Abstract
The title ionic compound, (C21H27N2)[AuCl4]·CH2Cl2, was obtained from the reaction of 1,3-dimesitylimidazolidinium chloride with t-BuOK and a solution of AuCl3 in tetrahydrofuran. In the crystal structure, numerous weak C—H⋯Cl hydrogen bonds form double layers parallel to (100), which are further stabilized by π–π interactions between mesitylene rings [centroid–centroid distance = 4.308 (4) Å], resulting in the formation of a three-dimensional supramolecular assembly.
Related literature
For related literature, see: Arduengo et al. (1995 ▶); da Costa et al. (2007 ▶); Adé et al. (2004 ▶); Asaji et al. (2004 ▶); Makotchenko et al. (2006 ▶); Brammer et al. (2001 ▶).
Experimental
Crystal data
(C21H27N2)[AuCl4]·CH2Cl2
M r = 731.14
Monoclinic,
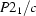
a = 19.590 (3) Å
b = 8.9986 (13) Å
c = 15.306 (2) Å
β = 96.601 (2)°
V = 2680.4 (7) Å3
Z = 4
Mo Kα radiation
μ = 6.10 mm−1
T = 100 (2) K
0.30 × 0.25 × 0.10 mm
Data collection
Bruker APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1997 ▶) T min = 0.146, T max = 0.546
15546 measured reflections
6083 independent reflections
4516 reflections with I > 2σ(I)
R int = 0.094
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.086
S = 0.91
6083 reflections
286 parameters
H-atom parameters constrained
Δρmax = 2.12 e Å−3
Δρmin = −2.24 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: X-SEED.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808031115/hk2528sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808031115/hk2528Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °) (with cut-off parameters as in Brammer et al., 2001 ▶).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9⋯Cl1i | 0.95 | 2.78 | 3.706 (6) | 164 |
| C12—H12C⋯Cl4ii | 0.98 | 2.87 | 3.724 (6) | 147 |
| C15—H15⋯Cl3ii | 0.95 | 2.87 | 3.740 (7) | 152 |
| C15—H15⋯Cl4ii | 0.95 | 2.88 | 3.368 (6) | 113 |
| C17—H17A⋯Cl4 | 0.99 | 3.05 | 3.794 (6) | 133 |
| C17—H17B⋯Cl2iii | 0.99 | 2.95 | 3.736 (6) | 137 |
| C17—H17B⋯Cl3iii | 0.99 | 2.96 | 3.882 (6) | 155 |
| C18—H18B⋯Cl1 | 0.99 | 2.78 | 3.511 (6) | 131 |
| C25—H25A⋯Cl4ii | 0.98 | 2.89 | 3.832 (6) | 162 |
| C25—H25B⋯Cl29iv | 0.98 | 2.88 | 3.742 (7) | 148 |
| C25—H25C⋯Cl2iii | 0.98 | 2.97 | 3.901 (6) | 160 |
| C25—H25C⋯Cl3iii | 0.98 | 3.03 | 3.691 (6) | 126 |
| C26—H26B⋯Cl4 | 0.98 | 2.90 | 3.829 (7) | 158 |
| C27—H27C⋯Cl30v | 0.98 | 3.04 | 3.838 (8) | 140 |
| C28—H28A⋯Cl3 | 0.99 | 2.63 | 3.486 (7) | 145 |
Symmetry codes: (i) 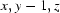 ; (ii)
; (ii) 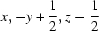 ; (iii)
; (iii) 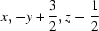 ; (iv)
; (iv) 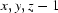 ; (v)
; (v) 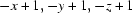 .
.
