Abstract
We have developed an in vitro mutation assay using primary hepatocytes from the transgenic Muta™Mouse. Primary hepatocytes were isolated using a two-step perfusion method with purification by Percoll, cultured, and treated with benzo[a]pyrene (BaP), 2-amino-1-methyl-6-phenyl- imidazo[4,5-b]pyridine (PhIP), 3-nitrobenzoanthrone (3-NBA), and cigarette smoke condensate (CSC). The mean lacZ mutant frequency (MF) for the solvent control was approximately twofold greater than the spontaneous MF observed in liver tissue. A concentration-dependent increase in MF (up to 3.7-fold above control) was observed following exposure to BaP. Fourfold and twofold increases in mutant frequency were observed for 3-NBA and PhIP exposures, respectively, without the addition of any exogenous metabolic activation. A slight but statistically significant increase in lacZ MF was observed for CSC, but only at the lowest concentration. This is the first report demonstrating that mutations can be detected in cultured primary hepatocytes from Muta™Mouse. The preliminary results presented suggest that the Muta™Mouse primary hepatocyte mutagenicity assay can be used as a cost-effective tool for screening of environmental mutagens and therapeutic products. Environ. Mol. Mutagen. 51:330–337, 2010. © 2009 Wiley-Liss, Inc.
Keywords: primary hepatocytes, mutation, Muta™Mouse
Transgenic rodent (TGR) mutation models such as Muta™Mouse and Big Blue® rat/mouse provide efficient methods for quantitative assessments of in vivo gene mutation. Such transgenic mutation assays involve scoring of mutations at transgenic lacZ or lacI sequences carried on a lambda phage shuttle vector that has been stably integrated into the rodent genome. The shuttle vectors containing the transgenic targets exist in every cell of the transgenic animal and are easily recovered from genomic DNA using a convenient in vitro packaging system [Gossen et al., 1989; Kohler et al., 1991; Douglas et al 1996]. A major advantage of the transgenic mutation system lies in its ability to provide reliable and reproducible assessments of in vivo mutagenicity in any organ or tissue [Heddle et al., 2000; Nohmi et al., 2000; Thybaud et al., 2003]. In their detailed review paper, Lambert et al. concluded that TGR mutation models showed excellent concordance (77%) with rodent carcinogenicity that meets or exceeds what has been observed for other genotoxicity assays commonly employed for regulatory decision-making (e.g., bone marrow micronuclei or unscheduled DNA synthesis in liver) [Thybaud et al., 2003; Lambert et al., 2005].
Although in vivo TGR mutagenicity assays offer the advantages of utility for regulatory screening, matching in vitro versions provide an opportunity for high-throughput analyses of test mutagens (e.g., new chemicals or drug candidates). A number of approaches have been employed to establish cell lines derived from TGRs. For example, a Big Blue® mouse embryonic fibroblast cell line was derived from primary embryo cells immortalized and transformed by X-ray irradiation and benzo[a]pyrene (BaP) exposure [Erexson et al., 1998]. BBR1 and BBM1 cells were derived from the primary skin fibroblasts of the Big Blue® rodents [Erexson et al., 1999]. Several epithelial and fibroblast cell lines have been derived from the rat mammary gland and oral cavity, and these cells were immortalized by exposure to the alkylating agent N-ethyl-N-nitrosourea [McDiarmid et al., 2001; Papp-Szabó et al., 2003]. Watanabe et al. [2001] established two mammary carcinoma cell lines derived from 2-amino-1-methyl-6-phenyl-imidazo[4,5-b]pyridine (PhIP)-induced Big Blue® rat mammary adenocarcinomas. Finally, a spontaneously immortalized epithelial cell line, known as FE1, was derived from Muta™Mouse lung tissue. The FE1 line has proved to be a useful tool for rapid and effective screening of environmental mutagens [White et al., 2003; Jacobsen et al., 2007, 2008a,b; Berndt-Weis et al., 2009].
The aforementioned cell lines, and indeed all cell lines derived from nonhepatic tissue, have a limited endogenous capacity to metabolize test mutagens. In general, transformed cell lines lose their capacity to metabolize or activate promutagens. Some researchers have even reported a lack of sensitivity for the widely used hepatic HepG2 cells, in comparison with primary human hepatocytes [Wilkening et al., 2003]. Consequently, an exogenous metabolic activation mixture (e.g., postmitochondrial supernatant from Aroclor-induced rat liver) is often required to permit Phase I metabolism and conversion of promutagens into reactive metabolites. For example, an exogenous S9 mixture from rat liver was required in a study that investigated the muta-genic activity of PhIP in the BBR/MFib fibroblast system [McDiarmid et al., 2002].
The liver is the primary organ for the metabolism of xenobiotic substances by Phase I and Phase II biotransformation enzymes. Cultured primary mammalian hepatocytes can retain the characteristics of liver cells and have been shown to contain a broad spectrum of xenobiotic metabolizing enzymes [Ulrich et al., 1995]. The metabolic capacity of cultured primary mammalian hepatocytes suggests that they should be ideal for the evaluation and screening of suspected environmental mutagens. Indeed, the utility of cultured primary hepatocytes has already been definitively demonstrated in general toxicology and
for early screening of drug candidates [Ulrich et al., 1995]. However, the established hepatic assays for genotoxicity screening (e.g., unscheduled DNA synthesis, DNA adducts/repair) do not require the property of cell proliferation [Casciano 2000].
