Abstract
The title compound, C18H17NO2, was prepared from 1-hydroxy-8-methylcarbazole and 3,3-dimethylacrylic acid with trifluoroacetic acid as the cyclization catalyst. Due to the –CMe2– group, the molecule is not quite planar. The packing is dominated by the strong N—H⋯O hydrogen bonds and some weaker C—H⋯O and C—H⋯π interactions. π–π Stacking interactions [centroid–centroid separation = 3.806 (2) Å] join neighboring molecules into loosely connected inversion dimers.
Related literature
Knölker & Reddy (2002 ▶) report on the isolation of pyranocarbazoles from various plant species. Sridharan et al. (2007 ▶) describe the synthesis of compounds related to the title compound. Sridharan, Rajendra Prasad & Zeller (2008 ▶) report the structure of the 9-methyl derivative of the title compound. Sridharan, Rajendra Prasad, Ngendahimana et al. (2008 ▶) report the structure of the 10-H derivative of the title compound.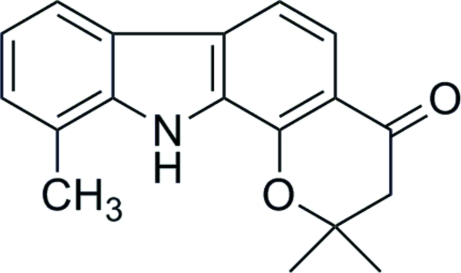
Experimental
Crystal data
C18H17NO2
M r = 279.33
Monoclinic,

a = 12.9740 (16) Å
b = 9.4195 (12) Å
c = 12.8444 (16) Å
β = 114.733 (2)°
V = 1425.7 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 100 (2) K
0.53 × 0.43 × 0.19 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2007 ▶) T min = 0.886, T max = 0.984
13755 measured reflections
3526 independent reflections
2941 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.109
S = 1.03
3526 reflections
193 parameters
H-atom parameters constrained
Δρmax = 0.31 e Å−3
Δρmin = −0.26 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033862/hb2805sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033862/hb2805Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.88 | 1.99 | 2.8634 (13) | 173 |
| C15—H15A⋯O1ii | 0.99 | 2.59 | 3.5411 (15) | 161 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors acknowledge UGC, New Delhi, India, for the award of a Major Research Project (grant No. F31-122/2005). MS thanks UGC, New Delhi, India, for the award of a research fellowship. The diffractometer was funded by the NSF (grant No. 0087210), the Ohio Board of Regents (grant No. CAP-491) and YSU.
supplementary crystallographic information
Comment
Carbazole alkaloids have been isolated from the taxonomically related higher plants of the genus Murraya, Glycosmis, and Clausena from the family Rutaceae. Among the carbazole alkaloids pyranocarbazole alkaloids play a very important role. In this class girinimbine was the first member of the pyrano[3,2-a]carbazole alkaloid family to be isolated from M. Koenigii Spreng (Knölker & Reddy, 2002, and references therein). The isolation of these classes of compounds became an active area of study since these compounds possess high levels of biological and pharmacological activity. Hence we attempted to synthesize pyranocarbazoles in a simple and efficient route.
Using trifluoroacetic acid as the acylating agent we had been able to synthezize in high yields a range of pyranocarbazolones and we recently reported (Sridharan et al., 2007) the synthesis and crystallographic behaviour of 2,3-dihydro-2,2,8-trimethylpyrano[2,3-a]carbazol-4-(11H)-one. As an extension of this reasearch, and to further proof the credibility of trifluoroacetic acid as a good acylating agent, we further extended this synthetic route with a series of substituted 1-hydroxycarbazoles. The components thus synthesized were used as starting synthons to develop routes towards substituted pyranocarbazole derivatives. Herein we report the crystal structures of two of the compounds thus obtained: 2,3-dihydro-2,2,9-trimethylpyrano[2,3-a]carbazol-4-(11H)-one (Sridharan, Rajendra Prasad & Zeller, 2008), the title compound of the preceeding article in this journal) and of the title compound 2,3-dihydro-2,2,10-trimethylpyrano[2,3-a]carbazol-4-(11H)-one (Figure 1).
