 | 2.788 (4) | 180 |
| O3—H5W⋯O7iv | 0.84 | 1.85 | 2.691 (4) | 175 |
| O3—H6W⋯O1iv | 0.85 | 1.92 | 2.730 (4) | 159 |
| O4—H8W⋯O1i | 0.85 | 2.61 | 3.457 (4) | 180 |
| O4—H7W⋯O1vii | 0.84 | 1.86 | 2.691 (4) | 168 |
| O4—H8W⋯O2vii | 0.85 | 2.37 | 2.788 (4) | 111 |
Symmetry codes: (i) 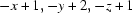 ; (iii)
; (iii) 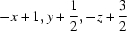 ; (iv)
; (iv) 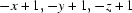 ; (v)
; (v) 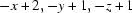 ; (vi)
; (vi) 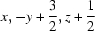 ; (vii)
; (vii) 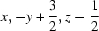 .
.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant No. 20471026) and the Natural Science Foundation of Henan Province (grant No. 0311021200).
supplementary crystallographic information
Comment
Pyridine-2,3-dicarboxylic acid (H2PDC), being a potential multidentate bridging ligand, has aroused considerable interests in recent decades and a number of metal complexes have been reported. In these complexes, pyridine-2,3-dicarboxylic acid is partly or fully deprotonated and shows diverse coordination modes, such as non-coordinate (Takusagawa & Koetzle, 1978; Manteghi et al., 2007), monodentate (Drew et al., 1971; Ghoer & Youssef, 1993; Patrick et al., 2003; Baruah et al., 2007), µ2-bridging (Zhang et al., 2003; Aghabozorg et al., 2007; Sun et al., 2006; Turner & Batten, 2007; Kang et al., 2006), µ3-bridging (Zhang & You, 2003; Li et al., 2006). Here we describe another new compound in which the PDC is µ3-bridging, (I),(Fig. 1).
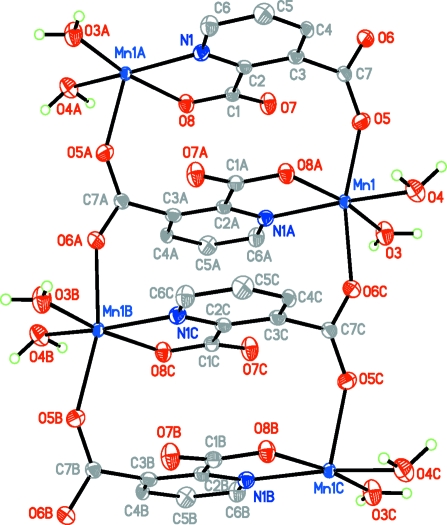
Complex (I) is composed of {[Mn(C7H3NO4)(H2O)2].2H2O}n units, in which the MnII ion is six-coordinated in a distorted octahedral geometry (Table 1) formed by two coordinated water molecules, one N and one O atoms of a PDC dianion and two different carboxylate O atoms in the axial position from another two adjacent PDC ligands. The deprotonated PDC are µ3-bridging ligands and they join the neighbouring MnII ions to form this one-dimensional linear chain structure along a axis. This kind of µ3-bridging mode is different from that have been published (Zhang & You, 2003; Li et al., 2006) as in this paper one bridging carboxylate O atom only links one metal ions, while in the latter one bridging carboxylate O atom simultaneously links two metal ions,respectively.
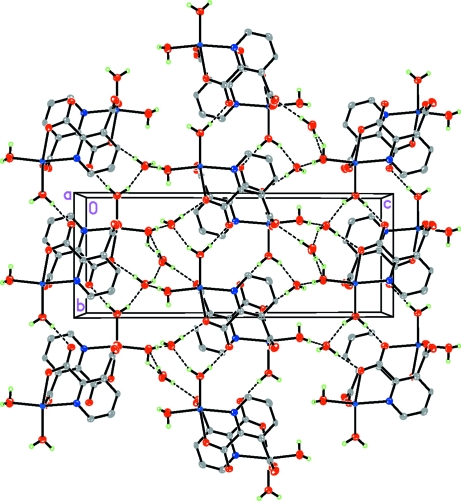
The carboxylate O atoms of PDC dianion and coordinate and non coordinated water molecules are all involved in rich O—H···O intermolecular hydrogen bonds (Table 2) and they connect polymetric chains into a three-dimensional framework (Fig. 2).