In vitro gene mutation assays are generally carried out with continuously dividing cells, despite their distinct metabolic insufficiency. The lack of mitogenesis, and thus, the limited capacity for cell division of primary hepatocytes, has prevented their use for the scoring of gene mutations. However, recent advances in cell culture techniques can permit limited proliferation of primary hepatocytes, and primary hepatocyte cultures have been employed to assess induction of sister chromatid exchanges and micronuclei [Eckl and Raffelsberger, 1997; Müller-Tegethoff et al., 1997]. Several studies have shown that the addition of selected growth factors and hormones (e.g., insulin, epidermal growth factor [EGF] or hepatocyte growth factor [HGF]) can induce proliferation of primary hepatocytes in vitro [Matsumoto and Nakamura, 1991; Block et al., 1996; Müller-Tegethoff et al., 1997]. In this pilot study, we demonstrate that cultured primary hepatocytes derived from the Muta™Mouse can be employed to assess the mutagenic activity of selected test mutagens that require metabolic activation by cytochrome P450 isozymes.
MATERIALS AND METHODS
Materials and Reagents
All cell culture media and reagents were purchased from Sigma-Aldrich (Oakville, ON, Canada). 3-Nitrobenzoanthrone (3-NBA) was obtained from the Sigma Library of Rare Chemicals (Oakville, ON, Canada). BaP was obtained from Supelco Canada (Mississauga, ON, Canada) and PhIP was obtained from Toronto Research Chemicals (Downsview, ON, Canada). Preparation of the cigarette smoke condensate (CSC) was performed at Labstat International Inc. (Kitchener, ON, Canada). Combustion (i.e., smoking) of commercially available full flavor cigarettes was carried out on a 20-port rotary smoking machine (see Moir et al. [2008] for details). The smoking parameters and smoking machine specifications followed the International Organization for Standardization's standard ISO 3308 (i.e., Routine Analytical Cigarette-Smoking Machines Definitions and Standard Conditions) (see Moir et al. [2008]). Mainstream smoke was passed through a 92-mm glass fiber filter disc for particulate matter collection. To prepare tobacco smoke condensates, filter pads were placed in a flask containing dimethyl sulfoxide (DMSO) (ACS spectrophotometric grade, >99.9%) and shaken on a wrist-action shaker (Barnstead International, Melrose Park, IL) for 20 min. Each sample was standardized to a concentration of 30 mg total particulate material (TPM) per ml of DMSO.
Transgenic Muta™Mouse
The transgenic Muta™Mouse (BALB/c × DBA2, mouse strain 40.6) was developed using a bacteriophage lambda shuttle vector containing the bacterial lacZ gene as a target for mutation detection [Gossen et al., 1989]. The transgenic mice were bred and maintained at Health Canada facilities under conditions approved by the Health Canada Animal Care Committee.
Isolation of Primary Hepatocyte Cell Culture and Chemical Treatment
Two 18–22-week-old male lacZ transgenic mice were used in this pilot study. Primary hepatocytes were isolated from Muta™Mouse by an adaptation of a two-step collagenase perfusion technique that involves enrichment prior to culturing using Percoll isodensity purification [Seglen, 1976; Kreamer et al., 1986; Tateno et al., 2000; Chen and Bunce, 2003]. A schematic of the procedure employed for isolation of parenchymal hepatocytes is provided in Figure 1. In brief, the mice were anesthetized by an i.p. injection of 100 mg/kg pentobarbital. The caudal vena cava was catheterized, the liver perfused with Hank's balanced salt solution (HBSS; pH 7.4, without Ca2+, Mg2+, 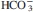 , or phenol red) containing 1 mM ethylene glycol tetraacetic acid and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for ∼2 min, followed by hepatocyte-qualified collagenase (0.3 mg/ml) in William's E medium (pH 7.4), supplemented with 10 mM HEPES and 0.1 mg/ml albumin, for ∼10 min. The digested liver was then excised, rinsed, and disaggregated in a 150-mm polystyrene Petri dish. The material was filtered through sterile gauze, and the filtrate was gently centrifuged for 3 min at 50g. The pellet was resuspended in 10 ml attachment medium (William's E medium supplemented with 10 mM HEPES, 2 mM l-gluta-mine, and 10% fetal bovine serum) combined with 10 ml of Percoll in HBSS and recentrifuged at 50g for 10 min. After the enrichment by Percoll isodensity purification, the cells were washed and gently centrifuged, and the pellets were resuspended in ∼20 ml of attachment media. The cells were counted using a hemocytometer. The viability of the cells was >90% as assessed by the trypan blue dye exclusion method.
, or phenol red) containing 1 mM ethylene glycol tetraacetic acid and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for ∼2 min, followed by hepatocyte-qualified collagenase (0.3 mg/ml) in William's E medium (pH 7.4), supplemented with 10 mM HEPES and 0.1 mg/ml albumin, for ∼10 min. The digested liver was then excised, rinsed, and disaggregated in a 150-mm polystyrene Petri dish. The material was filtered through sterile gauze, and the filtrate was gently centrifuged for 3 min at 50g. The pellet was resuspended in 10 ml attachment medium (William's E medium supplemented with 10 mM HEPES, 2 mM l-gluta-mine, and 10% fetal bovine serum) combined with 10 ml of Percoll in HBSS and recentrifuged at 50g for 10 min. After the enrichment by Percoll isodensity purification, the cells were washed and gently centrifuged, and the pellets were resuspended in ∼20 ml of attachment media. The cells were counted using a hemocytometer. The viability of the cells was >90% as assessed by the trypan blue dye exclusion method.
The cells were placed (2.5 × 105 cells/3.0 ml attachment media) in 60-mm polystyrene tissue culture dishes (Corning, Corning, NY) pre-coated with collagen. After 2 hr, the attachment medium was removed and 3.0 ml serum-free medium (William's E medium supplemented with 10 mM HEPES, 2 mM l-glutamine, 10 mM pyruvate, 0.35 mM proline, 20 units/l insulin, 100 units/ml penicillin G, 100 mg/ml streptomycin sulphate) containing 1 ng/ml murine EGF was added to each plate. The cells were then incubated at 37°C (95% relative humidity, 5% CO2). After 12 hr the cells were treated with various concentrations of test mutagens in serum-free medium containing 1 ng/ml EGF for 6 hr. After treatment, the cells were washed with phosphate-buffered saline (pH 7.6) and incubated in serum-free medium containing 1 ng/ml EGF for 48 hr before mutation scoring.