The single-crystal structure confirmed the formation of the dihydropyrano-[2,3-a]carbazol-4(11H)-one framework as shown in Figure 2. Data collection and structure refinement were unproblematic and all structural parameters (bond lengths, angles, etc) are in the expected ranges. The molecules crystallize in a monoclinic setting in P21/c with four largely planar molecules per unit cell. The plane defined by the sp2 hybridized carbon atoms, the C1 methyl and C15 methylene carbon atoms, and the N and O atoms has an r.m.s. deviation from planarity of only 0.0754 Å. Of all the ring C atoms only C14 of the pyran C(Me)2 unit is significately out of plane with the atoms of the four fused rings, its deviation being 0.534 (1) Å. The pyran ring thus exhibits a half chair conformation.
One of the methyl groups of the C(Me)2 unit is also located close to the average plane of the molecule (C18 with a deviation of 0.125 (2) Å). The other, C17, is however located 2.039 (2) Å away from this plane and thus makes the molecule as a whole not planar and prevents it form forming extensive π-π stacked entities in the solid state. The packing is thus indeed dominated by strong N—H···O hydrogen bonds (Figure 3, Table 1) and some weaker C—H···O (Table 1, Figure 4) and C—H···π interactions (e.g. C18—H18b···Cg1ii = 2.94 Å with Cg1 being the ring C8 to C13 and ii = -x, -1/2 + y, 1/2 - z). The only significant π···π stacking interaction with a centroid to centroid distance of 3.806 (2) Å is found between the pyrrole ring and the the aromatic ring made up of C2 to C7 (Figure 4). Two neighboring molecules related by an inversion center are forming loosly connected dimers via two sets of these π-π interactions (symmetry operator 1 - x, 2 - y, 1 - z).
The structures of the 2,2-dimethyl and the 2,2,10-methyl derivatives of the title compound are described in Sridharan, Rajendra Prasad, Ngendahimana et al. (2008) and Sridharan, Rajendra Prasad & Zeller (2008), the two preceeding articles in this journal. For a more detailed comparison of structures and packing of the three two derivatives please see in Sridharan, Rajendra Prasad & Zeller (2008).
Experimental
1-hydroxy-8-methylcarbazole (0.001 mol) dissolved in 10 ml of trifluroaceticacid and was heated with 3,3-dimethylacrylicacid (0.001 mol) at 323 K for 5 h. The reaction was monitored by TLC. After completion of the reaction, the excess trifluroacetic acid was removed using rotary evaporation. The solid that precipitated out was poured onto ice water, then extracted using ethyl acetate and dried over anhydrous sodium sulfate and filtered. Then the solvent was removed under vacuum and the residue was purified by column chromatography on silica gel using petroleum ether/ethyl acetate (95:5 v/v) as eluant to yield yellow plates of (I) (0.239 g, 86%), m.p. 475–477 K.
Refinement
All hydrogen atoms were added in calculated positions with C—H = 0.99Å (methylene), 0.95Å (aromatic) and 0.98 Å (methyl) and N—H = 0.88 Å. They were refined as riding with Uiso(H) = 1.2Ueq(C,N) or 1.5Ueq(methyl C).
Figures
Fig. 1.
Reaction sequence
Fig. 2.
View of (I) showing xx% displacement ellipsoids. H atoms are represented in stick mode.
Fig. 3.
Packing view of (I) down the a axis showing chains built by the N—H···O hydrogen bonds (indicated by blue dashed lines).
Fig. 4.
Packing view of (I) showing the secondary C—H···π and C—H···O interactions indicated by green lines. Numbers given are distances in Å. N—H···O hydrogen bonds are omitted for clarity.