Experimental
The ligand H2PDC (1 mmol, 0.17 g) and NaOH (2 mmol, 0.08 g) were dissolved in water and methanol mixed solvent (20 ml, v/v 1:1). To this solution, Mn(CH3COO)2.4H2O (1 mmol, 0.25 g) was added and the resulting mixture was stirred and refluxed at 343 K for 5 h, then cooled to room temperature. After filtration and evaporation in air for a week, pink block-shaped crystals were obtained in a yield of 37%. Analysis, found (%): C, 28.70; H, 3.80; N, 4.71. C7H11Mn N O8 requires (%): C,28.75; H,3.76; N,4.79. (CCDC number 668395)
Refinement
H atoms bonded to C atoms were positioned geometrically with C—H distance of 0.93 Å, and treated as riding atoms, with Uiso(H) = 1.2Ueq. H atoms bonded to O atoms were located in a difference Fourier map and refined isotropically.
Figures
Fig. 1.
The structure of the building unit of the one-dimensional of (I), with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. Uncoordinate water molecules and H atoms on C atoms have been omitted. [Symmetry codes: (A) 2 - x, 2 - y, 1 - z; (B) 1 - x, 2 - y, 1 - z; (C) -1 + x, y, z.]
Fig. 2.
The crystal packing of (I), showing hydrogen bonds as dashed lines. For the sake of clarity, H atoms on C atoms have been omitted.
Crystal data
| [Mn(C7H3NO4)(H2O)2]·2H2O | F(000) = 596 |
| Mr = 292.11 | Dx = 1.875 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4039 reflections |
| a = 6.5719 (8) Å | θ = 2.8–28.1° |
| b = 7.6703 (9) Å | µ = 1.31 mm−1 |
| c = 20.566 (3) Å | T = 291 K |
| β = 93.354 (1)° | Block, pink |
| V = 1034.9 (2) Å3 | 0.38 × 0.15 × 0.11 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 1921 independent reflections |
| Radiation source: fine-focus sealed tube | 1774 reflections with I > 2σ(I) |
| graphite | Rint = 0.015 |
| φ and ω scans | θmax = 25.5°, θmin = 2.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −7→7 |
| Tmin = 0.640, Tmax = 0.865 | k = −9→9 |
| 6428 measured reflections | l = −23→24 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.095 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.042P)2 + 2.0761P] where P = (Fo2 + 2Fc2)/3 |
| 1921 reflections | (Δ/σ)max = 0.001 |
| 154 parameters | Δρmax = 0.49 e Å−3 |
| 3 restraints | Δρmin = −0.74 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.69176 (6) | 0.76827 (6) | 0.38821 (2) | 0.01743 (17) | |
| O1 | 0.3592 (5) | 0.7546 (3) | 0.70493 (14) | 0.0449 (7) | |
| H1W | 0.3455 | 0.8191 | 0.6719 | 0.067* | |
| H2W | 0.2695 | 0.7814 | 0.7307 | 0.067* | |
| O2 | 0.9617 (5) | 0.4500 (4) | 0.74324 (14) | 0.0543 (8) | |
| H3W | 0.8800 | 0.5269 | 0.7561 | 0.081* | |
| H4W | 0.9878 | 0.3937 | 0.7099 | 0.081* | |