Isolation of Genomic DNA
Genomic DNA was isolated as previously described [Vijg and Douglas, 1996; Douglas et al., 1999], with modifications for cultured cells [White et al., 2003]. Briefly, treated cells were digested overnight in lysis buffer at 37°C (10 mM Tris, pH 7.6, 150 mM NaCl, 10 mM ethylenediaminetetraacetic acid [EDTA], with 1% sodium dodecyl sulphate and 1 mg/ml fresh proteinase K), and lysates extracted with phenol/chloroform (1:1), followed by chloroform. Potassium acetate was added to a final concentration of 1.6 M and the DNA was precipitated in ethanol. DNA was spooled onto a sealed Pasteur pipette, washed with 70% ethanol, placed in 15–25 μl of Tris-EDTA buffer (10 mM Tris, pH 7.6, 0.1 mM EDTA), and stored at 4°C for further analysis.
lacZ Mutant Frequency Analysis
Transgene mutant frequency (MF) was determined using the phenyl-β-d-galactopyranoside (P-gal)–positive selection assay [Vijg and Douglas, 1996; Lambert et al., 2005]. The method employs galE− host bacteria to facilitate the isolation and enumeration of mutant copies of the lacZ transgene [Gossen et al., 1992]. λgt10lacZ DNA copies were rescued from genomic Muta™Mouse DNA (4 μl aliquots) using the Transpack™ lambda packaging system (Stratagene, La Jolla, CA). Packaged phage particles were mixed with host bacteria (Escherichia coli ΔlacZ−, galE−, recA−, pAA119 with galT and galK) [Gossen et al., 1992] and plated on minimal agar with 0.3% (w/v) P-gal. Concurrently, bacteria were plated on nonselective minimal agar to enumerate total plaque-forming units (pfu) or titer. All plates were incubated overnight at 37°C. MF was expressed as the ratio of mutant plaques to total pfu. The data presented are summaries across numerous experimental replicates. MF and pfu values are readily available from the corresponding author.
Statistical Analysis
MF data were analyzed by Poisson regression using SAS version 9.1 (SAS Institute, Cary, NC), and the data were fit to the model log(E(Yi)) = log ti + βxi, where E(Yi) is the expected value for the ith observation, β is the vector of regressions coefficients, xi is a vector of covariates for the ith observation, and ti is the offset variable used to account for differences in observation count period (i.e., pfu). The offset (i.e., natural log of pfu) was given a constant coefficient of 1.0 for each observation, and log-linear relationships between mutant count and test mutagen concentration were specified by a natural log link function. Type 1, or sequential analysis, was employed to examine the statistical significance of the chemical treatment, and custom contrasts were employed to evaluate the statistical significance of responses at selected concentrations. Custom contrasts were accomplished by specifying an L matrix, and computing statistics for pairwise comparisons based on the asymptotic chi-square distribution of the likelihood ratio.
RESULTS
Morphological Changes
Figure 2 illustrates the phenotypic changes of hepatocytes cultured in the presence of EGF. The freshly isolated mouse hepatocytes display typical cubic, nonproliferating morphology 2 hr after being plated (Fig. 2A). Both mononuclear and binuclear parenchymal hepatocytes were observed in the isolated cell populations. After 48 hr culture in serum-free medium supplemented with EGF, the hepatocytes display a more scattered proliferating morphology (Fig. 2B).
Fig. 2.
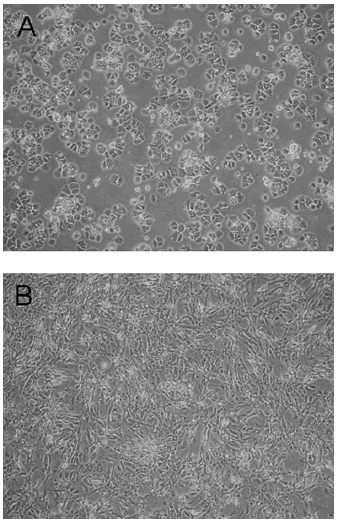
Phase-contrast photomicrographs of cultured primary hepatocytes. (A) Typical cubic hepatocytes shortly after isolation; (B) scattered, elongated hepatocytes after 48 hr (magnification, 40×).
Mutagenic Activity of Promutagens in Muta™Mouse Primary Hepatocytes
The overall MF for the vehicle control was 14.2 ± 5.6 × 10−5. This value is approximately twofold greater than that commonly observed in Muta™Mouse tissues (i.e., 5.9 × 10−5) [White et al., 2003]. In the initial pilot experiment (hepatocytes from two male mice), 1 ng/ml HGF was employed to stimulate hepatocyte growth, and the results revealed substantial inductions of lacZ mutants by 1.58 μM BaP (29.7 × 10−5) and 4 μM PhIP (30.8 × 10−5). These results confirmed the feasibility of the assay system. However, all subsequent experiments were conducted using EGF, a culture reagent that is far less expensive than HGF.
Muta™Mouse primary hepatocytes cultured in the presence of EGF were treated with several known environmental promutagens including BaP, PhIP, 3-NBA, and CSC. A summary of the MF values for these agents is presented in Table I. A concentration-dependent increase in MF (up to 3.7-fold above the concurrent control) was observed for primary hepatocytes exposed to BaP concentrations between 1.58 and 6.34 μM. A concentration-dependent increase in MF was also observed for PhIP, with the maximum response about 2.6-fold above control at the highest concentration tested (8 μM). At the low and middle concentrations (i.e., 1.45 and 3.63 μM), 3-NBA exposure induced a more than fourfold increase in MF; however, at the highest concentration (18.15 μM), cytotoxicity contributed to a low DNA recovery, and reliable MF scoring was not possible. For the CSC exposure, a statistically significant increase in MF was observed at the low concentration (80 μg TPM/ml); however, no significant increase was observed for the higher concentrations (i.e., 120, 160 μg/ml).