Acknowledgments
The authors thank the National Research Foundation of South Africa and the University of Stellenbosch for financial support.
supplementary crystallographic information
Comment
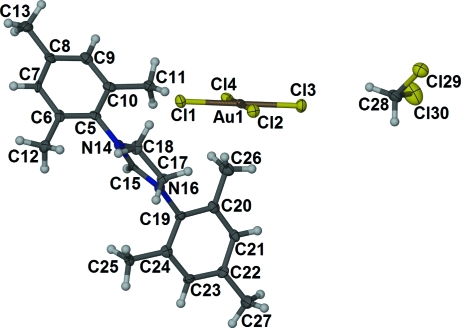
During the course of ongoing studies on imine compounds of gold(III), we have isolated the title ionic compound, (I). The asymmetric unit (Fig. 1) consists of 1,3-dimesitylimidazolidinium cation, tetrachloro-gold(III) anion, and a dichloromethane molecule. To the best of our knowledge, there have been only two reports on crystal structures containing the title carbenium ion, presenting the structure of the 1,3-dimesitylimidazolidinium chloride acetonitrile solvate, (II), (Arduengo et al., 1995) and the imidazolidinium salt of a ruthenium(III) complex, (III) (da Costa et al., 2007), respectively. The structural parameters associated with the carbenium ion are similar to the reported ones. The only difference is the orientation of one of the mesitylene rings (C19-C24), which is almost perpendicular with respect to the plane of the imidazolidinium ring. The dihedral angle between those two planes is 89.5 (3)°, whereas in previous reports both the mesitylene rings were more or less twisted with respect to the plane of the imidazolidinium ring [66.0 (3)° and 75.1 (3)°] for (II) and 82.0 (3)° for (III). This corresponds with the orientation of the other mesitylene ring (C5-C10) [72.1 (3)°] described here.
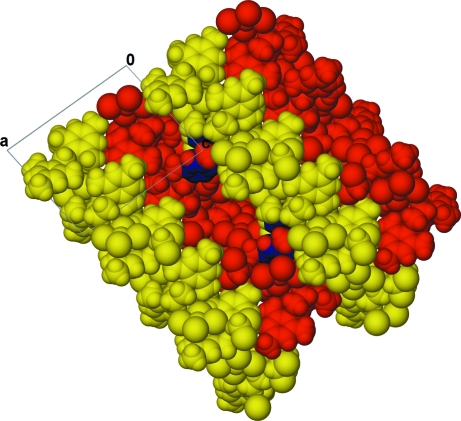
The anionic part displays a typical square-planar geometry around Au and the Au-Cl distances compare well with previously reported values (Adé et al., 2004; Asaji et al., 2004; Makotchenko et al., 2006). All Cl atoms participate in the formation of weak C-H···Cl hydrogen bonds (Table 1) forming double layers in the bc plane [individual layers are linked by C27-H27C···Cl30 bonds with C···Cl distance of 3.838 (8) Å] which are further extended in the third dimension by face-to-face π-π interactions between mesitylene rings (C5-C10) of neighbouring double layers [symmetry code: 2 - x, -y, 1 - z] with centroid-centroid distance of 4.308 (4) Å (Fig. 2).
Experimental
For the preparation of the title compound, 1,3-dimesitylimidazolidinium chloride (0.04 g, 1.2 mmol) in THF (20 ml) was treated with t-BuOK (0.13 g, 1.2 mmol) at room temperature, and then filtered through Celite into a solution of AuCl3 (0.35 g, 1.2 mmol) in THF (20 ml). The solvent was removed under reduced pressure. Orange crystals suitable for single crystal X-ray analysis were obtained from a dichloromethane solution layered with hexane at 253 K.
Refinement
H atoms were positioned geometrically, with C-H = 0.95, 0.99 and 0.98 Å for aromatic, methylene and methyl H, respectively, and constrained to ride on their parent atoms with Uiso(H) = xUeq(C), where x = 1.5 for methyl H and x = 1.2 for all other H atoms.
Figures
Fig. 1.
The molecular structure of the title compound, with the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
Space-filling representation of double layers (yellow-red) extended in the third dimension by π-π interactions (shown in blue).
