Abstract
In the title compound, C28H24N2O4, the dihedral angle between the two rings of the biphenyl unit is 75.34 (9)°. The outer aromatic rings form dihedral angles of 66.96 (1) and 85.69 (8)° with the rings to which they are attached . The molecular structure is stabilized by intramolecular C—H⋯O and N—H⋯O hydrogen bonds. In the crystal structure, intermolecular N—H⋯O interactions are observed.
Related literature
For the synthesis, see: Gao & Gao (2002 ▶). For related structures, see: Wang & Han (2004 ▶); Wang & Jiang (2004 ▶); Huang & Yang (2008 ▶).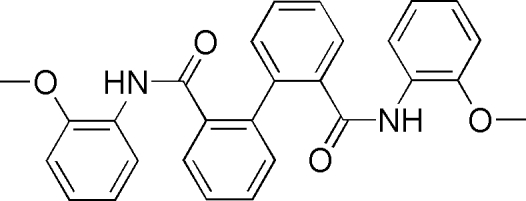
Experimental
Crystal data
C28H24N2O4
M r = 452.49
Monoclinic,

a = 18.184 (4) Å
b = 16.304 (3) Å
c = 7.9998 (16) Å
β = 108.90 (3)°
V = 2243.9 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 113 (2) K
0.16 × 0.14 × 0.10 mm
Data collection
Rigaku Saturn CCD diffractometer
Absorption correction: multi-scan (CrystalStructure; Rigaku/MSC, 2004 ▶) T min = 0.986, T max = 0.991
6422 measured reflections
1991 independent reflections
1858 reflections with I > 2σ(I)
R int = 0.054
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.093
S = 1.06
1991 reflections
309 parameters
2 restraints
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.20 e Å−3
Data collection: RAPID-AUTO (Rigaku/MSC, 2004 ▶); cell refinement: RAPID-AUTO; data reduction: RAPID-AUTO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808032352/gw2044sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808032352/gw2044Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O3i | 0.86 | 2.02 | 2.833 (3) | 157 |
| N2—H2N⋯O2 | 0.86 | 2.24 | 3.081 (4) | 167 |
| N2—H2N⋯O4 | 0.86 | 2.24 | 2.612 (3) | 106 |
| C22—H22⋯O3 | 0.93 | 2.30 | 2.885 (4) | 120 |
Symmetry code: (i)  .
.
Acknowledgments
The authors thank the Scientific Research Fund Projects of China West Normal University (grant No. 06B003) and the Youth Fund Projects of Sichuan Education Department (grant No. 2006B039).
supplementary crystallographic information
Comment
The distortion of diphenyl spacer about central bond not only endows dpa a peculiar characterization to link metal ions or metal clusters into macrocycles or helical chains, but also makes diphenic acid (H2dpa) can deprotonate partially forming hydrogen bonds of carboxylic groups to meet both geometric and energetic requirements. We here report the crystal structure of the title compound.
The C8—C13 and C14—C19 planes form the dihedral angle of 75.34 (9)°, and C1—C6 ring are nearly perpendicular to C14—C19 ring, with a dihedral angle of 85.69 (8)°.The molecular structure is stabilized by C—H···O and N—H···O intramolecular hydrogen bonds.In addition, weak C—H···O intermolecular hydrogen bonds are observed.
Experimental
The title compound was prepared according to the reported procedure of M. Z. Gao & Gao (2002). Colourless single crystals suitable for X-ray diffraction were obtained by recrystallization from dimethyl sulfoxide.
Refinement
H atoms were placed in calculated positions with C—H = 0.93 Å, and N—H = 0.86 Å,and refined in riding mode with Uiso(H) = 1.2Ueq(C,N).
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% probability displacement ellipsoids and the atomic numbering.
