Abstract
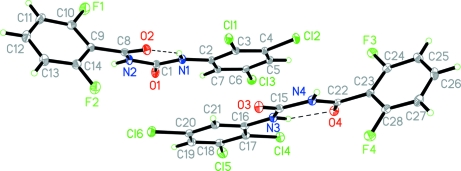
The asymmetric unit of the title compound, C14H7Cl3F2N2O2, contains two unique molecules. The 2,3,5-trichlorophenyl ring is almost coplanar with the urea group in both molecules, whereas the 2,6-difluorophenyl ring is twisted from the urea plane by 54.83 (10)° in one molecule and 60.58 (10)° in the other. An intramolecular N—H—O hydrogen bond stabilizes the molecular conformation. The crystal packing is formed by intermolecular N—H—O hydrogen bonds and F⋯F interactions [2.841 (2) Å].
Related literature
For general background, see: Yan et al. (2003 ▶). For synthetic details, see: Lin et al. (2003 ▶, 2005 ▶). 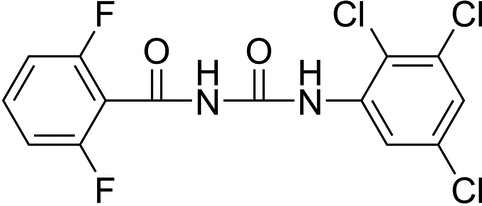
Experimental
Crystal data
C14H7Cl3F2N2O2
M r = 379.57
Monoclinic,
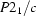
a = 7.1669 (4) Å
b = 22.8228 (12) Å
c = 18.2885 (10) Å
β = 94.768 (2)°
V = 2981.1 (3) Å3
Z = 8
Mo Kα radiation
μ = 0.65 mm−1
T = 113 (2) K
0.24 × 0.14 × 0.12 mm
Data collection
Rigaku Saturn diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2006 ▶) T min = 0.860, T max = 0.927
27779 measured reflections
7091 independent reflections
6011 reflections with I > 2σ(I)
R int = 0.050
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.089
S = 1.07
7091 reflections
431 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.29 e Å−3
Δρmin = −0.29 e Å−3
Data collection: CrystalClear (Rigaku, 2006 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: CrystalStructure (Rigaku, 2006 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808032029/jh2065sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808032029/jh2065Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯Cl1 | 0.89 (2) | 2.46 (2) | 2.9126 (15) | 111.8 (16) |
| N1—H1⋯O2 | 0.89 (2) | 1.88 (2) | 2.641 (2) | 141.5 (19) |
| N2—H2⋯O1i | 0.82 (2) | 2.00 (2) | 2.8205 (19) | 173 (2) |
| N3—H3⋯Cl4 | 0.80 (2) | 2.43 (2) | 2.8944 (16) | 118.2 (19) |
| N3—H3⋯O4 | 0.80 (2) | 1.99 (2) | 2.658 (2) | 140 (2) |
| N4—H4⋯O3ii | 0.91 (2) | 1.93 (2) | 2.8378 (18) | 176 (2) |
Symmetry codes: (i) 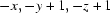 ; (ii)
; (ii) 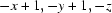 .
.
Acknowledgments
This project was supported by the 11th Five-Year Construction Item for Yunnan University Teachers (grant No. 0030-WX069051).
supplementary crystallographic information
Comment
Derivatives of benzoylphenylureas (BPUs) are kind of insect growth regulators (IGRs), interferes the chitin synthesis in target pests, causing death or abortive development. BPUs posses high selectivity, low acute toxicity for mammals. At the time, the different groups on the phenyl that have different bioactivity. So research the configuration of the different compound is important to find more potent insecticide. The title compound (I) (Fig. 1), C14H7Cl3F2N2O2, which possesses high bioactivity to pests (Yan et al., 2003).
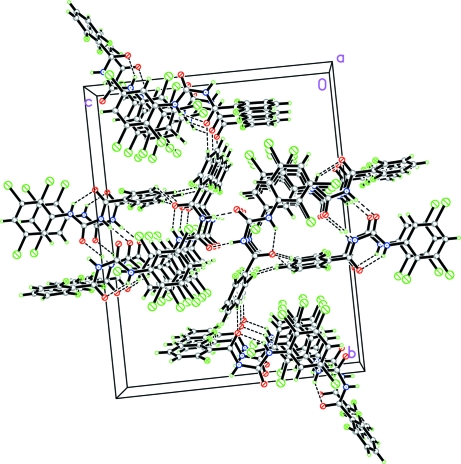
The geometrical parameters for (I) (Table 1) show the conjugation present: the length of the C1═O1 and C═O2 double bond is greater than that of a normal C═O double bond. The lengths of the C1—N1, C1—N2, C8—N2 bonds are shorter than that of normal C—N single bonds. The 2,3,5-trichlorophenyl ring of the title compound is almost coplanar with the urea group, whereas the 2,6-difluorophenyl ring is twisted from the urea plane by 60.58 (10)°. An intramolecular N—H—O hydrogen bond stabilizes the molecular conformation. The crystal packing of the title compound formed by intermolecular N—H—O hydrogen bonds and F···F bond (Fig 2).