| O3 | 0.7081 (4) | 0.4951 (3) | 0.38849 (13) | 0.0437 (7) | |
| H5W | 0.7244 | 0.4234 | 0.4193 | 0.066* | |
| H6W | 0.6950 | 0.4383 | 0.3532 | 0.066* | |
| O4 | 0.6927 (4) | 0.7979 (4) | 0.28481 (13) | 0.0420 (6) | |
| H7W | 0.5997 | 0.7749 | 0.2562 | 0.063* | |
| H8W | 0.6803 | 0.9080 | 0.2873 | 0.063* | |
| O5 | 0.3643 (4) | 0.7440 (3) | 0.37627 (14) | 0.0374 (6) | |
| O6 | 0.0271 (4) | 0.7556 (3) | 0.37392 (13) | 0.0362 (6) | |
| O7 | 0.2349 (5) | 0.7489 (3) | 0.51820 (13) | 0.0393 (6) | |
| O8 | 0.3057 (4) | 0.9571 (3) | 0.59238 (11) | 0.0313 (5) | |
| N1 | 0.2620 (4) | 1.2081 (4) | 0.50222 (14) | 0.0272 (6) | |
| C1 | 0.2629 (5) | 0.9023 (4) | 0.53482 (15) | 0.0251 (7) | |
| C2 | 0.2440 (4) | 1.0408 (4) | 0.48204 (15) | 0.0234 (6) | |
| C3 | 0.2117 (5) | 1.0010 (4) | 0.41594 (16) | 0.0257 (7) | |
| C4 | 0.1928 (4) | 1.1277 (4) | 0.37076 (14) | 0.0212 (6) | |
| H4 | 0.1690 | 1.1020 | 0.3268 | 0.025* | |
| C5 | 0.2096 (6) | 1.2911 (5) | 0.39172 (18) | 0.0374 (9) | |
| H5 | 0.1971 | 1.3810 | 0.3614 | 0.045* | |
| C6 | 0.2452 (6) | 1.3332 (5) | 0.45735 (17) | 0.0331 (8) | |
| H6 | 0.2574 | 1.4495 | 0.4699 | 0.040* | |
| C7 | 0.2003 (5) | 0.8171 (5) | 0.38797 (15) | 0.0255 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0193 (3) | 0.0159 (3) | 0.0170 (3) | −0.00007 (16) | 0.00015 (17) | −0.00174 (16) |
| O1 | 0.0536 (18) | 0.0450 (16) | 0.0354 (15) | 0.0076 (13) | −0.0033 (13) | 0.0040 (12) |
| O2 | 0.0584 (19) | 0.0623 (19) | 0.0426 (16) | 0.0277 (16) | 0.0062 (13) | −0.0098 (14) |
| O3 | 0.0653 (19) | 0.0273 (13) | 0.0376 (15) | −0.0005 (12) | −0.0048 (13) | 0.0011 (11) |
| O4 | 0.0416 (15) | 0.0498 (16) | 0.0339 (14) | −0.0068 (13) | −0.0048 (11) | 0.0014 (12) |
| O5 | 0.0277 (13) | 0.0371 (14) | 0.0475 (16) | 0.0024 (10) | 0.0035 (11) | −0.0113 (11) |
| O6 | 0.0259 (13) | 0.0410 (15) | 0.0413 (15) | −0.0027 (10) | −0.0015 (11) | −0.0103 (11) |
| O7 | 0.0592 (18) | 0.0225 (13) | 0.0353 (14) | −0.0028 (11) | −0.0030 (13) | 0.0002 (10) |
| O8 | 0.0405 (14) | 0.0286 (12) | 0.0242 (12) | 0.0002 (10) | −0.0024 (10) | 0.0017 (10) |
| N1 | 0.0272 (14) | 0.0247 (13) | 0.0297 (15) | 0.0008 (11) | 0.0024 (11) | −0.0001 (11) |
| C1 | 0.0223 (16) | 0.0250 (16) | 0.0279 (16) | 0.0009 (12) | 0.0010 (12) | −0.0004 (13) |
| C2 | 0.0182 (15) | 0.0249 (16) | 0.0273 (16) | 0.0004 (12) | 0.0016 (12) | 0.0000 (13) |
| C3 | 0.0183 (15) | 0.0292 (17) | 0.0298 (17) | −0.0003 (12) | 0.0016 (12) | −0.0001 (13) |
| C4 | 0.0262 (16) | 0.0241 (15) | 0.0133 (13) | 0.0011 (12) | −0.0004 (11) | 0.0015 (11) |
| C5 | 0.046 (2) | 0.0328 (19) | 0.033 (2) | 0.0036 (16) | 0.0031 (16) | 0.0099 (15) |
| C6 | 0.043 (2) | 0.0233 (16) | 0.0325 (18) | −0.0003 (14) | 0.0019 (15) | 0.0017 (14) |