Crystal data
| C28H24N2O4 | F(000) = 952 |
| Mr = 452.49 | Dx = 1.339 Mg m−3 |
| Monoclinic, Cc | Mo Kα radiation, λ = 0.71073 Å |
| a = 18.184 (4) Å | Cell parameters from 3190 reflections |
| b = 16.304 (3) Å | θ = 1.7–27.9° |
| c = 7.9998 (16) Å | µ = 0.09 mm−1 |
| β = 108.90 (3)° | T = 113 K |
| V = 2243.9 (8) Å3 | Block, colourless |
| Z = 4 | 0.16 × 0.14 × 0.10 mm |
Data collection
| Rigaku Saturn diffractometer | 1991 independent reflections |
| Radiation source: rotating anode | 1858 reflections with I > 2σ(I) |
| confocal | Rint = 0.054 |
| ω scans | θmax = 25.0°, θmin = 1.7° |
| Absorption correction: multi-scan (CrystalStructure; Rigaku/MSC, 2004) | h = −21→20 |
| Tmin = 0.986, Tmax = 0.991 | k = −17→19 |
| 6422 measured reflections | l = −9→9 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.093 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0537P)2] where P = (Fo2 + 2Fc2)/3 |
| 1991 reflections | (Δ/σ)max = 0.007 |
| 309 parameters | Δρmax = 0.17 e Å−3 |
| 2 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −0.01240 (15) | 0.26417 (14) | 0.9137 (3) | 0.0338 (6) | |
| O2 | 0.21854 (13) | 0.17729 (12) | 1.0081 (2) | 0.0234 (5) | |
| O3 | 0.45976 (12) | 0.32159 (13) | 1.0239 (2) | 0.0239 (5) | |
| O4 | 0.31213 (13) | 0.07137 (13) | 0.8380 (3) | 0.0273 (5) | |
| N1 | 0.09211 (15) | 0.17216 (15) | 0.8317 (3) | 0.0222 (6) | |
| H1N | 0.0615 | 0.1801 | 0.7258 | 0.027* | |
| N2 | 0.37809 (15) | 0.21050 (14) | 0.9645 (3) | 0.0216 (6) | |
| H2N | 0.3350 | 0.1932 | 0.9743 | 0.026* | |
| C1 | 0.00360 (19) | 0.19053 (18) | 0.9988 (4) | 0.0229 (7) | |
| C2 | −0.0328 (2) | 0.16090 (19) | 1.1154 (4) | 0.0277 (7) | |
| H2 | −0.0694 | 0.1928 | 1.1441 | 0.033* | |
| C3 | −0.0139 (2) | 0.0831 (2) | 1.1889 (4) | 0.0298 (8) | |
| H3 | −0.0378 | 0.0632 | 1.2674 | 0.036* | |
| C4 | 0.0399 (2) | 0.0355 (2) | 1.1457 (4) | 0.0305 (8) | |
| H4 | 0.0512 | −0.0170 | 1.1926 | 0.037* | |
| C5 | 0.07701 (19) | 0.06580 (18) | 1.0327 (3) | 0.0258 (7) | |
| H5 | 0.1148 | 0.0345 | 1.0072 | 0.031* | |
| C6 | 0.05817 (18) | 0.14248 (18) | 0.9576 (3) | 0.0210 (7) | |
| C7 | 0.16777 (18) | 0.18877 (17) | 0.8643 (3) | 0.0188 (6) | |
| C8 | 0.18756 (18) | 0.22120 (18) | 0.7085 (3) | 0.0201 (7) | |
| C9 | 0.16253 (18) | 0.17982 (18) | 0.5469 (4) | 0.0219 (7) | |
| H9 | 0.1304 | 0.1342 | 0.5336 | 0.026* | |