Experimental
A solution of 2,6-difluorobenzoyl isocyanate (II) (10 mmol, 1.0 equiv.) in 1,2-dichloroethane (10 ml) was added to a stirred solution of 2,3,5-trichloroaniline (III) (10 mmol, 1.0 equiv.) in dry 1,2-dichloroethane (20 ml) and stirred at room temperature for 24 hrs, the solvent was removed in vacuo and the residue was recrystallized with ethyl acetate to give desired compounds as white needle-crystals (I) in 93% yield (Lin et al., 2003; Lin et al., 2005). The desire product recrystallized from acetone (m.p. 517 K).
Refinement
In the absence of significant anomalous dispersion effects, Freidel pairs were merged; the absolute configuration was assigned on the basis of the known configuration of the starting material. All H atoms were placed in idealized positions and refined with riding constraints, with C—H distances in the range 0.93–0.96 Å and with Uiso(H) = 1.2 or 1.5 times Ueq(C).
Figures
Fig. 1.
View of (I) showing 30% displacement ellipsoids (arbitrary spheres for the H atoms).
Fig. 2.
The crystal packing of complex 1 showing the hydrogen bonds as broken lines. Symmetry code: (i) -x, -y + 1, -z + 1; (ii) -x + 1, -y + 1, -z.
Fig. 3.
The formation of the title compound.
Crystal data
| C14H7Cl3F2N2O2 | Dx = 1.691 Mg m−3 |
| Mr = 379.57 | Melting point: 517 K |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71070 Å |
| a = 7.1669 (4) Å | Cell parameters from 6247 reflections |
| b = 22.8228 (12) Å | θ = 1.8–27.9° |
| c = 18.2885 (10) Å | µ = 0.65 mm−1 |
| β = 94.768 (2)° | T = 113 K |
| V = 2981.1 (3) Å3 | Prism, colourless |
| Z = 8 | 0.24 × 0.14 × 0.12 mm |
| F(000) = 1520 |
Data collection
| Rigaku Saturn diffractometer | 7091 independent reflections |
| Radiation source: rotating anode | 6011 reflections with I > 2σ(I) |
| confocal | Rint = 0.050 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.9°, θmin = 1.8° |
| ω and φ scans | h = −9→9 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2006) | k = −30→30 |
| Tmin = 0.860, Tmax = 0.927 | l = −24→24 |
| 27779 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.089 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0419P)2 + 0.1727P] where P = (Fo2 + 2Fc2)/3 |
| 7091 reflections | (Δ/σ)max = 0.001 |
| 431 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.03364 (7) | 0.50654 (2) | 0.15961 (2) | 0.02733 (12) | |
| Cl2 | −0.07937 (7) | 0.40202 (2) | 0.05408 (2) | 0.03038 (12) | |
| Cl3 | −0.17311 (7) | 0.26475 (2) | 0.28607 (3) | 0.03149 (12) | |
| Cl4 | 0.34943 (7) | 0.283841 (19) | 0.19434 (2) | 0.02671 (11) | |