Crystal data
| (C21H27N2)[AuCl4]·CH2Cl2 | F(000) = 1424 |
| Mr = 731.14 | Dx = 1.812 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3906 reflections |
| a = 19.590 (3) Å | θ = 2.5–27.6° |
| b = 8.9986 (13) Å | µ = 6.10 mm−1 |
| c = 15.306 (2) Å | T = 100 K |
| β = 96.601 (2)° | Plate, orange |
| V = 2680.4 (7) Å3 | 0.30 × 0.25 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker APEX CCD area-detector diffractometer | 6083 independent reflections |
| Radiation source: fine-focus sealed tube | 4516 reflections with I > 2σ(I) |
| graphite | Rint = 0.094 |
| ω scans | θmax = 28.3°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1997) | h = −21→25 |
| Tmin = 0.146, Tmax = 0.546 | k = −9→11 |
| 15546 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.087 | H-atom parameters constrained |
| S = 0.91 | w = 1/[σ2(Fo2) + (0.0211P)2] where P = (Fo2 + 2Fc2)/3 |
| 6083 reflections | (Δ/σ)max = 0.002 |
| 286 parameters | Δρmax = 2.12 e Å−3 |
| 0 restraints | Δρmin = −2.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Au1 | 0.816928 (12) | 0.61599 (3) | 0.706473 (15) | 0.01554 (8) | |
| Cl1 | 0.90506 (8) | 0.57587 (17) | 0.62476 (10) | 0.0221 (4) | |
| Cl2 | 0.86227 (8) | 0.84160 (17) | 0.74770 (10) | 0.0229 (4) | |
| Cl3 | 0.72796 (8) | 0.65262 (18) | 0.78862 (11) | 0.0240 (4) | |
| Cl4 | 0.76938 (8) | 0.39359 (17) | 0.65952 (10) | 0.0220 (3) | |
| C5 | 0.8618 (3) | 0.1217 (7) | 0.3991 (4) | 0.0168 (13) | |
| C6 | 0.9177 (3) | 0.1185 (7) | 0.3509 (4) | 0.0184 (13) | |
| C7 | 0.9615 (3) | −0.0040 (7) | 0.3640 (4) | 0.0179 (14) | |
| H7 | 0.9997 | −0.0101 | 0.3311 | 0.022* | |
| C8 | 0.9517 (3) | −0.1156 (7) | 0.4218 (4) | 0.0223 (14) | |
| C9 | 0.8956 (3) | −0.1060 (7) | 0.4701 (4) | 0.0234 (15) | |
| H9 | 0.8881 | −0.1829 | 0.5106 | 0.028* | |
| C10 | 0.8503 (3) | 0.0132 (7) | 0.4605 (4) | 0.0222 (15) | |
| C11 | 0.7906 (3) | 0.0228 (8) | 0.5168 (4) | 0.0259 (16) | |
| H11C | 0.7899 | 0.1217 | 0.5434 | 0.039* | |
| H11A | 0.7965 | −0.0526 | 0.5632 | 0.039* | |
| H11B | 0.7471 | 0.0055 | 0.4796 | 0.039* | |
| C12 | 0.9314 (3) | 0.2390 (7) | 0.2867 (4) | 0.0228 (15) | |
| H12C | 0.8938 | 0.2418 | 0.2386 | 0.034* | |