| C10 | 0.18522 (19) | 0.20637 (19) | 0.4066 (3) | 0.0251 (7) | |
| H10 | 0.1695 | 0.1779 | 0.3000 | 0.030* | |
| C11 | 0.2317 (2) | 0.27591 (19) | 0.4260 (4) | 0.0275 (7) | |
| H11 | 0.2471 | 0.2940 | 0.3321 | 0.033* | |
| C12 | 0.25501 (19) | 0.31821 (19) | 0.5843 (4) | 0.0252 (7) | |
| H12 | 0.2853 | 0.3651 | 0.5956 | 0.030* | |
| C13 | 0.23333 (17) | 0.29096 (18) | 0.7281 (3) | 0.0190 (6) | |
| C14 | 0.25474 (18) | 0.34128 (17) | 0.8950 (3) | 0.0204 (6) | |
| C15 | 0.1978 (2) | 0.39160 (18) | 0.9223 (4) | 0.0270 (7) | |
| H15 | 0.1475 | 0.3901 | 0.8428 | 0.032* | |
| C16 | 0.2152 (2) | 0.4441 (2) | 1.0670 (4) | 0.0307 (8) | |
| H16 | 0.1763 | 0.4766 | 1.0847 | 0.037* | |
| C17 | 0.2897 (2) | 0.44812 (19) | 1.1841 (4) | 0.0290 (8) | |
| H17 | 0.3015 | 0.4837 | 1.2800 | 0.035* | |
| C18 | 0.34668 (19) | 0.39879 (18) | 1.1576 (4) | 0.0244 (7) | |
| H18 | 0.3972 | 0.4021 | 1.2355 | 0.029* | |
| C19 | 0.32979 (19) | 0.34399 (18) | 1.0156 (3) | 0.0211 (6) | |
| C20 | 0.39544 (18) | 0.29130 (17) | 1.0011 (3) | 0.0210 (7) | |
| C21 | 0.42452 (18) | 0.15199 (18) | 0.9115 (3) | 0.0212 (7) | |
| C22 | 0.5013 (2) | 0.1627 (2) | 0.9232 (4) | 0.0317 (8) | |
| H22 | 0.5276 | 0.2099 | 0.9756 | 0.038* | |
| C23 | 0.5398 (2) | 0.1035 (2) | 0.8573 (5) | 0.0407 (9) | |
| H23 | 0.5918 | 0.1111 | 0.8674 | 0.049* | |
| C24 | 0.5015 (2) | 0.0338 (2) | 0.7773 (4) | 0.0354 (8) | |
| H24 | 0.5271 | −0.0048 | 0.7305 | 0.042* | |
| C25 | 0.4250 (2) | 0.0217 (2) | 0.7669 (4) | 0.0275 (8) | |
| H25 | 0.3988 | −0.0252 | 0.7127 | 0.033* | |
| C26 | 0.38714 (18) | 0.07931 (18) | 0.8372 (3) | 0.0225 (7) | |
| C28 | 0.2734 (2) | −0.0051 (2) | 0.7829 (5) | 0.0366 (9) | |
| H28A | 0.2719 | −0.0168 | 0.6642 | 0.055* | |
| H28B | 0.2214 | −0.0019 | 0.7873 | 0.055* | |
| H28C | 0.3011 | −0.0480 | 0.8601 | 0.055* | |
| C27 | −0.0779 (3) | 0.3085 (2) | 0.9290 (5) | 0.0458 (10) | |
| H27A | −0.1234 | 0.2746 | 0.8892 | 0.069* | |
| H27B | −0.0856 | 0.3571 | 0.8578 | 0.069* | |
| H27C | −0.0684 | 0.3234 | 1.0502 | 0.069* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0436 (16) | 0.0284 (13) | 0.0359 (11) | 0.0087 (11) | 0.0218 (11) | 0.0078 (9) |
| O2 | 0.0200 (12) | 0.0303 (12) | 0.0181 (10) | −0.0008 (9) | 0.0038 (9) | 0.0020 (8) |
| O3 | 0.0182 (13) | 0.0273 (12) | 0.0252 (10) | −0.0060 (9) | 0.0054 (10) | −0.0002 (8) |