Crystal data
| C18H17NO2 | F(000) = 592 |
| Mr = 279.33 | Dx = 1.301 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4194 reflections |
| a = 12.9740 (16) Å | θ = 2.8–31.5° |
| b = 9.4195 (12) Å | µ = 0.09 mm−1 |
| c = 12.8444 (16) Å | T = 100 K |
| β = 114.733 (2)° | Plate, yellow |
| V = 1425.7 (3) Å3 | 0.53 × 0.43 × 0.19 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 3526 independent reflections |
| Radiation source: fine-focus sealed tube | 2941 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| ω scans | θmax = 28.3°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2007) | h = −17→17 |
| Tmin = 0.886, Tmax = 0.984 | k = −12→12 |
| 13755 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.109 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0532P)2 + 0.5237P] where P = (Fo2 + 2Fc2)/3 |
| 3526 reflections | (Δ/σ)max = 0.002 |
| 193 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.30161 (12) | 1.23026 (14) | 0.61446 (11) | 0.0287 (3) | |
| H1A | 0.3274 | 1.1580 | 0.6749 | 0.043* | |
| H1B | 0.2199 | 1.2193 | 0.5680 | 0.043* | |
| H1C | 0.3168 | 1.3249 | 0.6493 | 0.043* | |
| C2 | 0.36377 (10) | 1.21263 (12) | 0.53998 (10) | 0.0205 (2) | |
| C3 | 0.44870 (10) | 1.30290 (13) | 0.54197 (11) | 0.0235 (3) | |
| H3 | 0.4698 | 1.3806 | 0.5939 | 0.028* | |
| C4 | 0.50503 (11) | 1.28432 (13) | 0.47042 (11) | 0.0244 (3) | |
| H4 | 0.5636 | 1.3484 | 0.4757 | 0.029* | |
| C5 | 0.47647 (10) | 1.17431 (13) | 0.39258 (10) | 0.0217 (2) | |
| H5 | 0.5142 | 1.1624 | 0.3437 | 0.026* | |
| C6 | 0.39071 (10) | 1.08061 (12) | 0.38720 (10) | 0.0186 (2) | |
| C7 | 0.33727 (9) | 1.09998 (12) | 0.46184 (10) | 0.0178 (2) | |
| C8 | 0.25796 (9) | 0.90879 (12) | 0.35603 (9) | 0.0165 (2) | |
| C9 | 0.33879 (9) | 0.95775 (12) | 0.31816 (10) | 0.0176 (2) | |
| C10 | 0.35259 (10) | 0.88720 (13) | 0.22829 (10) | 0.0197 (2) | |
| H10 | 0.4078 | 0.9184 | 0.2029 | 0.024* | |
| C11 | 0.28477 (10) | 0.77237 (13) | 0.17803 (10) | 0.0199 (2) | |
| H11 | 0.2932 | 0.7246 | 0.1169 | 0.024* | |
| C12 | 0.20236 (10) | 0.72342 (12) | 0.21537 (10) | 0.0176 (2) | |
| C13 | 0.19004 (9) | 0.79075 (12) | 0.30620 (10) | 0.0165 (2) | |
| C14 | 0.06708 (10) | 0.60905 (12) | 0.32183 (10) | 0.0208 (2) | |
| C15 | 0.03119 (10) | 0.58064 (13) | 0.19434 (10) | 0.0199 (2) | |
| H15A | 0.0054 | 0.4810 | 0.1773 | 0.024* | |
| H15B | −0.0338 | 0.6428 | 0.1493 | 0.024* | |
| C16 | 0.12561 (10) | 0.60623 (12) | 0.15718 (10) | 0.0186 (2) | |
| C17 | 0.15681 (12) | 0.50413 (14) | 0.39605 (11) | 0.0288 (3) | |
| H17A | 0.2229 | 0.5095 | 0.3778 | 0.043* | |