| C7 | 0.0232 (17) | 0.0311 (17) | 0.0220 (15) | 0.0000 (13) | 0.0002 (12) | −0.0007 (13) |
Geometric parameters (Å, °)
| Mn1—O3 | 2.098 (3) | O6—Mn1iii | 2.243 (2) |
| Mn1—O4 | 2.139 (3) | O7—C1 | 1.236 (4) |
| Mn1—O8i | 2.144 (2) | O8—C1 | 1.272 (4) |
| Mn1—O5 | 2.160 (3) | O8—Mn1i | 2.144 (2) |
| Mn1—O6ii | 2.242 (3) | N1—C6 | 1.332 (4) |
| Mn1—N1i | 2.263 (3) | N1—C2 | 1.352 (4) |
| O1—H1W | 0.8416 | N1—Mn1i | 2.263 (3) |
| O1—H2W | 0.8409 | C1—C2 | 1.519 (4) |
| O2—H3W | 0.8502 | C2—C3 | 1.397 (4) |
| O2—H4W | 0.8369 | C3—C4 | 1.346 (4) |
| O3—H5W | 0.8410 | C3—C7 | 1.523 (5) |
| O3—H6W | 0.8474 | C4—C5 | 1.327 (5) |
| O4—H7W | 0.8414 | C4—H4 | 0.9300 |
| O4—H8W | 0.8497 | C5—C6 | 1.394 (5) |
| O5—C7 | 1.250 (4) | C5—H5 | 0.9300 |
| O6—C7 | 1.250 (4) | C6—H6 | 0.9300 |
| O3—Mn1—O4 | 96.07 (11) | C1—O8—Mn1i | 119.8 (2) |
| O3—Mn1—O8i | 168.80 (10) | C6—N1—C2 | 118.0 (3) |
| O4—Mn1—O8i | 94.60 (10) | C6—N1—Mn1i | 129.3 (2) |
| O3—Mn1—O5 | 87.98 (10) | C2—N1—Mn1i | 112.7 (2) |
| O4—Mn1—O5 | 87.54 (10) | O7—C1—O8 | 126.4 (3) |
| O8i—Mn1—O5 | 95.89 (9) | O7—C1—C2 | 117.6 (3) |
| O3—Mn1—O6ii | 84.63 (10) | O8—C1—C2 | 116.0 (3) |
| O4—Mn1—O6ii | 79.30 (10) | N1—C2—C3 | 120.8 (3) |
| O8i—Mn1—O6ii | 94.02 (9) | N1—C2—C1 | 116.3 (3) |
| O5—Mn1—O6ii | 164.11 (10) | C3—C2—C1 | 122.9 (3) |
| O3—Mn1—N1i | 94.21 (10) | C4—C3—C2 | 121.1 (3) |
| O4—Mn1—N1i | 166.71 (11) | C4—C3—C7 | 114.1 (3) |
| O8i—Mn1—N1i | 74.75 (9) | C2—C3—C7 | 124.8 (3) |
| O5—Mn1—N1i | 101.24 (10) | C5—C4—C3 | 117.1 (3) |
| O6ii—Mn1—N1i | 93.33 (10) | C5—C4—H4 | 121.5 |
| H1W—O1—H2W | 108.6 | C3—C4—H4 | 121.5 |
| H3W—O2—H4W | 140.9 | C4—C5—C6 | 122.6 (3) |
| Mn1—O3—H5W | 131.3 | C4—C5—H5 | 118.7 |
| Mn1—O3—H6W | 120.6 | C6—C5—H5 | 118.7 |
| H5W—O3—H6W | 108.1 | N1—C6—C5 | 120.4 (3) |
| Mn1—O4—H7W | 128.9 | N1—C6—H6 | 119.8 |
| Mn1—O4—H8W | 92.3 | C5—C6—H6 | 119.8 |
| H7W—O4—H8W | 100.4 | O6—C7—O5 | 124.7 (3) |
| C7—O5—Mn1 | 143.5 (2) | O6—C7—C3 | 117.4 (3) |
| C7—O6—Mn1iii | 147.3 (2) | O5—C7—C3 | 117.6 (3) |
| O3—Mn1—O5—C7 | 151.0 (4) | N1—C2—C3—C7 | 176.7 (3) |
| O4—Mn1—O5—C7 | −112.9 (4) | C1—C2—C3—C7 | −2.7 (5) |
| O8i—Mn1—O5—C7 | −18.5 (4) | C2—C3—C4—C5 | 1.1 (5) |
| O6ii—Mn1—O5—C7 | −146.8 (4) | C7—C3—C4—C5 | −177.4 (3) |
| N1i—Mn1—O5—C7 | 57.1 (4) | C3—C4—C5—C6 | 0.0 (5) |
| Mn1i—O8—C1—O7 | −171.8 (3) | C2—N1—C6—C5 | 0.2 (5) |
| Mn1i—O8—C1—C2 | 7.9 (4) | Mn1i—N1—C6—C5 | −177.7 (3) |
| C6—N1—C2—C3 | 0.9 (5) | C4—C5—C6—N1 | −0.7 (6) |
| Mn1i—N1—C2—C3 | 179.1 (2) | Mn1iii—O6—C7—O5 | 165.6 (3) |
| C6—N1—C2—C1 | −179.7 (3) | Mn1iii—O6—C7—C3 | −20.0 (6) |
| Mn1i—N1—C2—C1 | −1.4 (3) | Mn1—O5—C7—O6 | −177.6 (3) |