| O4 | 0.0194 (12) | 0.0223 (12) | 0.0408 (11) | −0.0041 (9) | 0.0104 (10) | −0.0049 (9) |
| N1 | 0.0164 (14) | 0.0333 (15) | 0.0163 (11) | −0.0017 (11) | 0.0045 (11) | 0.0036 (9) |
| N2 | 0.0172 (14) | 0.0205 (14) | 0.0279 (13) | 0.0000 (10) | 0.0084 (11) | −0.0017 (10) |
| C1 | 0.0267 (18) | 0.0191 (15) | 0.0228 (14) | −0.0017 (13) | 0.0076 (13) | 0.0001 (11) |
| C2 | 0.030 (2) | 0.0306 (19) | 0.0255 (14) | 0.0003 (14) | 0.0135 (15) | −0.0028 (12) |
| C3 | 0.039 (2) | 0.0303 (19) | 0.0251 (14) | −0.0089 (15) | 0.0176 (16) | 0.0009 (12) |
| C4 | 0.042 (2) | 0.0234 (18) | 0.0272 (16) | 0.0002 (14) | 0.0133 (16) | 0.0033 (12) |
| C5 | 0.030 (2) | 0.0248 (17) | 0.0225 (14) | 0.0023 (14) | 0.0082 (14) | −0.0017 (12) |
| C6 | 0.0203 (17) | 0.0250 (17) | 0.0163 (12) | −0.0035 (12) | 0.0042 (13) | −0.0008 (11) |
| C7 | 0.0183 (17) | 0.0183 (15) | 0.0201 (14) | 0.0011 (12) | 0.0066 (14) | −0.0023 (11) |
| C8 | 0.0134 (16) | 0.0294 (18) | 0.0170 (12) | 0.0040 (12) | 0.0045 (12) | 0.0016 (11) |
| C9 | 0.0174 (17) | 0.0245 (17) | 0.0219 (14) | −0.0004 (13) | 0.0036 (13) | −0.0011 (11) |
| C10 | 0.0213 (19) | 0.0347 (18) | 0.0182 (13) | 0.0025 (14) | 0.0050 (13) | −0.0018 (11) |
| C11 | 0.028 (2) | 0.0348 (19) | 0.0219 (14) | 0.0019 (14) | 0.0117 (14) | 0.0081 (12) |
| C12 | 0.0236 (19) | 0.0249 (17) | 0.0278 (15) | −0.0002 (13) | 0.0094 (14) | 0.0035 (12) |
| C13 | 0.0123 (16) | 0.0232 (16) | 0.0209 (13) | 0.0025 (11) | 0.0045 (12) | 0.0027 (11) |
| C14 | 0.0205 (17) | 0.0175 (15) | 0.0238 (14) | −0.0011 (12) | 0.0079 (13) | 0.0020 (11) |
| C15 | 0.0231 (18) | 0.0254 (17) | 0.0305 (15) | 0.0006 (14) | 0.0057 (14) | 0.0010 (12) |
| C16 | 0.032 (2) | 0.0278 (19) | 0.0365 (17) | 0.0041 (14) | 0.0167 (17) | −0.0022 (13) |
| C17 | 0.037 (2) | 0.0222 (17) | 0.0277 (15) | −0.0017 (14) | 0.0105 (15) | −0.0026 (12) |
| C18 | 0.0267 (19) | 0.0206 (17) | 0.0239 (14) | −0.0020 (13) | 0.0054 (13) | −0.0010 (11) |
| C19 | 0.0221 (17) | 0.0192 (16) | 0.0234 (14) | 0.0007 (12) | 0.0091 (14) | 0.0030 (11) |
| C20 | 0.0223 (19) | 0.0250 (17) | 0.0143 (13) | −0.0018 (13) | 0.0041 (13) | 0.0012 (10) |
| C21 | 0.0207 (18) | 0.0212 (16) | 0.0222 (14) | 0.0009 (12) | 0.0076 (14) | −0.0005 (11) |
| C22 | 0.0246 (19) | 0.0289 (18) | 0.0423 (17) | −0.0024 (14) | 0.0118 (16) | −0.0068 (14) |
| C23 | 0.0207 (19) | 0.042 (2) | 0.063 (2) | −0.0016 (15) | 0.0182 (18) | −0.0110 (17) |