Table I.
lacZ Mutant Frequency in Cultured Muta™Mouse Primary Hepatocytes
| Chemicals | Concentration (μM)a | nb | Total mutants | Total plaques | Mean MF (×10−5)c | SDd | P valuee |
|---|---|---|---|---|---|---|---|
| Solvent control | 0 | 11 | 66 | 503,461 | 14.2 | 5.6 | |
| BaP | 1.58 | 5 | 78 | 286,935 | 27.5 | 11.3 | 0.0006 |
| 3.17 | 5 | 71 | 217,852 | 35.4 | 17.9 | <0.0001 | |
| 6.34 | 5 | 149 | 286,106 | 53.1 | 8.0 | <0.0001 | |
| Poisson regression chi-square for test mutagen concentration effect = 61.7, P < 0.0001 | |||||||
| PhIP | 2.0 | 5 | 38 | 168,483 | 22.1 | 6.8 | 0.015 |
| 4.0 | 5 | 75 | 241,211 | 30.7 | 5.9 | <0.0001 | |
| 8.0 | 5 | 39 | 130,380 | 36.8 | 17.5 | 0.0002 | |
| Poisson regression chi-square for test mutagen concentration effect = 28.2, P < 0.0001 | |||||||
| 3-NBA | 1.45 | 5 | 62 | 119,777 | 56.6 | 14.8 | <0.0001 |
| 3.63 | 3 | 26 | 32,139 | 79.4 | 19.5 | <0.0001 | |
| Poisson regression chi-square for test mutagen concentration effect = 70.6, P < 0.0001 | |||||||
| CSC | 80 | 5 | 46 | 198,966 | 23.1 | 6.6 | 0.005 |
| 120 | 5 | 44 | 300,188 | 15.1 | 7.4 | NSf | |
| 160 | 4 | 21 | 110,831 | 18.8 | 4.5 | NS | |
| Poisson regression chi-square for test mutagen concentration effect = 8.8, P = 0.03 | |||||||
All concentrations in μM, except CSC, which is expressed as μg TPM/ml.
n, the number of assays for mutation scoring.
Mean lacZ mutant frequency per 105 pfu.
SD, standard deviation of the mean.
Poisson regression with custom contrasts against the solvent control.
NS, not significant.
DISCUSSION
This study introduces a novel in vitro assay system for mutagenicity assessment that takes simultaneous advantage of the P-gal-positive selection system to score mutations at the lacZ transgene, and the metabolic capacity of primary hepatocytes. Although hepatocytes have a limited capacity for cell proliferation, EGF supplementation was employed to stimulate growth and division, and microscopic observations showed cell elongation and proliferation. Earlier works by Ichihara et al. [1982] and Nakamura and Ichihara [1985] have shown that mature hepatocytes, which are usually quiescent, will synthesize DNA and show density-dependent growth when cultured in the presence of insulin and EGF. Moreover, the work by Müller-Tegethoff et al. demonstrated the utility of cultured primary hepatocytes for the assay of micronuclei [Müller-Tegethoff et al., 1997]. This work extends the application of hepatocytes for genetic toxicity assessment.
Hepatocytes are the main functional liver cells, and they make up at least 60% of the cytoplasmic mass of the liver [Seglen, 1976]. In addition to hepatocytes, liver tissue contains endothelial cells, bile duct cells, Stellate cells, Kupffer cells, as well as supporting tissues. The standard two-step collagenase perfusion method employed in this study has been shown to yield 98% parenchymal hepatocytes, and the remaining 2% consisting of nonpar-enchymal cells (e.g., endothelial cells, bile duct cells, Stellate cells, Kupffer cells) can be separated by centrifu-gation (Fig. 1) [Seglen, 1976; Block et al., 1996; Tateno et al., 2000]. In our experiment, the isolated cells showed a homogeneous cubic morphology typical of nonproliferating hepatocytes (Fig. 2A); however, after 48 hr incubation with EGF the cultured cells displayed a scattered morphology (Fig. 2B), and this morphology is consistent with the observations of Block et al. [1996]. Moreover, Tateno et al. have shown that isolated hepatocytes, such as those shown in Figure 2, can be highly heterogeneous with respect to size and proliferation potential [Tateno et al., 2000]. Although it is possible that some fibroblasts coexist with the isolated hepatocytes and contribute to the measured MF values, it is important to note that liver fibroblasts are present mainly in the hepatic hilum, and the isolation of fibroblasts from the hilum requires the use of a different enzyme (i.e., pronase) [Kruglov et al., 2002]. Furthermore, the first centrifugation step employed in this study will effectively separate fibroblasts from hepatocytes. Thus, if fibroblasts are present in the isolated cell population, they would be expected to occur in trace amounts (see Fig. 1). Consequently, the isolated DNA employed for mutation scoring is mainly from hepatocytes, and not fibroblast contamination. Nevertheless, subsequent analyses should employ biochemical methods to investigate the composition of the isolated cell population (see discussion below).
Fig. 1.