| Cl5 | 0.36147 (7) | 0.28306 (2) | 0.36548 (2) | 0.03282 (13) | |
| Cl6 | 0.54168 (7) | 0.51085 (2) | 0.37218 (2) | 0.02918 (12) | |
| F1 | −0.17544 (16) | 0.64246 (6) | 0.45899 (7) | 0.0423 (3) | |
| F2 | 0.44718 (15) | 0.65048 (6) | 0.39926 (7) | 0.0414 (3) | |
| F3 | 0.04346 (14) | 0.34365 (5) | −0.10568 (6) | 0.0303 (3) | |
| F4 | 0.67828 (15) | 0.39222 (5) | −0.10684 (6) | 0.0327 (3) | |
| O1 | −0.02709 (19) | 0.44667 (6) | 0.42993 (6) | 0.0291 (3) | |
| O2 | 0.14058 (17) | 0.59069 (6) | 0.31730 (6) | 0.0248 (3) | |
| O3 | 0.51390 (18) | 0.48843 (5) | 0.09749 (6) | 0.0247 (3) | |
| O4 | 0.33939 (17) | 0.32720 (5) | 0.01436 (6) | 0.0240 (3) | |
| N1 | 0.0186 (2) | 0.48145 (7) | 0.31516 (8) | 0.0214 (3) | |
| N2 | 0.0531 (2) | 0.54179 (7) | 0.41771 (8) | 0.0225 (3) | |
| N3 | 0.4215 (2) | 0.39641 (7) | 0.12945 (8) | 0.0202 (3) | |
| N4 | 0.4259 (2) | 0.42336 (7) | 0.00736 (8) | 0.0201 (3) | |
| C1 | 0.0123 (3) | 0.48592 (8) | 0.38878 (9) | 0.0221 (4) | |
| C2 | −0.0276 (2) | 0.43225 (8) | 0.27132 (9) | 0.0197 (4) | |
| C3 | −0.0277 (2) | 0.43974 (8) | 0.19488 (9) | 0.0212 (4) | |
| C4 | −0.0753 (2) | 0.39311 (8) | 0.14825 (9) | 0.0225 (4) | |
| C5 | −0.1203 (2) | 0.33913 (8) | 0.17552 (10) | 0.0246 (4) | |
| H5 | −0.1524 | 0.3073 | 0.1434 | 0.029* | |
| C6 | −0.1176 (2) | 0.33237 (8) | 0.25079 (10) | 0.0226 (4) | |
| C7 | −0.0722 (2) | 0.37795 (8) | 0.29909 (9) | 0.0215 (4) | |
| H7 | −0.0717 | 0.3721 | 0.3505 | 0.026* | |
| C8 | 0.1103 (2) | 0.59011 (8) | 0.38219 (9) | 0.0205 (4) | |
| C9 | 0.1360 (2) | 0.64407 (8) | 0.42805 (9) | 0.0207 (4) | |
| C10 | −0.0058 (3) | 0.66965 (9) | 0.46427 (10) | 0.0270 (4) | |
| C11 | 0.0158 (3) | 0.72142 (9) | 0.50213 (11) | 0.0331 (5) | |
| H11 | −0.0859 | 0.7382 | 0.5250 | 0.040* | |
| C12 | 0.1879 (3) | 0.74862 (9) | 0.50626 (10) | 0.0314 (5) | |
| H12 | 0.2058 | 0.7840 | 0.5334 | 0.038* | |
| C13 | 0.3350 (3) | 0.72521 (8) | 0.47147 (10) | 0.0291 (4) | |
| H13 | 0.4537 | 0.7440 | 0.4742 | 0.035* | |
| C14 | 0.3047 (3) | 0.67426 (8) | 0.43304 (10) | 0.0252 (4) | |
| C15 | 0.4574 (2) | 0.43923 (8) | 0.08133 (9) | 0.0193 (4) | |
| C16 | 0.4364 (2) | 0.39918 (8) | 0.20631 (9) | 0.0193 (4) | |
| C17 | 0.4017 (2) | 0.34714 (8) | 0.24369 (9) | 0.0205 (4) | |
| C18 | 0.4090 (3) | 0.34667 (8) | 0.31982 (9) | 0.0227 (4) | |
| C19 | 0.4524 (2) | 0.39688 (8) | 0.36001 (9) | 0.0241 (4) | |
| H19 | 0.4574 | 0.3966 | 0.4121 | 0.029* | |
| C20 | 0.4882 (2) | 0.44752 (8) | 0.32218 (9) | 0.0219 (4) | |