| H12A | 0.9747 | 0.2184 | 0.2628 | 0.034* | |
| H12B | 0.9345 | 0.3352 | 0.3169 | 0.034* | |
| C13 | 0.9994 (4) | −0.2463 (8) | 0.4345 (4) | 0.0299 (17) | |
| H13B | 0.9829 | −0.3258 | 0.3937 | 0.045* | |
| H13C | 1.0010 | −0.2824 | 0.4952 | 0.045* | |
| H13A | 1.0456 | −0.2161 | 0.4231 | 0.045* | |
| N14 | 0.8162 (3) | 0.2491 (6) | 0.3895 (3) | 0.0171 (12) | |
| C15 | 0.7516 (3) | 0.2431 (7) | 0.3605 (4) | 0.0161 (13) | |
| H15 | 0.7305 | 0.1576 | 0.3327 | 0.019* | |
| N16 | 0.7176 (3) | 0.3665 (5) | 0.3729 (3) | 0.0168 (11) | |
| C17 | 0.7660 (3) | 0.4782 (7) | 0.4162 (4) | 0.0170 (14) | |
| H17A | 0.7505 | 0.5136 | 0.4719 | 0.020* | |
| H17B | 0.7707 | 0.5644 | 0.3772 | 0.020* | |
| C18 | 0.8333 (3) | 0.3929 (7) | 0.4333 (4) | 0.0210 (14) | |
| H18A | 0.8707 | 0.4441 | 0.4069 | 0.025* | |
| H18B | 0.8471 | 0.3795 | 0.4971 | 0.025* | |
| C19 | 0.6464 (3) | 0.4010 (7) | 0.3447 (4) | 0.0165 (13) | |
| C20 | 0.5985 (3) | 0.3755 (7) | 0.4036 (4) | 0.0207 (14) | |
| C21 | 0.5323 (3) | 0.4244 (7) | 0.3781 (4) | 0.0191 (14) | |
| H21 | 0.4983 | 0.4097 | 0.4168 | 0.023* | |
| C22 | 0.5134 (3) | 0.4949 (7) | 0.2971 (4) | 0.0209 (15) | |
| C23 | 0.5620 (3) | 0.5118 (7) | 0.2400 (4) | 0.0210 (15) | |
| H23 | 0.5492 | 0.5574 | 0.1845 | 0.025* | |
| C24 | 0.6297 (3) | 0.4639 (7) | 0.2613 (4) | 0.0202 (15) | |
| C25 | 0.6811 (3) | 0.4777 (7) | 0.1960 (4) | 0.0230 (15) | |
| H25C | 0.7222 | 0.5291 | 0.2236 | 0.034* | |
| H25B | 0.6609 | 0.5347 | 0.1449 | 0.034* | |
| H25A | 0.6938 | 0.3785 | 0.1771 | 0.034* | |
| C26 | 0.6168 (4) | 0.2988 (8) | 0.4893 (4) | 0.0303 (17) | |
| H26C | 0.5780 | 0.3048 | 0.5243 | 0.045* | |
| H26B | 0.6571 | 0.3467 | 0.5212 | 0.045* | |
| H26A | 0.6272 | 0.1942 | 0.4787 | 0.045* | |
| C27 | 0.4414 (3) | 0.5523 (9) | 0.2740 (5) | 0.0317 (18) | |
| H27C | 0.4317 | 0.5619 | 0.2100 | 0.048* | |
| H27A | 0.4369 | 0.6496 | 0.3015 | 0.048* | |
| H27B | 0.4087 | 0.4826 | 0.2955 | 0.048* | |
| C28 | 0.6210 (4) | 0.7360 (8) | 0.9489 (4) | 0.0303 (17) | |
| H28B | 0.6054 | 0.8168 | 0.9075 | 0.036* | |
| H28A | 0.6592 | 0.6833 | 0.9251 | 0.036* | |
| Cl29 | 0.65120 (9) | 0.8137 (2) | 1.05262 (12) | 0.0335 (4) | |
