Abstract
The title compound, [Ni(C5O5)(C10H8N2)2], lies across a crystallographic twofold axis, around which two 2,2′-bipyridine (2,2′-bipy) ligands are arranged in a propeller manner. The local geometry of the NiN4O2 coordination core basically adopts an octahedral geometry. The molecular twofold axis is along the direction of the molecular dipole moment, and the complex is packed with its dipole moment alternately along the +b and −b directions. The crystal structure is stabilized by intermolecular C—H⋯O hydrogen bonds.
Related literature
For the synthesis, see: Chen et al. (2008 ▶). For related structures, see: Chen et al. (2005 ▶, 2007 ▶). For other related literature, see: Coronado et al. (2007 ▶); Wang et al. (2002 ▶).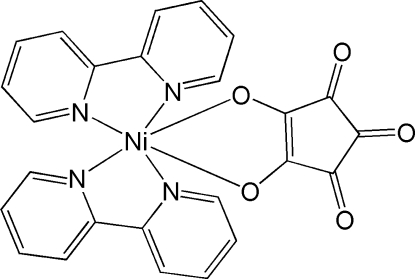
Experimental
Crystal data
[Ni(C5O5)(C10H8N2)2]
M r = 511.13
Orthorhombic,

a = 12.725 (5) Å
b = 10.752 (5) Å
c = 15.733 (5) Å
V = 2152.6 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.95 mm−1
T = 293 (2) K
0.20 × 0.19 × 0.10 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (APEX2; Bruker, 2005 ▶) T min = 0.821, T max = 0.902
10055 measured reflections
2466 independent reflections
1536 reflections with I > 2σ(I)
R int = 0.089
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.102
S = 1.02
2466 reflections
160 parameters
H-atom parameters constrained
Δρmax = 0.26 e Å−3
Δρmin = −0.38 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033771/lx2071sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033771/lx2071Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C4—H4⋯O1i | 0.93 | 2.57 | 3.340 (4) | 141 |
| C8—H8⋯O2ii | 0.93 | 2.24 | 3.114 (4) | 156 |
| C9—H9⋯O3iii | 0.93 | 2.57 | 3.222 (4) | 127 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by the PhD Foundation of the Ministry of Education of China and by the National Natural Science Foundation of China (grant No. 50673054).
supplementary crystallographic information
Comment
The croconate C5O52- anion has attracted increasing attention in recent years because this polydentate ligand gave rise to a variety of interesting complexes (Chen et al., 2008; Coronado et al., 2007; Chen et al., 2005; Wang et al., 2002;). Typically, the C5O52- anion serves as a terminal bidentate chelate ligand or a bridging ligand utilizing more than two O atoms for coordination. We previously reported a mixing-coordinated complex [Ni(C5O5)(phen)2] with 1,10-phenanthroline (phen) as the first ligand and the C5O52- anion as the second ligand (Chen et al., 2007). The similar chelating behavior of 2,2'-bipy and 1,10-phen prompted us to replace phen ligand by 2,2'-bipy. In this report, the structure of this mixing-coordinated complex is reported.
The title compound crystalizes to the same space group as [Ni(C5O5)(phen)2]. Both crystals have very similar cell parameters and show many common features. The chiral molecule lies across twofold axis which is along the direction of the molecular dipole moment. Around the molecular axis, two 2,2'-bipy ligands are arranged in a propeller manner. The Ni2+ is coordinated by four N atoms of the two 2,2'-bipy ligands and two O atoms of a croconate ligand to furnish a slightly distored octahedral NiN4O2 coordination core. The dihedral angle between the croconate plane and a 2,2'-bipy plane is 88.7 (1)°, and that between the two 2,2'-bipy planes is 81.9 (1)° in [Ni(C5O5) (2,2'-bipy)2]. These are close to the corresponding dihedral angles (86.9 (1)° and 86.6 (1)°) in [Ni(C5O5)(phen)2]. The C—O bond lengths involving coordinated O atoms are longer than those of other C—O bonds. In both crystals, molecules packed alternately along +b and-b directions.
However, we can not fail to notice some differences between the two crystals. The Ni—O band length of [Ni(C5O5)(2,2'-bipy)2] {2.102 (2) Å} is longer than that {2.098 (3) Å} of [Ni(C5O5)(phen)2]. Meanwhile, the Ni—N band lengths of [Ni(C5O5)(2,2'-bipy)2] {2.059 (2), 2.066 (2) Å} is considerably shorter than those {2.071 (3), 2.088 (3) Å} of [Ni(C5O5)(phen)2]. It seems that 2,2'-bipyridine is a stronger ligand to Ni2+ in comparison with 1,10-phenanthroline. The crystal structure is stabilized by intermolecular C—H···O hydrogen bonds (Table 1).