| C21 | 0.4809 (2) | 0.44985 (8) | 0.24629 (9) | 0.0208 (4) | |
| H21 | 0.5058 | 0.4854 | 0.2220 | 0.025* | |
| C22 | 0.3743 (2) | 0.37040 (7) | −0.02176 (9) | 0.0181 (4) | |
| C23 | 0.3613 (2) | 0.36860 (7) | −0.10363 (9) | 0.0185 (4) | |
| C24 | 0.1951 (3) | 0.35418 (8) | −0.14347 (9) | 0.0219 (4) | |
| C25 | 0.1753 (3) | 0.35137 (8) | −0.21846 (10) | 0.0265 (4) | |
| H25 | 0.0583 | 0.3416 | −0.2438 | 0.032* | |
| C26 | 0.3300 (3) | 0.36319 (8) | −0.25635 (10) | 0.0290 (4) | |
| H26 | 0.3189 | 0.3617 | −0.3084 | 0.035* | |
| C27 | 0.5015 (3) | 0.37721 (8) | −0.21950 (10) | 0.0281 (4) | |
| H27 | 0.6082 | 0.3848 | −0.2456 | 0.034* | |
| C28 | 0.5121 (2) | 0.37969 (8) | −0.14440 (10) | 0.0224 (4) | |
| H1 | 0.050 (3) | 0.5150 (9) | 0.2941 (11) | 0.033 (6)* | |
| H2 | 0.053 (3) | 0.5433 (10) | 0.4626 (12) | 0.039 (6)* | |
| H3 | 0.392 (3) | 0.3651 (9) | 0.1119 (12) | 0.036 (7)* | |
| H4 | 0.441 (3) | 0.4529 (10) | −0.0251 (12) | 0.045 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0356 (3) | 0.0278 (3) | 0.0190 (2) | −0.0018 (2) | 0.00479 (18) | −0.00021 (17) |
| Cl2 | 0.0346 (3) | 0.0393 (3) | 0.0171 (2) | 0.0015 (2) | 0.00166 (18) | −0.00678 (18) |
| Cl3 | 0.0359 (3) | 0.0224 (3) | 0.0360 (3) | −0.0003 (2) | 0.0023 (2) | −0.00089 (19) |
| Cl4 | 0.0352 (3) | 0.0200 (2) | 0.0249 (2) | −0.00355 (19) | 0.00249 (19) | 0.00437 (17) |
| Cl5 | 0.0422 (3) | 0.0311 (3) | 0.0256 (2) | −0.0054 (2) | 0.0058 (2) | 0.01259 (19) |
| Cl6 | 0.0362 (3) | 0.0271 (3) | 0.0243 (2) | 0.0022 (2) | 0.00324 (19) | −0.00380 (18) |
| F1 | 0.0232 (6) | 0.0534 (9) | 0.0519 (8) | −0.0052 (6) | 0.0122 (5) | −0.0179 (6) |
| F2 | 0.0237 (6) | 0.0504 (8) | 0.0518 (8) | −0.0071 (6) | 0.0141 (5) | −0.0213 (6) |
| F3 | 0.0211 (6) | 0.0405 (7) | 0.0298 (6) | −0.0052 (5) | 0.0058 (5) | 0.0026 (5) |
| F4 | 0.0204 (6) | 0.0436 (7) | 0.0345 (6) | −0.0041 (5) | 0.0055 (5) | −0.0001 (5) |
| O1 | 0.0437 (8) | 0.0258 (7) | 0.0185 (6) | −0.0046 (6) | 0.0073 (6) | −0.0011 (5) |
| O2 | 0.0277 (7) | 0.0292 (7) | 0.0178 (6) | −0.0046 (6) | 0.0037 (5) | −0.0016 (5) |
| O3 | 0.0349 (8) | 0.0192 (7) | 0.0204 (6) | −0.0058 (6) | 0.0052 (5) | 0.0014 (5) |
| O4 | 0.0319 (7) | 0.0177 (7) | 0.0229 (6) | −0.0010 (5) | 0.0050 (5) | 0.0041 (5) |
| N1 | 0.0256 (9) | 0.0223 (9) | 0.0166 (7) | −0.0023 (7) | 0.0035 (6) | −0.0022 (6) |
| N2 | 0.0290 (9) | 0.0241 (9) | 0.0149 (7) | −0.0038 (7) | 0.0048 (6) | −0.0033 (6) |
| N3 | 0.0258 (8) | 0.0179 (8) | 0.0172 (7) | −0.0012 (7) | 0.0031 (6) | 0.0026 (6) |