| Cl30 | 0.55384 (12) | 0.6129 (3) | 0.95642 (15) | 0.0580 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Au1 | 0.01694 (12) | 0.01813 (12) | 0.01117 (12) | −0.00007 (12) | 0.00005 (8) | 0.00093 (11) |
| Cl1 | 0.0243 (8) | 0.0229 (8) | 0.0202 (8) | −0.0012 (7) | 0.0073 (7) | −0.0006 (6) |
| Cl2 | 0.0252 (9) | 0.0197 (8) | 0.0236 (9) | −0.0021 (7) | 0.0019 (7) | −0.0038 (6) |
| Cl3 | 0.0232 (8) | 0.0241 (9) | 0.0263 (9) | 0.0015 (7) | 0.0089 (7) | 0.0007 (7) |
| Cl4 | 0.0268 (8) | 0.0220 (8) | 0.0167 (8) | −0.0060 (7) | 0.0004 (6) | −0.0012 (7) |
| C5 | 0.019 (3) | 0.019 (3) | 0.010 (3) | −0.002 (3) | −0.004 (2) | −0.002 (3) |
| C6 | 0.014 (3) | 0.027 (4) | 0.014 (3) | −0.002 (3) | −0.002 (2) | −0.008 (3) |
| C7 | 0.011 (3) | 0.028 (4) | 0.015 (3) | 0.004 (3) | 0.002 (3) | −0.009 (3) |
| C8 | 0.032 (4) | 0.024 (3) | 0.010 (3) | 0.007 (3) | −0.003 (3) | −0.004 (3) |
| C9 | 0.029 (4) | 0.024 (4) | 0.017 (3) | −0.003 (3) | 0.001 (3) | −0.001 (3) |
| C10 | 0.024 (4) | 0.027 (4) | 0.015 (4) | 0.000 (3) | −0.004 (3) | 0.001 (3) |
| C11 | 0.027 (4) | 0.031 (4) | 0.020 (4) | 0.006 (3) | 0.006 (3) | 0.007 (3) |
| C12 | 0.015 (3) | 0.027 (4) | 0.027 (4) | 0.001 (3) | 0.005 (3) | 0.002 (3) |
| C13 | 0.036 (4) | 0.039 (4) | 0.014 (4) | 0.013 (4) | −0.003 (3) | −0.003 (3) |
| N14 | 0.014 (3) | 0.025 (3) | 0.012 (3) | 0.001 (2) | 0.000 (2) | −0.002 (2) |
| C15 | 0.021 (3) | 0.019 (3) | 0.008 (3) | −0.005 (3) | 0.002 (3) | 0.006 (2) |
| N16 | 0.017 (3) | 0.015 (3) | 0.018 (3) | 0.000 (2) | 0.002 (2) | −0.004 (2) |
| C17 | 0.015 (3) | 0.017 (3) | 0.017 (3) | −0.002 (3) | −0.004 (3) | −0.003 (3) |
| C18 | 0.013 (3) | 0.025 (4) | 0.025 (3) | −0.007 (3) | 0.002 (3) | −0.006 (3) |
| C19 | 0.018 (3) | 0.014 (3) | 0.017 (3) | 0.003 (3) | 0.001 (3) | 0.001 (3) |
| C20 | 0.025 (3) | 0.019 (3) | 0.018 (3) | −0.003 (3) | 0.003 (3) | −0.004 (3) |
| C21 | 0.017 (3) | 0.024 (4) | 0.017 (3) | −0.006 (3) | 0.004 (3) | 0.000 (3) |
| C22 | 0.011 (3) | 0.025 (4) | 0.026 (4) | −0.002 (3) | 0.000 (3) | −0.001 (3) |
| C23 | 0.022 (4) | 0.023 (4) | 0.018 (4) | 0.008 (3) | −0.002 (3) | 0.000 (3) |