| C24 | 0.034 (2) | 0.029 (2) | 0.0465 (19) | 0.0069 (15) | 0.0177 (17) | −0.0012 (14) |
| C25 | 0.031 (2) | 0.0201 (17) | 0.0321 (16) | −0.0004 (13) | 0.0110 (15) | −0.0015 (12) |
| C26 | 0.0204 (19) | 0.0219 (16) | 0.0241 (14) | −0.0011 (12) | 0.0059 (13) | 0.0019 (11) |
| C28 | 0.033 (2) | 0.031 (2) | 0.0477 (19) | −0.0132 (15) | 0.0152 (17) | −0.0088 (14) |
| C27 | 0.062 (3) | 0.037 (2) | 0.049 (2) | 0.0233 (19) | 0.032 (2) | 0.0113 (16) |
Geometric parameters (Å, °)
| O1—C1 | 1.364 (4) | C12—C13 | 1.403 (4) |
| O1—C27 | 1.431 (4) | C12—H12 | 0.9300 |
| O2—C7 | 1.234 (3) | C13—C14 | 1.507 (4) |
| O3—C20 | 1.228 (4) | C14—C19 | 1.394 (4) |
| O4—C26 | 1.372 (4) | C14—C15 | 1.393 (4) |
| O4—C28 | 1.429 (4) | C15—C16 | 1.390 (4) |
| N1—C7 | 1.342 (4) | C15—H15 | 0.9300 |
| N1—C6 | 1.425 (4) | C16—C17 | 1.377 (5) |
| N1—H1N | 0.8600 | C16—H16 | 0.9300 |
| N2—C20 | 1.364 (4) | C17—C18 | 1.381 (5) |
| N2—C21 | 1.426 (4) | C17—H17 | 0.9300 |
| N2—H2N | 0.8600 | C18—C19 | 1.398 (4) |
| C1—C6 | 1.385 (5) | C18—H18 | 0.9300 |
| C1—C2 | 1.393 (4) | C19—C20 | 1.506 (4) |
| C2—C3 | 1.393 (5) | C21—C22 | 1.380 (5) |
| C2—H2 | 0.9300 | C21—C26 | 1.399 (4) |
| C3—C4 | 1.377 (5) | C22—C23 | 1.393 (5) |
| C3—H3 | 0.9300 | C22—H22 | 0.9300 |
| C4—C5 | 1.383 (5) | C23—C24 | 1.377 (5) |
| C4—H4 | 0.9300 | C23—H23 | 0.9300 |
| C5—C6 | 1.381 (4) | C24—C25 | 1.381 (5) |
| C5—H5 | 0.9300 | C24—H24 | 0.9300 |
| C7—C8 | 1.501 (4) | C25—C26 | 1.387 (4) |
| C8—C13 | 1.388 (4) | C25—H25 | 0.9300 |
| C8—C9 | 1.397 (4) | C28—H28A | 0.9600 |
| C9—C10 | 1.385 (4) | C28—H28B | 0.9600 |
| C9—H9 | 0.9300 | C28—H28C | 0.9600 |
| C10—C11 | 1.392 (5) | C27—H27A | 0.9600 |
| C10—H10 | 0.9300 | C27—H27B | 0.9600 |
| C11—C12 | 1.383 (4) | C27—H27C | 0.9600 |
| C11—H11 | 0.9300 | ||
| C1—O1—C27 | 116.8 (3) | C15—C14—C13 | 117.9 (3) |
| C26—O4—C28 | 118.1 (2) | C16—C15—C14 | 120.8 (3) |
| C7—N1—C6 | 125.6 (2) | C16—C15—H15 | 119.6 |
| C7—N1—H1N | 117.2 | C14—C15—H15 | 119.6 |
| C6—N1—H1N | 117.2 | C17—C16—C15 | 120.3 (3) |
| C20—N2—C21 | 126.3 (3) | C17—C16—H16 | 119.9 |
| C20—N2—H2N | 116.9 | C15—C16—H16 | 119.9 |
| C21—N2—H2N | 116.9 | C16—C17—C18 | 119.3 (3) |
| O1—C1—C6 | 115.6 (3) | C16—C17—H17 | 120.3 |
| O1—C1—C2 | 124.6 (3) | C18—C17—H17 | 120.3 |
| C6—C1—C2 | 119.7 (3) | C17—C18—C19 | 121.2 (3) |