Experimental
[K2(C5O5)] (0.050 g, 0.23 mmol) and NiCl2.6H2O (0.060 g, 0.25 mmol) were dissolved in mixed solvent of water (15 ml) and dimethylformamide (10 ml). Then 2,2'-bipy (0.080 g, 0.51 mmol) was added. The mixture was heated to 340–350 K under continuous stirring for 20 min and then filtered. The green-yellow prisms crystals were obtained by slow evaporation at 313 K.
Refinement
All H atoms were positioned geometrically and allowed to ride on their attached atom. The C—H bond lengths for aromatic groups were set to 0.93 Å.
Figures
Fig. 1.
The molecular structure of [Ni(C5O5) (2,2'-bipy)2]. Displacement ellipsoids are drawn at the 30% probability level and H atoms have been omitted. [symmetry code: (i) -x + 1, y, -z + 3/2.]
Crystal data
| [Ni(C5O5)(C10H8N2)2] | F(000) = 1048 |
| Mr = 511.13 | Dx = 1.577 Mg m−3 |
| Orthorhombic, Pbcn | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P_2n 2ab | Cell parameters from 2518 reflections |
| a = 12.725 (5) Å | θ = 2.5–23.4° |
| b = 10.752 (5) Å | µ = 0.95 mm−1 |
| c = 15.733 (5) Å | T = 293 K |
| V = 2152.6 (15) Å3 | Prism, green-yellow |
| Z = 4 | 0.20 × 0.19 × 0.10 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2466 independent reflections |
| Radiation source: fine-focus sealed tube | 1536 reflections with I > 2σ(I) |
| graphite | Rint = 0.089 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 27.5°, θmin = 2.5° |
| φ and ω scans | h = −15→16 |
| Absorption correction: multi-scan (APEX2; Bruker, 2005) | k = −13→10 |
| Tmin = 0.821, Tmax = 0.902 | l = −20→19 |
| 10055 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.102 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0372P)2 + 0.0108P] where P = (Fo2 + 2Fc2)/3 |
| 2466 reflections | (Δ/σ)max < 0.001 |
| 160 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.38 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 0.5000 | 0.46011 (4) | 0.2500 | 0.03546 (17) | |
| O1 | 0.56048 (15) | 0.31353 (15) | 0.17633 (10) | 0.0439 (5) | |
| O2 | 0.6016 (2) | 0.0512 (2) | 0.12408 (16) | 0.1053 (10) | |
| O3 | 0.5000 | −0.1097 (3) | 0.2500 | 0.0700 (9) | |
| N1 | 0.45364 (18) | 0.58822 (19) | 0.34037 (12) | 0.0397 (5) | |
| N2 | 0.63184 (17) | 0.46676 (19) | 0.32573 (12) | 0.0419 (6) | |
| C1 | 0.3650 (3) | 0.6535 (3) | 0.34092 (17) | 0.0531 (8) | |
| H1 | 0.3173 | 0.6410 | 0.2970 | 0.064* | |
| C2 | 0.3402 (3) | 0.7384 (3) | 0.4029 (2) | 0.0717 (10) | |
| H2 | 0.2774 | 0.7824 | 0.4013 | 0.086* | |
| C3 | 0.4114 (4) | 0.7561 (3) | 0.4673 (2) | 0.0742 (11) | |
| H3 | 0.3977 | 0.8142 | 0.5097 | 0.089* | |