| C24 | 0.021 (4) | 0.020 (3) | 0.019 (4) | −0.004 (3) | 0.001 (3) | −0.004 (3) |
| C25 | 0.026 (4) | 0.028 (4) | 0.016 (4) | 0.003 (3) | 0.006 (3) | −0.001 (3) |
| C26 | 0.030 (4) | 0.035 (4) | 0.026 (4) | −0.006 (3) | 0.002 (3) | 0.004 (3) |
| C27 | 0.020 (4) | 0.049 (5) | 0.026 (4) | 0.006 (4) | 0.005 (3) | 0.006 (4) |
| C28 | 0.039 (4) | 0.032 (4) | 0.020 (4) | 0.004 (4) | 0.005 (3) | 0.004 (3) |
| Cl29 | 0.0332 (10) | 0.0352 (10) | 0.0312 (10) | −0.0055 (9) | 0.0001 (8) | 0.0011 (8) |
| Cl30 | 0.0640 (15) | 0.0613 (15) | 0.0492 (13) | −0.0310 (13) | 0.0087 (11) | −0.0151 (12) |
Geometric parameters (Å, °)
| Au1—Cl1 | 2.2742 (16) | N16—C17 | 1.484 (7) |
| Au1—Cl2 | 2.2759 (16) | C17—C18 | 1.523 (8) |
| Au1—Cl3 | 2.2872 (16) | C17—H17A | 0.9900 |
| Au1—Cl4 | 2.2881 (16) | C17—H17B | 0.9900 |
| C5—C6 | 1.390 (8) | C18—H18A | 0.9900 |
| C5—C10 | 1.391 (8) | C18—H18B | 0.9900 |
| C5—N14 | 1.449 (8) | C19—C20 | 1.392 (9) |
| C6—C7 | 1.397 (8) | C19—C24 | 1.401 (8) |
| C6—C12 | 1.508 (8) | C20—C21 | 1.383 (9) |
| C7—C8 | 1.367 (9) | C20—C26 | 1.489 (9) |
| C7—H7 | 0.9500 | C21—C22 | 1.404 (9) |
| C8—C9 | 1.397 (9) | C21—H21 | 0.9500 |
| C8—C13 | 1.501 (9) | C22—C23 | 1.373 (9) |
| C9—C10 | 1.389 (9) | C22—C27 | 1.505 (8) |
| C9—H9 | 0.9500 | C23—C24 | 1.397 (8) |
| C10—C11 | 1.533 (9) | C23—H23 | 0.9500 |
| C11—H11C | 0.9800 | C24—C25 | 1.503 (9) |
| C11—H11A | 0.9800 | C25—H25C | 0.9800 |
| C11—H11B | 0.9800 | C25—H25B | 0.9800 |
| C12—H12C | 0.9800 | C25—H25A | 0.9800 |
| C12—H12A | 0.9800 | C26—H26C | 0.9800 |
| C12—H12B | 0.9800 | C26—H26B | 0.9800 |
| C13—H13B | 0.9800 | C26—H26A | 0.9800 |
| C13—H13C | 0.9800 | C27—H27C | 0.9800 |
| C13—H13A | 0.9800 | C27—H27A | 0.9800 |
| N14—C15 | 1.294 (7) | C27—H27B | 0.9800 |
| N14—C18 | 1.478 (8) | C28—Cl30 | 1.734 (7) |
| C15—N16 | 1.320 (8) | C28—Cl29 | 1.773 (7) |
| C15—H15 | 0.9500 | C28—H28B | 0.9900 |
| N16—C19 | 1.446 (7) | C28—H28A | 0.9900 |
| Cl1—Au1—Cl2 | 89.85 (6) | C18—C17—H17A | 111.1 |
| Cl1—Au1—Cl3 | 179.15 (6) | N16—C17—H17B | 111.1 |
| Cl2—Au1—Cl3 | 90.96 (6) | C18—C17—H17B | 111.1 |
| Cl1—Au1—Cl4 | 89.76 (6) | H17A—C17—H17B | 109.1 |
| Cl2—Au1—Cl4 | 177.57 (6) | N14—C18—C17 | 102.5 (4) |