| H17B | 0.1255 | 0.4078 | 0.3811 | 0.043* | |
| H17C | 0.1798 | 0.5275 | 0.4771 | 0.043* | |
| C18 | −0.03389 (12) | 0.61299 (15) | 0.35277 (12) | 0.0302 (3) | |
| H18A | −0.0085 | 0.6424 | 0.4329 | 0.045* | |
| H18B | −0.0681 | 0.5183 | 0.3425 | 0.045* | |
| H18C | −0.0902 | 0.6808 | 0.3029 | 0.045* | |
| N1 | 0.25730 (8) | 0.99440 (10) | 0.44254 (8) | 0.0176 (2) | |
| H1 | 0.2137 | 0.9838 | 0.4791 | 0.021* | |
| O1 | 0.11424 (7) | 0.75262 (9) | 0.34888 (7) | 0.01967 (19) | |
| O2 | 0.13335 (7) | 0.53564 (10) | 0.08062 (7) | 0.0245 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0343 (7) | 0.0268 (7) | 0.0265 (6) | −0.0040 (5) | 0.0145 (6) | −0.0059 (5) |
| C2 | 0.0223 (6) | 0.0185 (5) | 0.0189 (5) | 0.0003 (4) | 0.0068 (5) | 0.0011 (4) |
| C3 | 0.0256 (6) | 0.0190 (6) | 0.0220 (6) | −0.0024 (5) | 0.0061 (5) | 0.0003 (4) |
| C4 | 0.0227 (6) | 0.0229 (6) | 0.0251 (6) | −0.0052 (5) | 0.0076 (5) | 0.0031 (5) |
| C5 | 0.0201 (6) | 0.0238 (6) | 0.0215 (6) | −0.0018 (5) | 0.0090 (5) | 0.0040 (5) |
| C6 | 0.0187 (5) | 0.0185 (5) | 0.0182 (5) | 0.0004 (4) | 0.0074 (4) | 0.0025 (4) |
| C7 | 0.0173 (5) | 0.0168 (5) | 0.0185 (5) | 0.0008 (4) | 0.0068 (4) | 0.0031 (4) |
| C8 | 0.0177 (5) | 0.0170 (5) | 0.0160 (5) | 0.0018 (4) | 0.0083 (4) | 0.0024 (4) |
| C9 | 0.0175 (5) | 0.0179 (5) | 0.0182 (5) | 0.0002 (4) | 0.0083 (4) | 0.0032 (4) |
| C10 | 0.0192 (5) | 0.0229 (6) | 0.0209 (6) | −0.0001 (4) | 0.0122 (5) | 0.0022 (4) |
| C11 | 0.0214 (6) | 0.0230 (6) | 0.0191 (5) | 0.0014 (4) | 0.0121 (5) | 0.0002 (4) |
| C12 | 0.0181 (5) | 0.0185 (5) | 0.0182 (5) | 0.0008 (4) | 0.0096 (4) | 0.0011 (4) |
| C13 | 0.0163 (5) | 0.0176 (5) | 0.0177 (5) | 0.0015 (4) | 0.0090 (4) | 0.0024 (4) |
| C14 | 0.0255 (6) | 0.0189 (6) | 0.0219 (6) | −0.0073 (4) | 0.0137 (5) | −0.0034 (4) |
| C15 | 0.0200 (5) | 0.0218 (6) | 0.0200 (5) | −0.0038 (4) | 0.0104 (5) | −0.0035 (4) |
| C16 | 0.0199 (5) | 0.0197 (6) | 0.0177 (5) | 0.0019 (4) | 0.0093 (4) | 0.0019 (4) |
| C17 | 0.0388 (7) | 0.0219 (6) | 0.0242 (6) | −0.0039 (5) | 0.0117 (6) | 0.0022 (5) |
| C18 | 0.0349 (7) | 0.0347 (7) | 0.0312 (7) | −0.0148 (6) | 0.0237 (6) | −0.0103 (6) |
| N1 | 0.0196 (5) | 0.0175 (5) | 0.0183 (5) | −0.0013 (4) | 0.0106 (4) | −0.0008 (4) |
| O1 | 0.0229 (4) | 0.0189 (4) | 0.0231 (4) | −0.0050 (3) | 0.0155 (4) | −0.0034 (3) |
| O2 | 0.0277 (5) | 0.0264 (5) | 0.0239 (4) | −0.0031 (4) | 0.0153 (4) | −0.0064 (3) |
Geometric parameters (Å, °)
| C1—C2 | 1.4963 (18) | C11—C12 | 1.4195 (16) |
| C1—H1A | 0.9800 | C11—H11 | 0.9500 |
| C1—H1B | 0.9800 | C12—C13 | 1.3945 (16) |