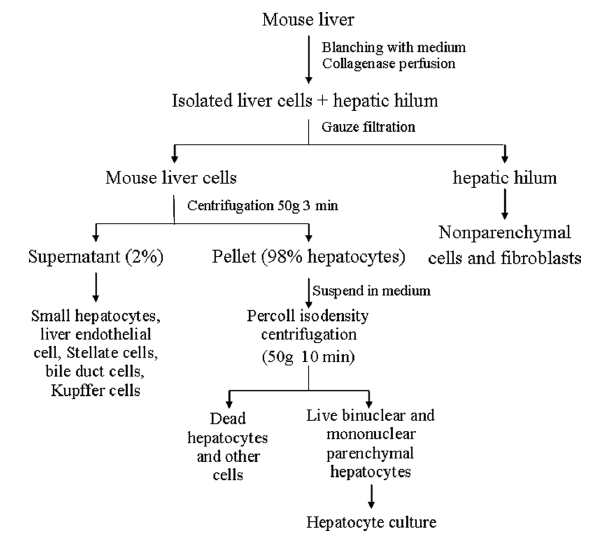
Scheme showing the procedure employed to isolate nearly pure hepatocytes via two-step perfusion and Percoll isodensity centrifugation (Adapted from the works of Seglen [1976], Kreamer et al. [1986], Block et al. [1996], Tateno et al. [2000], and Kruglov et al. [2002]).
BaP has been frequently used as a prototypical promu-tagenic carcinogen, and the observed increase in MF in the cultured Muta™Mouse hepatocytes employed in this study (i.e., 3.7-fold) is consistent with the results of in vivo studies. Although liver is not necessarily the target organ for BaP-induced neoplasia, up to a fivefold increase of lacZ gene MF (i.e., 22 × 10−5 vs. 4.1 × 10−5 in corn oil control) has been observed in the liver of Muta™Mouse orally exposed to BaP at 125 mg/kg/day for 5 days, followed by a 14-day manifestation time [Hakura et al., 1998]. And threefold induction of MF (i.e., 62.5 × 10−5 vs. 22.8 × 10−5 in the corn oil control) was seen in a subsequent study employing a 6-month manifestation period [Hakura et al., 1999]. Although differences in the exposure kinetics of in vitro and in vivo systems prohibit direct comparisons of MF values, the similarity in the trends highlights the utility of the in vitro system based on cultured primary hepatocytes.
PhIP, a heterocyclic aromatic amine identified in cooked foods, is a potent mutagen and animal carcinogen. Cytochrome P450 isozymes 1A1, 1A2, and 1B1 are believed to be involved in the metabolism and activation of PhIP via N-hydroxylation, followed by esterification to form N-acetoxy-PhIP that ultimately yields the highly reactive nitrenium ion [Boobis et al., 1994; Crofts et al., 1998]. PhIP is mutagenic in Salmonella and induces DNA damage, gene mutations, and cytogenetic abnormalities in cultured mammalian cells in the presence of an exogenous S9 metabolic activation system [IARC, 1993; Felton et al., 1994]. It has been documented that PhIP induced increases in MF in the liver of transgenic animals [Lynch et al., 1996; Masumura et al., 1999; Klein et al., 2001]. Our earlier in vitro work with the Muta™Mouse FE1 epithelial cell line showed PhIP-induced increases in lacZ MF only in the presence of exogenous S9 [White et al., 2003]. In this study, a concentration-dependent increase in MF was observed in cultured Muta™Mouse hepatocytes exposed to PhIP, and the observed increase (i.e., 2.6-fold) is consistent with the aforementioned results.
3-NBA is one of the most potent mutagens isolated from diesel emission particulates. Metabolism and activation of 3-NBA in mammalian systems is complex and believed to involve nitroreduction by NAD(P)H:quinone oxidoreductase and/or xanthine oxidase [Arlt et al., 2005; Chen et al., 2008]. TGR mutagenicity assessments revealed up to 4.8-fold induction in cII MF in Muta™Mouse liver after intraperitoneal treatment with 3-NBA (25 mg/kg body weight, administered once per week for 4 weeks) [Arlt et al., 2004]. Similarly, our earlier work showed a 4.2-fold induction in lacZ mutant frequency in Muta™Mouse liver following oral administration of 2 mg/kg/day for 28 days [Chen et al., 2008]. The increase in lacZ gene MF observed in this study (i.e., 5.6-fold) is consistent with these results.
Tobacco smoke is the most extreme example of a “systemic human mutagen” [DeMarini, 2004]. CSC has been shown to induce mutations at the tk locus in mouse lym-phoma cells [Clive et al., 1979] and Hprt mutations in CHO cells [Jongen et al., 1985] in the presence of exogenous metabolic activation. Hprt mutations have also been observed in a human lymphoblastoid cell line (MCL-5) that carries two recombinant plasmids expressing xenobiotic metabolizing enzymes [Krause et al., 1999]. There are more than 60 carcinogens in cigarette smoke, including several tobacco-specific nitrosamines such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N′-nitrosonornicotine, several polycyclic aromatic hydrocarbons (e.g., BaP), and aromatic amines such as 4-aminobiphenyl [Hecht, 2003]. Numerous studies have established that these carcinogens require metabolic activation by several cytochrome P450 isozymes [Hecht, 2008]. The results obtained here (Table I) demonstrate that 80 lg TPM/ml induced a significant increase in lacZ MF in cultured Muta™Mouse hepatocytes. The lack of mutagenicity observed at higher concentrations (i.e., 120, 160 μg/ml) was likely the result of cytotoxicity, which was evidenced by reduced cell survival and low yield of extractable DNA.
The results presented here confirm the utility of a lacZ gene mutation assay in cultured primary hepatocytes derived from the Muta™Mouse for in vitro screening of suspected environmental mutagens. This in vitro mammalian cell assay system has several noteworthy advantages: (1) significant reduction in the number of animals required for mutagen screening relative to in vivo studies; (2) the metabolic competence of primary hepatocytes and concomitant ability to metabolize and activate several types of promutagens; (3) nearly pure populations of fresh hepatocytes are relatively easy to obtain and culture; (4) the Muta™Mouse system for scoring lacZ mutations is well established and validated. In addition, the use of primary hepatocytes readily permits comparisons of the metabolism of chemicals across species, thus increasing the confidence of extrapolations from animals to humans [NRC, 2007].