| Cl3—Au1—Cl4 | 89.44 (6) | N14—C18—H18A | 111.3 |
| C6—C5—C10 | 122.8 (6) | C17—C18—H18A | 111.3 |
| C6—C5—N14 | 118.4 (5) | N14—C18—H18B | 111.3 |
| C10—C5—N14 | 118.6 (6) | C17—C18—H18B | 111.3 |
| C5—C6—C7 | 116.7 (6) | H18A—C18—H18B | 109.2 |
| C5—C6—C12 | 122.7 (6) | C20—C19—C24 | 123.6 (6) |
| C7—C6—C12 | 120.6 (6) | C20—C19—N16 | 118.1 (5) |
| C8—C7—C6 | 122.9 (6) | C24—C19—N16 | 118.3 (5) |
| C8—C7—H7 | 118.6 | C21—C20—C19 | 116.4 (6) |
| C6—C7—H7 | 118.6 | C21—C20—C26 | 121.3 (6) |
| C7—C8—C9 | 118.4 (6) | C19—C20—C26 | 122.4 (6) |
| C7—C8—C13 | 121.9 (6) | C20—C21—C22 | 122.5 (6) |
| C9—C8—C13 | 119.7 (6) | C20—C21—H21 | 118.7 |
| C10—C9—C8 | 121.5 (6) | C22—C21—H21 | 118.7 |
| C10—C9—H9 | 119.2 | C23—C22—C21 | 118.7 (6) |
| C8—C9—H9 | 119.2 | C23—C22—C27 | 120.8 (6) |
| C9—C10—C5 | 117.7 (6) | C21—C22—C27 | 120.5 (6) |
| C9—C10—C11 | 120.2 (6) | C22—C23—C24 | 121.8 (6) |
| C5—C10—C11 | 122.2 (6) | C22—C23—H23 | 119.1 |
| C10—C11—H11C | 109.5 | C24—C23—H23 | 119.1 |
| C10—C11—H11A | 109.5 | C23—C24—C19 | 116.9 (6) |
| H11C—C11—H11A | 109.5 | C23—C24—C25 | 120.7 (6) |
| C10—C11—H11B | 109.5 | C19—C24—C25 | 122.4 (6) |
| H11C—C11—H11B | 109.5 | C24—C25—H25C | 109.5 |
| H11A—C11—H11B | 109.5 | C24—C25—H25B | 109.5 |
| C6—C12—H12C | 109.5 | H25C—C25—H25B | 109.5 |
| C6—C12—H12A | 109.5 | C24—C25—H25A | 109.5 |
| H12C—C12—H12A | 109.5 | H25C—C25—H25A | 109.5 |
| C6—C12—H12B | 109.5 | H25B—C25—H25A | 109.5 |
| H12C—C12—H12B | 109.5 | C20—C26—H26C | 109.5 |
| H12A—C12—H12B | 109.5 | C20—C26—H26B | 109.5 |
| C8—C13—H13B | 109.5 | H26C—C26—H26B | 109.5 |
| C8—C13—H13C | 109.5 | C20—C26—H26A | 109.5 |
| H13B—C13—H13C | 109.5 | H26C—C26—H26A | 109.5 |
| C8—C13—H13A | 109.5 | H26B—C26—H26A | 109.5 |
| H13B—C13—H13A | 109.5 | C22—C27—H27C | 109.5 |
| H13C—C13—H13A | 109.5 | C22—C27—H27A | 109.5 |
| C15—N14—C5 | 124.6 (5) | H27C—C27—H27A | 109.5 |
| C15—N14—C18 | 110.7 (5) | C22—C27—H27B | 109.5 |
| C5—N14—C18 | 122.8 (5) | H27C—C27—H27B | 109.5 |
| N14—C15—N16 | 113.9 (6) | H27A—C27—H27B | 109.5 |
| N14—C15—H15 | 123.0 | Cl30—C28—Cl29 | 111.7 (4) |
| N16—C15—H15 | 123.0 | Cl30—C28—H28B | 109.3 |