| N4 | 0.0257 (8) | 0.0174 (8) | 0.0176 (7) | −0.0017 (6) | 0.0039 (6) | 0.0034 (6) |
| C1 | 0.0236 (10) | 0.0244 (10) | 0.0184 (8) | −0.0008 (8) | 0.0027 (7) | −0.0035 (7) |
| C2 | 0.0163 (9) | 0.0248 (10) | 0.0182 (8) | 0.0019 (7) | 0.0023 (7) | −0.0053 (7) |
| C3 | 0.0180 (9) | 0.0248 (10) | 0.0212 (8) | 0.0025 (7) | 0.0032 (7) | −0.0002 (7) |
| C4 | 0.0177 (9) | 0.0317 (11) | 0.0178 (8) | 0.0041 (8) | 0.0007 (7) | −0.0059 (7) |
| C5 | 0.0216 (10) | 0.0263 (10) | 0.0259 (9) | 0.0025 (8) | 0.0026 (7) | −0.0067 (7) |
| C6 | 0.0179 (9) | 0.0216 (10) | 0.0286 (9) | 0.0031 (7) | 0.0031 (7) | −0.0017 (7) |
| C7 | 0.0182 (9) | 0.0265 (10) | 0.0198 (8) | 0.0029 (7) | 0.0015 (7) | −0.0006 (7) |
| C8 | 0.0155 (9) | 0.0257 (10) | 0.0203 (8) | 0.0000 (7) | 0.0017 (7) | 0.0000 (7) |
| C9 | 0.0220 (9) | 0.0226 (10) | 0.0173 (8) | 0.0009 (7) | 0.0005 (7) | 0.0004 (7) |
| C10 | 0.0212 (10) | 0.0313 (11) | 0.0287 (10) | 0.0002 (8) | 0.0040 (8) | −0.0021 (8) |
| C11 | 0.0374 (12) | 0.0305 (12) | 0.0325 (11) | 0.0085 (9) | 0.0101 (9) | −0.0058 (8) |
| C12 | 0.0482 (13) | 0.0213 (10) | 0.0249 (9) | 0.0001 (9) | 0.0043 (9) | −0.0016 (8) |
| C13 | 0.0349 (12) | 0.0274 (11) | 0.0251 (9) | −0.0068 (9) | 0.0028 (8) | 0.0001 (8) |
| C14 | 0.0235 (10) | 0.0278 (11) | 0.0250 (9) | −0.0001 (8) | 0.0052 (7) | −0.0012 (7) |
| C15 | 0.0188 (9) | 0.0206 (10) | 0.0190 (8) | 0.0016 (7) | 0.0039 (7) | 0.0029 (7) |
| C16 | 0.0171 (9) | 0.0221 (9) | 0.0190 (8) | 0.0025 (7) | 0.0027 (7) | 0.0039 (7) |
| C17 | 0.0175 (9) | 0.0213 (10) | 0.0231 (9) | 0.0011 (7) | 0.0034 (7) | 0.0036 (7) |
| C18 | 0.0219 (9) | 0.0244 (10) | 0.0221 (9) | 0.0023 (8) | 0.0047 (7) | 0.0090 (7) |
| C19 | 0.0227 (10) | 0.0316 (11) | 0.0185 (8) | 0.0034 (8) | 0.0049 (7) | 0.0044 (7) |
| C20 | 0.0184 (9) | 0.0243 (10) | 0.0232 (9) | 0.0040 (7) | 0.0027 (7) | −0.0003 (7) |
| C21 | 0.0199 (9) | 0.0206 (9) | 0.0223 (9) | 0.0028 (7) | 0.0039 (7) | 0.0047 (7) |
| C22 | 0.0156 (9) | 0.0177 (9) | 0.0214 (8) | 0.0036 (7) | 0.0032 (7) | −0.0001 (7) |
| C23 | 0.0217 (9) | 0.0147 (9) | 0.0197 (8) | 0.0030 (7) | 0.0049 (7) | 0.0013 (6) |
| C24 | 0.0237 (10) | 0.0179 (9) | 0.0250 (9) | −0.0003 (7) | 0.0075 (7) | 0.0028 (7) |
| C25 | 0.0301 (11) | 0.0239 (10) | 0.0249 (9) | −0.0028 (8) | −0.0017 (8) | 0.0020 (7) |
| C26 | 0.0461 (13) | 0.0223 (10) | 0.0192 (9) | −0.0019 (9) | 0.0056 (8) | −0.0004 (7) |
| C27 | 0.0328 (11) | 0.0251 (11) | 0.0283 (10) | −0.0008 (8) | 0.0142 (8) | 0.0011 (8) |
| C28 | 0.0209 (9) | 0.0203 (10) | 0.0265 (9) | 0.0010 (7) | 0.0040 (7) | −0.0014 (7) |