| C1—C2—C3 | 119.5 (3) | C17—C18—H18 | 119.4 |
| C1—C2—H2 | 120.2 | C19—C18—H18 | 119.4 |
| C3—C2—H2 | 120.2 | C14—C19—C18 | 119.4 (3) |
| C4—C3—C2 | 120.3 (3) | C14—C19—C20 | 123.5 (3) |
| C4—C3—H3 | 119.8 | C18—C19—C20 | 117.1 (3) |
| C2—C3—H3 | 119.8 | O3—C20—N2 | 124.3 (3) |
| C3—C4—C5 | 120.0 (3) | O3—C20—C19 | 120.0 (3) |
| C3—C4—H4 | 120.0 | N2—C20—C19 | 115.7 (3) |
| C5—C4—H4 | 120.0 | C22—C21—C26 | 118.6 (3) |
| C6—C5—C4 | 120.2 (3) | C22—C21—N2 | 125.3 (3) |
| C6—C5—H5 | 119.9 | C26—C21—N2 | 116.1 (3) |
| C4—C5—H5 | 119.9 | C21—C22—C23 | 120.5 (3) |
| C5—C6—C1 | 120.3 (3) | C21—C22—H22 | 119.7 |
| C5—C6—N1 | 120.8 (3) | C23—C22—H22 | 119.7 |
| C1—C6—N1 | 118.9 (3) | C24—C23—C22 | 120.5 (3) |
| O2—C7—N1 | 124.1 (3) | C24—C23—H23 | 119.7 |
| O2—C7—C8 | 121.3 (3) | C22—C23—H23 | 119.7 |
| N1—C7—C8 | 114.6 (2) | C23—C24—C25 | 119.6 (3) |
| C13—C8—C9 | 120.3 (3) | C23—C24—H24 | 120.2 |
| C13—C8—C7 | 119.4 (2) | C25—C24—H24 | 120.2 |
| C9—C8—C7 | 120.3 (3) | C24—C25—C26 | 120.1 (3) |
| C10—C9—C8 | 120.4 (3) | C24—C25—H25 | 120.0 |
| C10—C9—H9 | 119.8 | C26—C25—H25 | 120.0 |
| C8—C9—H9 | 119.8 | O4—C26—C25 | 124.3 (3) |
| C9—C10—C11 | 119.6 (3) | O4—C26—C21 | 115.1 (3) |
| C9—C10—H10 | 120.2 | C25—C26—C21 | 120.6 (3) |
| C11—C10—H10 | 120.2 | O4—C28—H28A | 109.5 |
| C12—C11—C10 | 120.2 (3) | O4—C28—H28B | 109.5 |
| C12—C11—H11 | 119.9 | H28A—C28—H28B | 109.5 |
| C10—C11—H11 | 119.9 | O4—C28—H28C | 109.5 |
| C11—C12—C13 | 120.6 (3) | H28A—C28—H28C | 109.5 |
| C11—C12—H12 | 119.7 | H28B—C28—H28C | 109.5 |
| C13—C12—H12 | 119.7 | O1—C27—H27A | 109.5 |
| C8—C13—C12 | 119.0 (3) | O1—C27—H27B | 109.5 |
| C8—C13—C14 | 121.3 (2) | H27A—C27—H27B | 109.5 |
| C12—C13—C14 | 119.6 (3) | O1—C27—H27C | 109.5 |
| C19—C14—C15 | 118.9 (3) | H27A—C27—H27C | 109.5 |
| C19—C14—C13 | 123.0 (3) | H27B—C27—H27C | 109.5 |
| C27—O1—C1—C6 | 169.2 (3) | C8—C13—C14—C15 | −75.3 (4) |
| C27—O1—C1—C2 | −8.3 (4) | C12—C13—C14—C15 | 99.9 (3) |
| O1—C1—C2—C3 | 177.3 (3) | C19—C14—C15—C16 | 0.3 (4) |
| C6—C1—C2—C3 | −0.2 (4) | C13—C14—C15—C16 | −175.3 (3) |
| C1—C2—C3—C4 | −0.4 (5) | C14—C15—C16—C17 | 1.2 (5) |
| C2—C3—C4—C5 | 1.8 (5) | C15—C16—C17—C18 | −0.8 (5) |
| C3—C4—C5—C6 | −2.5 (4) | C16—C17—C18—C19 | −1.0 (5) |
| C4—C5—C6—C1 | 1.8 (4) | C15—C14—C19—C18 | −2.0 (4) |
| C4—C5—C6—N1 | −175.7 (3) | C13—C14—C19—C18 | 173.3 (3) |