| C4 | 0.5025 (3) | 0.6888 (3) | 0.46926 (18) | 0.0570 (9) | |
| H4 | 0.5502 | 0.6989 | 0.5135 | 0.068* | |
| C5 | 0.5226 (2) | 0.6052 (2) | 0.40425 (15) | 0.0406 (7) | |
| C6 | 0.6200 (2) | 0.5320 (2) | 0.39824 (16) | 0.0431 (7) | |
| C7 | 0.6964 (3) | 0.5264 (3) | 0.4613 (2) | 0.0638 (9) | |
| H7 | 0.6872 | 0.5696 | 0.5119 | 0.077* | |
| C8 | 0.7854 (3) | 0.4569 (4) | 0.4486 (2) | 0.0770 (12) | |
| H8 | 0.8367 | 0.4526 | 0.4906 | 0.092* | |
| C9 | 0.7982 (2) | 0.3940 (3) | 0.3737 (2) | 0.0679 (10) | |
| H9 | 0.8587 | 0.3479 | 0.3633 | 0.081* | |
| C10 | 0.7195 (2) | 0.4008 (3) | 0.31437 (19) | 0.0575 (8) | |
| H10 | 0.7276 | 0.3572 | 0.2637 | 0.069* | |
| C11 | 0.5300 (2) | 0.2129 (3) | 0.21214 (15) | 0.0393 (6) | |
| C12 | 0.5511 (3) | 0.0864 (3) | 0.18605 (18) | 0.0530 (8) | |
| C13 | 0.5000 | 0.0035 (4) | 0.2500 | 0.0487 (10) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.0404 (3) | 0.0388 (3) | 0.0271 (2) | 0.000 | −0.0053 (2) | 0.000 |
| O1 | 0.0587 (13) | 0.0418 (11) | 0.0310 (9) | −0.0040 (10) | 0.0083 (9) | 0.0013 (8) |
| O2 | 0.167 (3) | 0.0586 (15) | 0.0903 (17) | −0.0026 (15) | 0.0769 (19) | −0.0177 (13) |
| O3 | 0.086 (2) | 0.0405 (17) | 0.084 (2) | 0.000 | 0.0118 (18) | 0.000 |
| N1 | 0.0510 (14) | 0.0360 (12) | 0.0322 (11) | −0.0008 (12) | −0.0020 (11) | 0.0015 (10) |
| N2 | 0.0437 (14) | 0.0480 (14) | 0.0340 (12) | 0.0003 (12) | −0.0082 (10) | 0.0017 (10) |
| C1 | 0.064 (2) | 0.0507 (18) | 0.0443 (16) | 0.0138 (17) | 0.0003 (16) | 0.0011 (14) |
| C2 | 0.098 (3) | 0.057 (2) | 0.060 (2) | 0.024 (2) | 0.022 (2) | −0.0018 (17) |
| C3 | 0.119 (3) | 0.048 (2) | 0.056 (2) | −0.002 (2) | 0.028 (2) | −0.0121 (16) |
| C4 | 0.084 (2) | 0.0514 (17) | 0.0359 (15) | −0.022 (2) | 0.0095 (16) | −0.0076 (13) |
| C5 | 0.0582 (19) | 0.0341 (14) | 0.0295 (13) | −0.0130 (14) | 0.0012 (12) | 0.0026 (11) |
| C6 | 0.0535 (18) | 0.0442 (16) | 0.0317 (14) | −0.0198 (15) | −0.0080 (13) | 0.0070 (13) |
| C7 | 0.076 (2) | 0.068 (2) | 0.0482 (18) | −0.023 (2) | −0.0272 (17) | 0.0038 (15) |
| C8 | 0.065 (2) | 0.090 (3) | 0.076 (3) | −0.023 (2) | −0.041 (2) | 0.033 (2) |
| C9 | 0.0442 (18) | 0.080 (3) | 0.079 (2) | 0.0036 (19) | −0.0144 (18) | 0.026 (2) |
| C10 | 0.0494 (19) | 0.067 (2) | 0.0555 (19) | 0.0078 (18) | −0.0081 (15) | 0.0055 (16) |
| C11 | 0.0431 (17) | 0.0439 (16) | 0.0309 (13) | −0.0025 (14) | −0.0005 (11) | −0.0019 (12) |
| C12 | 0.066 (2) | 0.0462 (17) | 0.0463 (17) | −0.0038 (17) | 0.0129 (16) | −0.0085 (14) |
| C13 | 0.052 (2) | 0.042 (2) | 0.052 (2) | 0.000 | 0.000 (2) | 0.000 |
Geometric parameters (Å, °)
| Ni1—N2 | 2.059 (2) | C3—C4 | 1.367 (4) |