Nevertheless, it should be noted that this work constitutes a pilot study, and follow-up work will be required to refine, validate, and optimize an assay based on cultured primary hepatocytes. There are numerous avenues for follow-up research. First, subsequent analyses should rigorously investigate the composition of the isolated cell population. To this end, biochemical tools, such as those described by Modriansky et al. [2000], could be employed to provide an enzymatic and proteomic profile of the isolated cells (e.g., total cytochrome P450 content, activity of selected P450 isozymes). As fibroblasts can proliferate in the absence of EGF, experiments conducted both in the presence and in the absence of EGF could permit an assessment of fibroblast contamination. Second, subsequent analyses should refine and optimize the assay protocol. For example, the magnitude and reproducibility of the response to selected mutagens could be assessed for cell populations derived from numerous animals, including very young animals (e.g., 14 days), as well as animals exposed to chemical inducers of liver enzymes (e.g., Aroclor). The former would be expected to maximize the proliferation potential of the isolated cells, and the latter would be expected to increase the metabolic capacity of the isolated cells. In addition, a larger culture surface could be employed to permit an increase in the number of exposed cells. Finally, subsequent analyses could employ established cytotoxicity-assessment tools (e.g., clonal survival) to reliably quantify effects that prevent or retard cell growth and proliferation.
In summary, we have developed and introduced an in vitro mutation bioassay based on cultured primary hepatocytes from the transgenic Muta™Mouse, and preliminary results indicate that the assay can be employed as a cost-effective complement to in vivo analyses for screening of environmental mutagens. The assay system can quantify mutations at the transgenic lacZ locus (this work), as well as the smaller cII locus. The latter can be more readily subjected to sequence analysis. Moreover, these endpoints can readily be combined with other genotoxicity end-points, including DNA strand breaks (i.e., comet), micronucleus formation, and unscheduled DNA synthesis.
Acknowledgments
The authors are grateful to Dr. Azam Tayabali, Alexandra Long, and two anonymous reviewers for their constructive comments.
Glossary
Abbreviations
- BaP
benzo[a]pyrene
- CSC
cigarette smoke condensate
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
- EGF
epidermal growth factor
- HBSS
Hank's balanced salt solution
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HGF
hepatocyte growth factor
- MF
mutant frequency
- 3-NBA
3-nitrobenzoanthrone
- pfu
plaque-forming units
- P-gal
phenyl-β-d-galactopyranoside
- PhIP
2-amino-1-methyl-6-phenyl-imidazo[4,5-b]pyridine
- TGR
transgenic rodent
- TPM
total particulate material
REFERENCES
- Arlt VM, Zhan L, Schmeiser HH, Honma M, Hayashi M, Phillips DH, Suzuki T. DNA adducts and mutagenic specificity of the ubiquitous environmental pollutant 3-nitrobenzanthrone in Muta™Mouse. Environ Mol Mutagen. 2004;43:186–195. doi: 10.1002/em.20014. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, Henderson CJ, Osborne MR, Bieler CA, Frei E, Martinek V, Sopko B, Wolf CR, Schmeiser HH, Phillips DH. Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H:quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Res. 2005;65:2644–2652. doi: 10.1158/0008-5472.CAN-04-3544. [DOI] [PubMed] [Google Scholar]
- Berndt-Weis ML, Kauri LM, Williams A, White P, Douglas G, Yauk C. Global transcriptional characterization of a mouse pulmonary epithelial cell line for use in genetic toxicology. Toxicol In Vitro. 2009;23:816–833. doi: 10.1016/j.tiv.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis AR, Lynch AM, Murray S, de la Torre R, Solans A, Farre M, Segura J, Gooderham NJ, Davies DS. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- Casciano DA. Development and utilization of primary hepatocyte culture systems to evaluate metabolism, DNA binding, and DNA repair of xenobiotics. Drug Metab Rev. 2000;32:1–13. doi: 10.1081/dmr-100100561. [DOI] [PubMed] [Google Scholar]
- Chen G, Bunce NJ. Polybrominated diphenyl ethers as Ah receptor agonists and antagonists. Toxicol Sci. 2003;76:310–320. doi: 10.1093/toxsci/kfg236. [DOI] [PubMed] [Google Scholar]
- Chen G, Gingerich J, Soper L, Douglas GR, White PA. Tissue-specific metabolic activation and mutagenicity of 3-nitrobenzanthrone in Muta™Mouse. Environ Mol Mutagen. 2008;49:602–613. doi: 10.1002/em.20410. [DOI] [PubMed] [Google Scholar]
- Clive D, Johnson KO, Spector JFS, Batson AG, Brown MMM. Validation and characterization of the L5178Y/TK+/− mouse lymphoma mutagen assay system. Mutat Res. 1979;59:61–108. doi: 10.1016/0027-5107(79)90195-7. [DOI] [PubMed] [Google Scholar]
- Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: A review. Mutat Res. 2004;567:447–474. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Douglas GR, Jiao J, Gingerich JD, Soper LM, Gossen JA. Temporal and molecular characteristics of lacZ mutations in somatic tissues of transgenic mice. Environ Mol Mutagen. 1996;28:317–324. doi: 10.1002/(SICI)1098-2280(1996)28:4<317::AID-EM4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Douglas GR, Gingerich JD, Soper LM, Potvin M, Bjarnason S. Evidence for the lack of base-change and small-deletion mutation induction by trichloroethylene in lacZ transgenic mice. Environ Mol Mutagen. 1999;34:190–194. [PubMed] [Google Scholar]
- Eckl PM, Raffelsberger I. The primary rat hepatocyte micronucleus assay: General features. Mutat Res. 1997;392:117–124. doi: 10.1016/s0165-1218(97)00050-5. [DOI] [PubMed] [Google Scholar]
- Erexson GL, Cunningham ML, Tindall KR. Cytogenetic characterization of the transgenic Big Blue Rat2 and Big Blue mouse embryonic fibroblast cell lines. Mutagen. 1998;13:649–653. doi: 10.1093/mutage/13.6.649. [DOI] [PubMed] [Google Scholar]
- Erexson GL, Watson DE, Tindall KR. Characterization of new transgenic Big Blue® mouse and rat primary fibroblast cell strains for use in molecular toxicology studies. Environ Mol Mutagen. 1999;34:90–96. [PubMed] [Google Scholar]
- Felton JS, Knize MG, Dolbeare FA, Wu R. Mutagenic activity of heterocyclic amines in cooked foods. Environ Health Perspect. 1994;102(6) Suppl:201–204. doi: 10.1289/ehp.94102s6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen JA, de Leeuw WJ, Tan CH, Zwarthoff EC, Berends F, Lohman PH, Knook DL, Vijg J. Efficient rescue of integrated shuttle vectors from transgenic mice: A model for studying mutations in vivo. Proc Natl Acad Sci USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen JA, Molijn AC, Douglas GR, Vijg J. Application of galactose-sensitive E. coli strains as selective hosts for LacZ-plasmids. Nucleic Acids Res. 1992;20:3254. doi: 10.1093/nar/20.12.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakura A, Tsutsui Y, Sonoda J, Kai J, Imade T, Shimada M, Sugihara Y, Mikami T. Comparison between in vivo mutagenicity and carcinogenicity in multiple organs by benzo[a]pyrene in the lacZ transgenic mouse (Muta™Mouse) Mutat Res. 1998;398:123–130. doi: 10.1016/s0027-5107(97)00248-0. [DOI] [PubMed] [Google Scholar]
- Hakura A, Tsutsui Y, Sonoda J, Mikami T, Tsukidate K, Sagami F, Kerns WD. Multiple organ mutation in the lacZ transgenic mouse (Muta™mouse) 6 months after oral treatment (5 days) with benzo[a]pyrene. Mutat Res. 1999;426:71–77. doi: 10.1016/s0027-5107(99)00046-9. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddle JA, Dean S, Nohmi T, Boerrigter M, Casciano D, Douglas GR, Glickman BW, Gorelick NJ, Mirsalis JC, Martus HJ, Skopek TR, Thybaud V, Tindall KR, Yajima N. In vivo transgenic mutation assays. Environ Mol Mutagen. 2000;35:253–259. doi: 10.1002/(sici)1098-2280(2000)35:3<253::aid-em11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- IARC. Lyon, France: International Agency for Research on Cancer; 1993. IARC Monographs on the Evaluation of Carcinogenic Risks to Human, Vol. 56: Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; pp. 229–242. [Google Scholar]
- Ichihara A, Nakamura T, Tanaka K. Use of hepatocytes in primary culture for biochemical studies on liver function. Mol Cell Biochem. 1982;43:145–160. doi: 10.1007/BF00223006. [DOI] [PubMed] [Google Scholar]
- Jacobsen NR, Saber AT, White P, Møller P, Pojana G, Vogel U, Loft S, Gingerich J, Soper L, Douglas GR, Wallin H. Increased mutant frequency by carbon black, but not quartz, in the lacZ and cII transgenes of muta mouse lung epithelial cells. Environ Mol Mutagen. 2007;48:451–461. doi: 10.1002/em.20300. [DOI] [PubMed] [Google Scholar]
- Jacobsen NR, Pojana G, White P, Møller P, Cohn CA, Korsholm KS, Vogel U, Marcomini A, Loft S, Wallin H. Genotoxicity, cytotoxicity, and reactive oxygen species induced by single-walled carbon nanotubes and C(60) fullerenes in the FE1- Muta™Mouse lung epithelial cells. Environ Mol Mutagen. 2008a;49:476–487. doi: 10.1002/em.20406. [DOI] [PubMed] [Google Scholar]
- Jacobsen NR, Møller P, Cohn CA, Loft S, Vogel U, Wallin H. Diesel exhaust particles are mutagenic in FE1-Muta™Mouse lung epithelial cells. Mutat Res. 2008b;641:54–57. doi: 10.1016/j.mrfmmm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Jongen WM, Hakkert BC, van der Hoeven JC. Genotoxicity testing of cigarette-smoke condensate in the SCE, Hgprt assays with V79 Chinese hamster cells. Food Chem Toxicol. 1985;23:603–607. doi: 10.1016/0278-6915(85)90186-3. [DOI] [PubMed] [Google Scholar]
- Klein JC, Beems RB, Zwart PE, Hamzink M, Zomer G, van Steeg H, van Kreijl CF. Intestinal toxicity and carcinogenic potential of the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in DNA repair deficient XPA−/− mice. Carcinogenesis. 2001;22:619–626. doi: 10.1093/carcin/22.4.619. [DOI] [PubMed] [Google Scholar]
- Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G, Garganta F, Vrieling H, Scherer G. Spontaneous and chemically induced point mutations in Hprt cDNA of the metabolically competent human lymphoblastoid cell line, MCL-5. Mutat Res. 1999;431:417–428. doi: 10.1016/s0027-5107(99)00183-9. [DOI] [PubMed] [Google Scholar]
- Kreamer BL, Staecker JL, Sawada N, Sattler GL, Hsia MT, Pitot HC. Use of a low-speed, iso-density Percoll centrifugation to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell Dev Biol. 1986;22:201–211. doi: 10.1007/BF02623304. [DOI] [PubMed] [Google Scholar]