Geometric parameters (Å, °)
| Cl1—C3 | 1.7265 (18) | C5—H5 | 0.9500 |
| Cl2—C4 | 1.7320 (18) | C6—C7 | 1.386 (2) |
| Cl3—C6 | 1.7317 (19) | C7—H7 | 0.9500 |
| Cl4—C17 | 1.7280 (18) | C8—C9 | 1.493 (2) |
| Cl5—C18 | 1.7228 (18) | C9—C10 | 1.387 (2) |
| Cl6—C20 | 1.7362 (19) | C9—C14 | 1.388 (3) |
| F1—C10 | 1.361 (2) | C10—C11 | 1.372 (3) |
| F2—C14 | 1.350 (2) | C11—C12 | 1.377 (3) |
| F3—C24 | 1.357 (2) | C11—H11 | 0.9500 |
| F4—C28 | 1.355 (2) | C12—C13 | 1.383 (3) |
| O1—C1 | 1.218 (2) | C12—H12 | 0.9500 |
| O2—C8 | 1.224 (2) | C13—C14 | 1.367 (3) |
| O3—C15 | 1.222 (2) | C13—H13 | 0.9500 |
| O4—C22 | 1.224 (2) | C16—C21 | 1.391 (2) |
| N1—C1 | 1.355 (2) | C16—C17 | 1.403 (2) |
| N1—C2 | 1.403 (2) | C17—C18 | 1.389 (2) |
| N1—H1 | 0.89 (2) | C18—C19 | 1.383 (3) |
| N2—C8 | 1.360 (2) | C19—C20 | 1.382 (2) |
| N2—C1 | 1.402 (2) | C19—H19 | 0.9500 |
| N2—H2 | 0.82 (2) | C20—C21 | 1.386 (2) |
| N3—C15 | 1.354 (2) | C21—H21 | 0.9500 |
| N3—C16 | 1.402 (2) | C22—C23 | 1.493 (2) |
| N3—H3 | 0.80 (2) | C23—C24 | 1.383 (2) |
| N4—C22 | 1.360 (2) | C23—C28 | 1.386 (2) |
| N4—C15 | 1.401 (2) | C24—C25 | 1.369 (2) |
| N4—H4 | 0.91 (2) | C25—C26 | 1.382 (3) |
| C2—C7 | 1.387 (2) | C25—H25 | 0.9500 |
| C2—C3 | 1.408 (2) | C26—C27 | 1.389 (3) |
| C3—C4 | 1.388 (2) | C26—H26 | 0.9500 |
| C4—C5 | 1.377 (3) | C27—C28 | 1.370 (2) |
| C5—C6 | 1.384 (2) | C27—H27 | 0.9500 |
| C1—N1—C2 | 127.03 (16) | C14—C13—C12 | 118.06 (19) |
| C1—N1—H1 | 113.2 (13) | C14—C13—H13 | 121.0 |
| C2—N1—H1 | 119.6 (13) | C12—C13—H13 | 121.0 |
| C8—N2—C1 | 128.23 (15) | F2—C14—C13 | 118.90 (17) |
| C8—N2—H2 | 117.9 (15) | F2—C14—C9 | 117.26 (16) |
| C1—N2—H2 | 113.4 (15) | C13—C14—C9 | 123.81 (18) |
| C15—N3—C16 | 128.03 (16) | O3—C15—N3 | 125.68 (16) |
| C15—N3—H3 | 116.0 (16) | O3—C15—N4 | 119.67 (15) |
| C16—N3—H3 | 115.9 (16) | N3—C15—N4 | 114.65 (15) |
| C22—N4—C15 | 128.67 (14) | C21—C16—N3 | 124.02 (15) |
| C22—N4—H4 | 116.5 (14) | C21—C16—C17 | 119.34 (16) |
| C15—N4—H4 | 114.8 (14) | N3—C16—C17 | 116.64 (15) |
| O1—C1—N1 | 125.96 (17) | C18—C17—C16 | 120.11 (16) |
| O1—C1—N2 | 119.17 (15) | C18—C17—Cl4 | 120.36 (14) |
| N1—C1—N2 | 114.85 (15) | C16—C17—Cl4 | 119.53 (13) |
| C7—C2—N1 | 123.82 (15) | C19—C18—C17 | 120.96 (16) |
| C7—C2—C3 | 119.40 (16) | C19—C18—Cl5 | 119.04 (13) |
| N1—C2—C3 | 116.79 (16) | C17—C18—Cl5 | 120.00 (14) |
| C4—C3—C2 | 119.77 (17) | C20—C19—C18 | 118.02 (16) |
| C4—C3—Cl1 | 120.30 (14) | C20—C19—H19 | 121.0 |
| C2—C3—Cl1 | 119.93 (14) | C18—C19—H19 | 121.0 |