| O7—C1—C2—N1 | 175.7 (3) | Mn1—O5—C7—C3 | 8.0 (6) |
| O8—C1—C2—N1 | −4.0 (4) | C4—C3—C7—O6 | −81.4 (4) |
| O7—C1—C2—C3 | −4.8 (5) | C2—C3—C7—O6 | 100.2 (4) |
| O8—C1—C2—C3 | 175.5 (3) | C4—C3—C7—O5 | 93.4 (4) |
| N1—C2—C3—C4 | −1.6 (5) | C2—C3—C7—O5 | −85.0 (4) |
| C1—C2—C3—C4 | 179.0 (3) |
Symmetry codes: (i) −x+1, −y+2, −z+1; (ii) x+1, y, z; (iii) x−1, y, z.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1W···O8 | 0.84 | 1.95 | 2.793 (4) | 177. |
| O1—H2W···O2iv | 0.84 | 2.09 | 2.845 (4) | 149. |
| O2—H4W···O6v | 0.84 | 2.07 | 2.884 (4) | 165. |
| O2—H4W···O4vi | 0.84 | 2.56 | 3.043 (4) | 118. |
| O2—H3W···O4vii | 0.85 | 1.94 | 2.788 (4) | 180. |
| O3—H5W···O7v | 0.84 | 1.85 | 2.691 (4) | 175. |
| O3—H6W···O1v | 0.85 | 1.92 | 2.730 (4) | 159. |
| O4—H8W···O1i | 0.85 | 2.61 | 3.457 (4) | 180. |
| O4—H7W···O1viii | 0.84 | 1.86 | 2.691 (4) | 168. |
| O4—H8W···O2viii | 0.85 | 2.37 | 2.788 (4) | 111. |
Symmetry codes: (iv) −x+1, y+1/2, −z+3/2; (v) −x+1, −y+1, −z+1; (vi) −x+2, −y+1, −z+1; (vii) x, −y+3/2, z+1/2; (i) −x+1, −y+2, −z+1; (viii) x, −y+3/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SG2261).
References
- Aghabozorg, H., Daneshvar, S., Motyeian, E., Ghadermazi, M. & Attar Gharamaleki, J. (2007). Acta Cryst.E63, m2468–m2469. [DOI] [PMC free article] [PubMed]
- Baruah, A. M., Karmakar, A. & Baruah, J. B. (2007). Polyhedron, 26, 4518–4524.
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Drew, M. B., Matthews, R. W. & Walton, R. A. (1971). J. Chem. Soc. A, pp. 2959–2962.
- Ghoer, M. S. & Youssef, A. A. (1993). Polyhedron, 12, 1871–1878.
- Kang, Y., Zhang, J., Li, Z. J., Cheng, J. K. & Yao, Y. G. (2006). Inorg. Chim. Acta, 359, 2201–2209.
- Li, M., Xiang, J. F., Yuan, L. J., Wu, S. M., Chen, S. P. & Sun, J. T. (2006). Cryst. Growth Des.6, 2036–2040.
- Manteghi, F., Ghadermazi, M. & Aghabozorg, H. (2007). Acta Cryst. E63, o2809. [DOI] [PMC free article] [PubMed]
- Patrick, B. O., Stevens, C. L., Storr, A. & Thompson, R. C. (2003). Polyhedron, 22, 3025–3035.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sun, L. P., Niu, S. Y., Jin, J., Yang, G. D. & Ye, L. (2006). Inorg. Chem. Commun.9, 679–682.
- Takusagawa, F. & Koetzle, T. F. (1978). Acta Cryst. B34, 1149–1154.
- Turner, D. R. & Batten, S. R. (2007). Acta Cryst. E63, m452–m454.
- Zhang, X.-M., Fang, R.-Q., Wu, H.-S. & Ng, S. W. (2003). Acta Cryst. E59, m1143–m1145.
- Zhang, H.-T. & You, X.-Z. (2003). Acta Cryst. C59, m313–m314. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808029413/sg2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808029413/sg2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report