| C1—H1C | 0.9800 | C12—C16 | 1.4649 (16) |
| C2—C3 | 1.3836 (17) | C13—O1 | 1.3594 (13) |
| C2—C7 | 1.4010 (16) | C14—O1 | 1.4650 (13) |
| C3—C4 | 1.4040 (19) | C14—C18 | 1.5202 (17) |
| C3—H3 | 0.9500 | C14—C17 | 1.5205 (18) |
| C4—C5 | 1.3787 (18) | C14—C15 | 1.5270 (16) |
| C4—H4 | 0.9500 | C15—C16 | 1.5086 (16) |
| C5—C6 | 1.3991 (16) | C15—H15A | 0.9900 |
| C5—H5 | 0.9500 | C15—H15B | 0.9900 |
| C6—C7 | 1.4103 (16) | C16—O2 | 1.2254 (14) |
| C6—C9 | 1.4420 (16) | C17—H17A | 0.9800 |
| C7—N1 | 1.3826 (14) | C17—H17B | 0.9800 |
| C8—N1 | 1.3759 (14) | C17—H17C | 0.9800 |
| C8—C13 | 1.3966 (16) | C18—H18A | 0.9800 |
| C8—C9 | 1.4057 (15) | C18—H18B | 0.9800 |
| C9—C10 | 1.4068 (16) | C18—H18C | 0.9800 |
| C10—C11 | 1.3732 (17) | N1—H1 | 0.8800 |
| C10—H10 | 0.9500 | ||
| C2—C1—H1A | 109.5 | C13—C12—C16 | 118.57 (10) |
| C2—C1—H1B | 109.5 | C11—C12—C16 | 121.31 (10) |
| H1A—C1—H1B | 109.5 | O1—C13—C12 | 124.94 (10) |
| C2—C1—H1C | 109.5 | O1—C13—C8 | 116.73 (10) |
| H1A—C1—H1C | 109.5 | C12—C13—C8 | 118.31 (10) |
| H1B—C1—H1C | 109.5 | O1—C14—C18 | 103.62 (9) |
| C3—C2—C7 | 115.69 (11) | O1—C14—C17 | 108.49 (10) |
| C3—C2—C1 | 123.92 (11) | C18—C14—C17 | 111.68 (11) |
| C7—C2—C1 | 120.39 (11) | O1—C14—C15 | 109.02 (9) |
| C2—C3—C4 | 122.68 (12) | C18—C14—C15 | 112.03 (10) |
| C2—C3—H3 | 118.7 | C17—C14—C15 | 111.62 (10) |
| C4—C3—H3 | 118.7 | C16—C15—C14 | 112.81 (9) |
| C5—C4—C3 | 120.85 (11) | C16—C15—H15A | 109.0 |
| C5—C4—H4 | 119.6 | C14—C15—H15A | 109.0 |
| C3—C4—H4 | 119.6 | C16—C15—H15B | 109.0 |
| C4—C5—C6 | 118.48 (11) | C14—C15—H15B | 109.0 |
| C4—C5—H5 | 120.8 | H15A—C15—H15B | 107.8 |
| C6—C5—H5 | 120.8 | O2—C16—C12 | 123.50 (11) |
| C5—C6—C7 | 119.46 (11) | O2—C16—C15 | 121.14 (11) |
| C5—C6—C9 | 133.93 (11) | C12—C16—C15 | 115.31 (10) |
| C7—C6—C9 | 106.61 (10) | C14—C17—H17A | 109.5 |
| N1—C7—C2 | 127.88 (11) | C14—C17—H17B | 109.5 |
| N1—C7—C6 | 109.30 (10) | H17A—C17—H17B | 109.5 |
| C2—C7—C6 | 122.81 (11) | C14—C17—H17C | 109.5 |
| N1—C8—C13 | 128.38 (10) | H17A—C17—H17C | 109.5 |
| N1—C8—C9 | 110.17 (10) | H17B—C17—H17C | 109.5 |
| C13—C8—C9 | 121.44 (10) | C14—C18—H18A | 109.5 |
| C8—C9—C10 | 119.89 (11) | C14—C18—H18B | 109.5 |
| C8—C9—C6 | 105.94 (10) | H18A—C18—H18B | 109.5 |
| C10—C9—C6 | 134.16 (11) | C14—C18—H18C | 109.5 |
| C11—C10—C9 | 118.78 (10) | H18A—C18—H18C | 109.5 |
| C11—C10—H10 | 120.6 | H18B—C18—H18C | 109.5 |
| C9—C10—H10 | 120.6 | C8—N1—C7 | 107.97 (10) |
| C10—C11—C12 | 121.47 (11) | C8—N1—H1 | 126.0 |
| C10—C11—H11 | 119.3 | C7—N1—H1 | 126.0 |
| C12—C11—H11 | 119.3 | C13—O1—C14 | 116.68 (9) |