- Kruglov EA, Jain D, Dranoff JA. Isolation of primary rat liver fibroblasts. J Invest Med. 2002;50:179–84. doi: 10.2310/6650.2002.33431. [DOI] [PubMed] [Google Scholar]
- Lambert IB, Singer TM, Boucher SE, Douglas GR. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lynch AM, Gooderham NJ, Boobis AR. Organ distinctive mutagenicity in Muta™Mouse after short-term exposure to PhIP. Mutagenesis. 1996;11:505–509. doi: 10.1093/mutage/11.5.505. [DOI] [PubMed] [Google Scholar]
- Masumura K, Matsui K, Yamada M, Horiguchi M, Ishida K, Watanabe M, Ueda O, Suzuki H, Kanke Y, Tindall KR, Wakabayashi K, Sofuni T, Nohmi T. Mutagenicity of 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine (PhIP) in the new gpt delta transgenic mouse. Cancer Lett. 1999;143:241–244. doi: 10.1016/s0304-3835(99)00132-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Nakamura T. Hepatocyte growth factor: Molecular structure and implications for a central role in liver regeneration. J Gastroenterol Hepatol. 1991;6:509–519. doi: 10.1111/j.1440-1746.1991.tb00897.x. [DOI] [PubMed] [Google Scholar]
- McDiarmid HM, Douglas GR, Coomber BL, Josephy PD. Epithelial and fibroblast cell lines cultured from the transgenic Big Blue rat: An in vitro mutagenesis assay. Mutat Res. 2001;497:39–47. doi: 10.1016/s1383-5718(01)00245-5. [DOI] [PubMed] [Google Scholar]
- McDiarmid HM, Douglas GR, Coomber BL, Josephy PD. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-induced mutagenesis in cultured Big Blue rat mammary epithelial and fibroblast cells. Environ Mol Mutagen. 2002;39:245–253. doi: 10.1002/em.10059. [DOI] [PubMed] [Google Scholar]
- Modriansky M, Ulrichova J, Bachleda P, Anzenbacher P, Anzenbacherova E, Walterova D, Simanek V. Human hepatocyte—A model for toxicological studies, functional and biochemical characterization. Gen Physiol Biophys. 2000;19:223–235. [PubMed] [Google Scholar]
- Moir D, Rickert WS, Levasseur G, Larose Y, Maertens R, White P, Desjardins S. A comparison of mainstream and sidestream marijuana and tobacco cigarette smoke produced under two machine smoking conditions. Chem Res Toxicol. 2008;21:494–502. doi: 10.1021/tx700275p. [DOI] [PubMed] [Google Scholar]
- Müller-Tegethoff K, Kersten B, Kasper P, Müller L. Application of the in vitro rat hepatocyte micronucleus assay in genetic toxicology testing. Mutat Res. 1997;392:125–138. doi: 10.1016/s0165-1218(97)00051-7. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Ichihara A. Control of growth and expression of differentiated functions of mature hepatocytes in primary culture. Cell Struct Funct. 1985;10:1–16. doi: 10.1247/csf.10.1. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC) Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academy Press; 2007. [Google Scholar]
- Nohmi T, Suzuki T, Masumura K. Recent advances in the protocols of transgenic mouse mutation assays. Mutat Res. 2000;455:191–215. doi: 10.1016/s0027-5107(00)00077-4. [DOI] [PubMed] [Google Scholar]
- Papp-Szabó E, Douglas GR, Coomber BL, Josephy PD. Mutagenicity of the oral carcinogen 4-nitroquinoline-1-oxide in cultured BigBlue rat tongue epithelial cells and fibroblasts. Mutat Res. 2003;522:107–117. doi: 10.1016/s0027-5107(02)00285-3. [DOI] [PubMed] [Google Scholar]
- Seglen PO. Preparation of isolated rat liver cells. Method Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- Tateno C, Takai-Kajihara K, Yamasaki C, Sato H, Yoshizato K. Heterogeneity of growth potential of adult rat hepatocytes in vitro. Hepatology. 2000;31:65–74. doi: 10.1002/hep.510310113. [DOI] [PubMed] [Google Scholar]
- Thybaud V, Dean S, Nohmi T, de Boer J, Douglas GR, Glickman BW, Gorelick NJ, Heddle JA, Heflich RH, Lambert IB, Martus HJ, Mirsalis JC, Suzuki T, Yajima N. In vivo transgenic mutation assays. Mutat Res. 2003;540:141–151. doi: 10.1016/j.mrgentox.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Ulrich RG, Bacon JA, Cramer CT, Peng GW, Petrella DK, Stryd RP, Sun EL. Cultured hepatocytes as investigational models for hepatic toxicity: Practical applications in drug discovery and development. Toxicol Lett. 1995;82/83:107–115. doi: 10.1016/0378-4274(95)03547-8. [DOI] [PubMed] [Google Scholar]
- Vijg J, Douglas G. Bacteriophage lambda and plasmid lacZ transgenic mice for studying mutations in vivo. In: Pfeifer G, editor. Technologies for Detection of DNA Damage and Mutations. New York: Plenum Press; 1996. pp. 391–410. [Google Scholar]
- Watanabe N, Okochi E, Hirayama Y, Shimada Y, Yanagihara K, Yoshida MC, Takahashi S, Mochizuki M, Sugimura T, Nagao M, Ushijima T. Single nucleotide instability without microsatellite instability in rat mammary carcinomas. Cancer Res. 2001;61:2632–2640. [PubMed] [Google Scholar]
- White PA, Douglas GR, Gingerich J, Parfett C, Shwed P, Seligy V, Soper L, Berndt L, Bayley J, Wagner S, Pound K, Blakey D. Development and characterization of a stable epithelial cell line from Muta™Mouse lung. Environ Mol Mutagen. 2003;42:166–184. doi: 10.1002/em.10185. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line HepG2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]