| Ni1—N2i | 2.059 (2) | C3—H3 | 0.9300 |
| Ni1—N1 | 2.066 (2) | C4—C5 | 1.385 (4) |
| Ni1—N1i | 2.066 (2) | C4—H4 | 0.9300 |
| Ni1—O1i | 2.1022 (18) | C5—C6 | 1.472 (4) |
| Ni1—O1 | 2.1022 (18) | C6—C7 | 1.389 (4) |
| O1—C11 | 1.280 (3) | C7—C8 | 1.371 (5) |
| O2—C12 | 1.227 (3) | C7—H7 | 0.9300 |
| O3—C13 | 1.217 (5) | C8—C9 | 1.369 (5) |
| N1—C1 | 1.328 (3) | C8—H8 | 0.9300 |
| N1—C5 | 1.347 (3) | C9—C10 | 1.370 (4) |
| N2—C10 | 1.334 (3) | C9—H9 | 0.9300 |
| N2—C6 | 1.347 (3) | C10—H10 | 0.9300 |
| C1—C2 | 1.372 (4) | C11—C11i | 1.415 (5) |
| C1—H1 | 0.9300 | C11—C12 | 1.446 (4) |
| C2—C3 | 1.373 (5) | C12—C13 | 1.493 (4) |
| C2—H2 | 0.9300 | C13—C12i | 1.493 (4) |
| N2—Ni1—N2i | 176.02 (12) | C3—C4—C5 | 118.9 (3) |
| N2—Ni1—N1 | 79.12 (9) | C3—C4—H4 | 120.6 |
| N2i—Ni1—N1 | 98.19 (8) | C5—C4—H4 | 120.6 |
| N2—Ni1—N1i | 98.19 (8) | N1—C5—C4 | 121.3 (3) |
| N2i—Ni1—N1i | 79.12 (9) | N1—C5—C6 | 115.4 (2) |
| N1—Ni1—N1i | 96.35 (11) | C4—C5—C6 | 123.3 (3) |
| N2—Ni1—O1i | 90.30 (8) | N2—C6—C7 | 120.2 (3) |
| N2i—Ni1—O1i | 92.68 (8) | N2—C6—C5 | 115.3 (2) |
| N1—Ni1—O1i | 90.91 (8) | C7—C6—C5 | 124.5 (3) |
| N1i—Ni1—O1i | 169.72 (7) | C8—C7—C6 | 119.8 (3) |
| N2—Ni1—O1 | 92.68 (8) | C8—C7—H7 | 120.1 |
| N2i—Ni1—O1 | 90.30 (8) | C6—C7—H7 | 120.1 |
| N1—Ni1—O1 | 169.72 (7) | C9—C8—C7 | 119.5 (3) |
| N1i—Ni1—O1 | 90.91 (8) | C9—C8—H8 | 120.2 |
| O1i—Ni1—O1 | 82.88 (10) | C7—C8—H8 | 120.2 |
| C11—O1—Ni1 | 106.29 (15) | C8—C9—C10 | 118.2 (3) |
| C1—N1—C5 | 118.5 (2) | C8—C9—H9 | 120.9 |
| C1—N1—Ni1 | 126.82 (19) | C10—C9—H9 | 120.9 |
| C5—N1—Ni1 | 114.72 (18) | N2—C10—C9 | 123.2 (3) |
| C10—N2—C6 | 118.9 (2) | N2—C10—H10 | 118.4 |
| C10—N2—Ni1 | 125.83 (19) | C9—C10—H10 | 118.4 |
| C6—N2—Ni1 | 114.66 (19) | O1—C11—C11i | 122.26 (14) |
| N1—C1—C2 | 123.5 (3) | O1—C11—C12 | 127.9 (2) |
| N1—C1—H1 | 118.3 | C11i—C11—C12 | 109.83 (15) |
| C2—C1—H1 | 118.3 | O2—C12—C11 | 127.8 (3) |
| C1—C2—C3 | 117.7 (3) | O2—C12—C13 | 125.4 (3) |
| C1—C2—H2 | 121.1 | C11—C12—C13 | 106.8 (2) |
| C3—C2—H2 | 121.1 | O3—C13—C12i | 126.64 (17) |
| C4—C3—C2 | 120.2 (3) | O3—C13—C12 | 126.64 (17) |
| C4—C3—H3 | 119.9 | C12i—C13—C12 | 106.7 (3) |
| C2—C3—H3 | 119.9 | ||
| N2—Ni1—O1—C11 | 90.49 (17) | C1—N1—C5—C6 | −178.6 (2) |
| N2i—Ni1—O1—C11 | −92.14 (18) | Ni1—N1—C5—C6 | 1.2 (3) |
| N1—Ni1—O1—C11 | 53.7 (5) | C3—C4—C5—N1 | −1.0 (4) |
| N1i—Ni1—O1—C11 | −171.27 (17) | C3—C4—C5—C6 | 177.3 (3) |
| O1i—Ni1—O1—C11 | 0.52 (13) | C10—N2—C6—C7 | −2.5 (4) |
| N2—Ni1—N1—C1 | 174.7 (2) | Ni1—N2—C6—C7 | 169.1 (2) |
| N2i—Ni1—N1—C1 | −2.3 (2) | C10—N2—C6—C5 | 178.1 (2) |