| C13—C12—C11 | 120.07 (11) | ||
| C7—C2—C3—C4 | 0.25 (18) | C11—C12—C13—O1 | −179.88 (10) |
| C1—C2—C3—C4 | −179.51 (12) | C16—C12—C13—O1 | 2.67 (17) |
| C2—C3—C4—C5 | 0.89 (19) | C11—C12—C13—C8 | 1.94 (17) |
| C3—C4—C5—C6 | −0.60 (18) | C16—C12—C13—C8 | −175.51 (10) |
| C4—C5—C6—C7 | −0.79 (17) | N1—C8—C13—O1 | −1.17 (17) |
| C4—C5—C6—C9 | 179.38 (12) | C9—C8—C13—O1 | −179.73 (10) |
| C3—C2—C7—N1 | 179.55 (11) | N1—C8—C13—C12 | 177.16 (11) |
| C1—C2—C7—N1 | −0.67 (19) | C9—C8—C13—C12 | −1.39 (16) |
| C3—C2—C7—C6 | −1.70 (17) | O1—C14—C15—C16 | 53.97 (13) |
| C1—C2—C7—C6 | 178.07 (11) | C18—C14—C15—C16 | 168.06 (10) |
| C5—C6—C7—N1 | −179.03 (10) | C17—C14—C15—C16 | −65.85 (13) |
| C9—C6—C7—N1 | 0.84 (13) | C13—C12—C16—O2 | −176.05 (11) |
| C5—C6—C7—C2 | 2.02 (17) | C11—C12—C16—O2 | 6.53 (18) |
| C9—C6—C7—C2 | −178.11 (10) | C13—C12—C16—C15 | 6.45 (15) |
| N1—C8—C9—C10 | −178.90 (10) | C11—C12—C16—C15 | −170.97 (11) |
| C13—C8—C9—C10 | −0.10 (17) | C14—C15—C16—O2 | 147.37 (11) |
| N1—C8—C9—C6 | 0.40 (12) | C14—C15—C16—C12 | −35.06 (14) |
| C13—C8—C9—C6 | 179.19 (10) | C13—C8—N1—C7 | −178.57 (11) |
| C5—C6—C9—C8 | 179.10 (12) | C9—C8—N1—C7 | 0.12 (12) |
| C7—C6—C9—C8 | −0.74 (12) | C2—C7—N1—C8 | 178.28 (11) |
| C5—C6—C9—C10 | −1.8 (2) | C6—C7—N1—C8 | −0.60 (12) |
| C7—C6—C9—C10 | 178.40 (12) | C12—C13—O1—C14 | 18.95 (16) |
| C8—C9—C10—C11 | 1.04 (17) | C8—C13—O1—C14 | −162.84 (10) |
| C6—C9—C10—C11 | −178.01 (12) | C18—C14—O1—C13 | −165.62 (10) |
| C9—C10—C11—C12 | −0.48 (17) | C17—C14—O1—C13 | 75.58 (12) |
| C10—C11—C12—C13 | −1.04 (18) | C15—C14—O1—C13 | −46.16 (13) |
| C10—C11—C12—C16 | 176.35 (11) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.88 | 1.99 | 2.8634 (13) | 173 |
| C15—H15A···O1ii | 0.99 | 2.59 | 3.5411 (15) | 161 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) −x, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB2805).
References
- Bruker (2007). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst.39, 453–457.
- Knölker, H. J. & Reddy, K. R. (2002). Chem. Rev.102, 4303–4427. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sridharan, M., Prasad, K. J. R. & Zeller, M. (2007). Acta Cryst. E63, o4344.
- Sridharan, M., Prasad, K. J. R. & Zeller, M. (2008). Acta Cryst. E64, o2156. [DOI] [PMC free article] [PubMed]
- Sridharan, M., Prasad, K. J. R., Ngendahimana, A. & Zeller, M. (2008). Acta Cryst. E64, o2155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033862/hb2805sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033862/hb2805Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report