| C5—C4—C3 | 121.06 (16) | C19—C20—C21 | 122.72 (17) |
| C5—C4—Cl2 | 118.68 (14) | C19—C20—Cl6 | 118.33 (13) |
| C3—C4—Cl2 | 120.25 (15) | C21—C20—Cl6 | 118.94 (14) |
| C4—C5—C6 | 118.41 (17) | C20—C21—C16 | 118.84 (16) |
| C4—C5—H5 | 120.8 | C20—C21—H21 | 120.6 |
| C6—C5—H5 | 120.8 | C16—C21—H21 | 120.6 |
| C5—C6—C7 | 122.24 (17) | O4—C22—N4 | 124.46 (16) |
| C5—C6—Cl3 | 119.04 (14) | O4—C22—C23 | 121.38 (15) |
| C7—C6—Cl3 | 118.72 (14) | N4—C22—C23 | 114.16 (14) |
| C6—C7—C2 | 119.11 (16) | C24—C23—C28 | 115.82 (16) |
| C6—C7—H7 | 120.4 | C24—C23—C22 | 120.91 (15) |
| C2—C7—H7 | 120.4 | C28—C23—C22 | 123.25 (16) |
| O2—C8—N2 | 123.81 (17) | F3—C24—C25 | 118.75 (16) |
| O2—C8—C9 | 120.80 (16) | F3—C24—C23 | 117.71 (15) |
| N2—C8—C9 | 115.39 (15) | C25—C24—C23 | 123.53 (17) |
| C10—C9—C14 | 115.37 (17) | C24—C25—C26 | 118.19 (17) |
| C10—C9—C8 | 123.65 (16) | C24—C25—H25 | 120.9 |
| C14—C9—C8 | 120.86 (16) | C26—C25—H25 | 120.9 |
| F1—C10—C11 | 119.38 (17) | C25—C26—C27 | 121.07 (17) |
| F1—C10—C9 | 117.38 (17) | C25—C26—H26 | 119.5 |
| C11—C10—C9 | 123.19 (18) | C27—C26—H26 | 119.5 |
| C10—C11—C12 | 118.59 (19) | C28—C27—C26 | 117.96 (17) |
| C10—C11—H11 | 120.7 | C28—C27—H27 | 121.0 |
| C12—C11—H11 | 120.7 | C26—C27—H27 | 121.0 |
| C11—C12—C13 | 120.94 (19) | F4—C28—C27 | 119.34 (16) |
| C11—C12—H12 | 119.5 | F4—C28—C23 | 117.21 (15) |
| C13—C12—H12 | 119.5 | C27—C28—C23 | 123.43 (17) |
| C2—N1—C1—O1 | 3.5 (3) | C16—N3—C15—O3 | 1.6 (3) |
| C2—N1—C1—N2 | −175.52 (16) | C16—N3—C15—N4 | −178.88 (16) |
| C8—N2—C1—O1 | 175.84 (18) | C22—N4—C15—O3 | 175.83 (17) |
| C8—N2—C1—N1 | −5.1 (3) | C22—N4—C15—N3 | −3.8 (3) |
| C1—N1—C2—C7 | −6.0 (3) | C15—N3—C16—C21 | 4.5 (3) |
| C1—N1—C2—C3 | 173.97 (17) | C15—N3—C16—C17 | −176.16 (17) |
| C7—C2—C3—C4 | 1.1 (3) | C21—C16—C17—C18 | 1.0 (3) |
| N1—C2—C3—C4 | −178.90 (16) | N3—C16—C17—C18 | −178.33 (16) |
| C7—C2—C3—Cl1 | −178.25 (13) | C21—C16—C17—Cl4 | −179.16 (13) |
| N1—C2—C3—Cl1 | 1.8 (2) | N3—C16—C17—Cl4 | 1.5 (2) |
| C2—C3—C4—C5 | −0.9 (3) | C16—C17—C18—C19 | −0.8 (3) |
| Cl1—C3—C4—C5 | 178.44 (14) | Cl4—C17—C18—C19 | 179.40 (14) |
| C2—C3—C4—Cl2 | 179.03 (13) | C16—C17—C18—Cl5 | 178.62 (14) |
| Cl1—C3—C4—Cl2 | −1.7 (2) | Cl4—C17—C18—Cl5 | −1.2 (2) |
| C3—C4—C5—C6 | 0.2 (3) | C17—C18—C19—C20 | 0.0 (3) |
| Cl2—C4—C5—C6 | −179.71 (13) | Cl5—C18—C19—C20 | −179.39 (13) |
| C4—C5—C6—C7 | 0.3 (3) | C18—C19—C20—C21 | 0.5 (3) |
| C4—C5—C6—Cl3 | −179.82 (13) | C18—C19—C20—Cl6 | 179.69 (14) |
| C5—C6—C7—C2 | −0.1 (3) | C19—C20—C21—C16 | −0.3 (3) |