| N1i—Ni1—N1—C1 | 77.5 (2) | Ni1—N2—C6—C5 | −10.3 (3) |
| O1i—Ni1—N1—C1 | −95.2 (2) | N1—C5—C6—N2 | 6.1 (3) |
| O1—Ni1—N1—C1 | −147.8 (4) | C4—C5—C6—N2 | −172.3 (2) |
| N2—Ni1—N1—C5 | −5.04 (17) | N1—C5—C6—C7 | −173.3 (2) |
| N2i—Ni1—N1—C5 | 177.93 (18) | C4—C5—C6—C7 | 8.3 (4) |
| N1i—Ni1—N1—C5 | −102.20 (19) | N2—C6—C7—C8 | 1.8 (4) |
| O1i—Ni1—N1—C5 | 85.09 (18) | C5—C6—C7—C8 | −178.8 (3) |
| O1—Ni1—N1—C5 | 32.5 (5) | C6—C7—C8—C9 | 0.2 (5) |
| N1—Ni1—N2—C10 | 179.3 (2) | C7—C8—C9—C10 | −1.5 (5) |
| N1i—Ni1—N2—C10 | −85.7 (2) | C6—N2—C10—C9 | 1.2 (4) |
| O1i—Ni1—N2—C10 | 88.5 (2) | Ni1—N2—C10—C9 | −169.4 (2) |
| O1—Ni1—N2—C10 | 5.6 (2) | C8—C9—C10—N2 | 0.8 (5) |
| N1—Ni1—N2—C6 | 8.43 (17) | Ni1—O1—C11—C11i | −1.5 (4) |
| N1i—Ni1—N2—C6 | 103.37 (17) | Ni1—O1—C11—C12 | −179.9 (2) |
| O1i—Ni1—N2—C6 | −82.44 (17) | O1—C11—C12—O2 | −0.7 (5) |
| O1—Ni1—N2—C6 | −165.32 (17) | C11i—C11—C12—O2 | −179.3 (3) |
| C5—N1—C1—C2 | 0.6 (4) | O1—C11—C12—C13 | 178.9 (2) |
| Ni1—N1—C1—C2 | −179.1 (2) | C11i—C11—C12—C13 | 0.3 (4) |
| N1—C1—C2—C3 | 0.1 (5) | O2—C12—C13—O3 | −0.6 (4) |
| C1—C2—C3—C4 | −1.2 (5) | C11—C12—C13—O3 | 179.89 (13) |
| C2—C3—C4—C5 | 1.7 (5) | O2—C12—C13—C12i | 179.4 (4) |
| C1—N1—C5—C4 | −0.1 (4) | C11—C12—C13—C12i | −0.11 (13) |
| Ni1—N1—C5—C4 | 179.63 (19) |
Symmetry codes: (i) −x+1, y, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C4—H4···O1ii | 0.93 | 2.57 | 3.340 (4) | 141 |
| C8—H8···O2iii | 0.93 | 2.24 | 3.114 (4) | 156 |
| C9—H9···O3iv | 0.93 | 2.57 | 3.222 (4) | 127 |
Symmetry codes: (ii) x, −y+1, z+1/2; (iii) −x+3/2, −y+1/2, z+1/2; (iv) x+1/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LX2071).
References
- Bruker (2005). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, H.-F., Chen, H.-Y., Chen, X., Batsanov, A. S. & Fang, Q. (2008). Acta Cryst. E64, m172. [DOI] [PMC free article] [PubMed]
- Chen, X., Chen, H.-F., Xue, G., Chen, H.-Y., Yu, W.-T. & Fang, Q. (2007). Acta Cryst. C63, m166–m168. [DOI] [PubMed]
- Chen, H.-Y., Fang, Q., Xue, G. & Yu, W.-T. (2005). Acta Cryst. C61, m535–m537. [DOI] [PubMed]
- Coronado, E., Curreli, S., Giménez-Saiz, C., Gómez-García, C.-J., Deplano, P., Mercuri, M.-L., Serpe, A., Pilia, L., Faulmann, C. & Canadell, E. (2007). Inorg. Chem.46, 4446–4457. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, C. C., Yang, C.-H. & Lee, G.-H. (2002). Inorg. Chem.41, 1015–1018. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033771/lx2071sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033771/lx2071Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