| O1—C1—C6—C5 | −178.2 (3) | C15—C14—C19—C20 | 178.8 (3) |
| C2—C1—C6—C5 | −0.5 (4) | C13—C14—C19—C20 | −5.9 (4) |
| O1—C1—C6—N1 | −0.6 (4) | C17—C18—C19—C14 | 2.4 (4) |
| C2—C1—C6—N1 | 177.1 (3) | C17—C18—C19—C20 | −178.4 (3) |
| C7—N1—C6—C5 | −65.1 (4) | C21—N2—C20—O3 | −13.6 (4) |
| C7—N1—C6—C1 | 117.3 (3) | C21—N2—C20—C19 | 167.3 (2) |
| C6—N1—C7—O2 | 2.9 (5) | C14—C19—C20—O3 | 135.1 (3) |
| C6—N1—C7—C8 | −178.7 (3) | C18—C19—C20—O3 | −44.0 (4) |
| O2—C7—C8—C13 | −50.2 (4) | C14—C19—C20—N2 | −45.7 (4) |
| N1—C7—C8—C13 | 131.3 (3) | C18—C19—C20—N2 | 135.1 (3) |
| O2—C7—C8—C9 | 127.4 (3) | C20—N2—C21—C22 | 14.1 (4) |
| N1—C7—C8—C9 | −51.1 (4) | C20—N2—C21—C26 | −163.9 (2) |
| C13—C8—C9—C10 | 2.1 (5) | C26—C21—C22—C23 | 2.0 (5) |
| C7—C8—C9—C10 | −175.5 (3) | N2—C21—C22—C23 | −175.9 (3) |
| C8—C9—C10—C11 | −1.6 (5) | C21—C22—C23—C24 | 0.9 (5) |
| C9—C10—C11—C12 | 0.0 (5) | C22—C23—C24—C25 | −1.8 (6) |
| C10—C11—C12—C13 | 1.1 (5) | C23—C24—C25—C26 | −0.2 (5) |
| C9—C8—C13—C12 | −1.1 (4) | C28—O4—C26—C25 | 8.0 (4) |
| C7—C8—C13—C12 | 176.6 (3) | C28—O4—C26—C21 | −173.0 (3) |
| C9—C8—C13—C14 | 174.1 (3) | C24—C25—C26—O4 | −177.9 (3) |
| C7—C8—C13—C14 | −8.2 (4) | C24—C25—C26—C21 | 3.1 (4) |
| C11—C12—C13—C8 | −0.5 (5) | C22—C21—C26—O4 | 176.9 (3) |
| C11—C12—C13—C14 | −175.8 (3) | N2—C21—C26—O4 | −4.9 (3) |
| C8—C13—C14—C19 | 109.4 (3) | C22—C21—C26—C25 | −4.0 (4) |
| C12—C13—C14—C19 | −75.4 (4) | N2—C21—C26—C25 | 174.2 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O3i | 0.86 | 2.02 | 2.833 (3) | 157 |
| N2—H2N···O2 | 0.86 | 2.24 | 3.081 (4) | 167 |
| N2—H2N···O4 | 0.86 | 2.24 | 2.612 (3) | 106 |
| C22—H22···O3 | 0.93 | 2.30 | 2.885 (4) | 120 |
Symmetry codes: (i) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GW2044).
References
- Gao, M. Z. & Gao, J. (2002). Tetrahedron Lett.43, 5001–5003.
- Huang, W.-W. & Yang, S.-P. (2008). Acta Cryst. E64, m525–m526. [DOI] [PMC free article] [PubMed]
- Rigaku/MSC (2004). RAPID-AUTO and CrystalStructure Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, R. H. & Han, L. (2004). J. Mol. Struct.694, 79–83.
- Wang, R. H. & Jiang, F. L. (2004). J. Mol. Struct.699, 79–84.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808032352/gw2044sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808032352/gw2044Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