| Cl3—C6—C7—C2 | −179.98 (13) | Cl6—C20—C21—C16 | −179.44 (13) |
| N1—C2—C7—C6 | 179.38 (16) | N3—C16—C21—C20 | 178.79 (16) |
| C3—C2—C7—C6 | −0.6 (3) | C17—C16—C21—C20 | −0.5 (3) |
| C1—N2—C8—O2 | −2.4 (3) | C15—N4—C22—O4 | 2.9 (3) |
| C1—N2—C8—C9 | 177.94 (17) | C15—N4—C22—C23 | −177.49 (16) |
| O2—C8—C9—C10 | 121.2 (2) | O4—C22—C23—C24 | 58.7 (2) |
| N2—C8—C9—C10 | −59.1 (2) | N4—C22—C23—C24 | −120.91 (18) |
| O2—C8—C9—C14 | −54.5 (2) | O4—C22—C23—C28 | −119.8 (2) |
| N2—C8—C9—C14 | 125.14 (18) | N4—C22—C23—C28 | 60.5 (2) |
| C14—C9—C10—F1 | 178.06 (16) | C28—C23—C24—F3 | −179.26 (15) |
| C8—C9—C10—F1 | 2.1 (3) | C22—C23—C24—F3 | 2.1 (2) |
| C14—C9—C10—C11 | 0.6 (3) | C28—C23—C24—C25 | −0.8 (3) |
| C8—C9—C10—C11 | −175.38 (18) | C22—C23—C24—C25 | −179.46 (17) |
| F1—C10—C11—C12 | −179.34 (17) | F3—C24—C25—C26 | 178.90 (16) |
| C9—C10—C11—C12 | −1.9 (3) | C23—C24—C25—C26 | 0.5 (3) |
| C10—C11—C12—C13 | 1.7 (3) | C24—C25—C26—C27 | 0.4 (3) |
| C11—C12—C13—C14 | −0.1 (3) | C25—C26—C27—C28 | −0.8 (3) |
| C12—C13—C14—F2 | −179.42 (17) | C26—C27—C28—F4 | 178.83 (16) |
| C12—C13—C14—C9 | −1.3 (3) | C26—C27—C28—C23 | 0.4 (3) |
| C10—C9—C14—F2 | 179.24 (16) | C24—C23—C28—F4 | −178.09 (15) |
| C8—C9—C14—F2 | −4.7 (3) | C22—C23—C28—F4 | 0.5 (3) |
| C10—C9—C14—C13 | 1.1 (3) | C24—C23—C28—C27 | 0.4 (3) |
| C8—C9—C14—C13 | 177.14 (17) | C22—C23—C28—C27 | 178.97 (17) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cl1 | 0.89 (2) | 2.46 (2) | 2.9126 (15) | 111.8 (16) |
| N1—H1···O2 | 0.89 (2) | 1.88 (2) | 2.641 (2) | 141.5 (19) |
| N2—H2···O1i | 0.82 (2) | 2.00 (2) | 2.8205 (19) | 173 (2) |
| N3—H3···Cl4 | 0.80 (2) | 2.43 (2) | 2.8944 (16) | 118.2 (19) |
| N3—H3···O4 | 0.80 (2) | 1.99 (2) | 2.658 (2) | 140 (2) |
| N4—H4···O3ii | 0.91 (2) | 1.93 (2) | 2.8378 (18) | 176 (2) |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x+1, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: JH2065).
References
- Lin, J., Yan, S. J., Mao, D. S., Xu, R., Yang, L. J. & Liu, F. C. (2003). Chin. Chem. Lett.14, 1219–1222.
- Lin, J., Yan, S. J., Yang, L. J., Li, J. F. & Liu, F. C. (2005). Chin. J. Org. Chem.25, 304–307.
- Rigaku (2006). CrystalClear and CrystalStructure Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Yan, S. J., Lin, J., Bi, F. C., Rang, L. J. & Cheng, Y. P. (2003). J. Yunnan Univ.25, 438–441.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808032029/jh2065sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808032029/jh2065Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report