| C15—N16—C19 | 128.6 (5) | Cl29—C28—H28B | 109.3 |
| C15—N16—C17 | 109.1 (5) | Cl30—C28—H28A | 109.3 |
| C19—N16—C17 | 122.2 (5) | Cl29—C28—H28A | 109.3 |
| N16—C17—C18 | 103.4 (5) | H28B—C28—H28A | 107.9 |
| N16—C17—H17A | 111.1 | ||
| C10—C5—C6—C7 | 2.7 (9) | C19—N16—C17—C18 | −180.0 (5) |
| N14—C5—C6—C7 | 178.1 (5) | C15—N14—C18—C17 | −5.3 (7) |
| C10—C5—C6—C12 | −177.8 (6) | C5—N14—C18—C17 | −170.6 (5) |
| N14—C5—C6—C12 | −2.4 (8) | N16—C17—C18—N14 | 5.5 (6) |
| C5—C6—C7—C8 | −1.1 (9) | C15—N16—C19—C20 | 94.1 (8) |
| C12—C6—C7—C8 | 179.4 (6) | C17—N16—C19—C20 | −91.1 (7) |
| C6—C7—C8—C9 | −0.2 (9) | C15—N16—C19—C24 | −88.5 (8) |
| C6—C7—C8—C13 | 179.9 (6) | C17—N16—C19—C24 | 86.3 (7) |
| C7—C8—C9—C10 | −0.1 (9) | C24—C19—C20—C21 | −3.9 (9) |
| C13—C8—C9—C10 | 179.9 (6) | N16—C19—C20—C21 | 173.3 (5) |
| C8—C9—C10—C5 | 1.6 (9) | C24—C19—C20—C26 | 175.6 (6) |
| C8—C9—C10—C11 | −177.9 (6) | N16—C19—C20—C26 | −7.2 (9) |
| C6—C5—C10—C9 | −3.0 (9) | C19—C20—C21—C22 | 0.6 (9) |
| N14—C5—C10—C9 | −178.4 (5) | C26—C20—C21—C22 | −178.9 (6) |
| C6—C5—C10—C11 | 176.6 (6) | C20—C21—C22—C23 | 2.0 (10) |
| N14—C5—C10—C11 | 1.1 (9) | C20—C21—C22—C27 | −177.2 (6) |
| C6—C5—N14—C15 | 118.6 (7) | C21—C22—C23—C24 | −1.4 (10) |
| C10—C5—N14—C15 | −65.8 (8) | C27—C22—C23—C24 | 177.8 (6) |
| C6—C5—N14—C18 | −78.2 (7) | C22—C23—C24—C19 | −1.6 (9) |
| C10—C5—N14—C18 | 97.4 (7) | C22—C23—C24—C25 | 177.3 (6) |
| C5—N14—C15—N16 | 167.8 (5) | C20—C19—C24—C23 | 4.5 (9) |
| C18—N14—C15—N16 | 2.9 (7) | N16—C19—C24—C23 | −172.8 (5) |
| N14—C15—N16—C19 | 176.4 (6) | C20—C19—C24—C25 | −174.5 (6) |
| N14—C15—N16—C17 | 1.0 (7) | N16—C19—C24—C25 | 8.3 (9) |
| C15—N16—C17—C18 | −4.3 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9···Cl1i | 0.95 | 2.78 | 3.706 (6) | 164 |
| C12—H12C···Cl4ii | 0.98 | 2.87 | 3.724 (6) | 147 |
| C15—H15···Cl3ii | 0.95 | 2.87 | 3.740 (7) | 152 |
| C15—H15···Cl4ii | 0.95 | 2.88 | 3.368 (6) | 113 |
| C17—H17A···Cl4 | 0.99 | 3.05 | 3.794 (6) | 133 |
| C17—H17B···Cl2iii | 0.99 | 2.95 | 3.736 (6) | 137 |
| C17—H17B···Cl3iii | 0.99 | 2.96 | 3.882 (6) | 155 |
| C18—H18B···Cl1 | 0.99 | 2.78 | 3.511 (6) | 131 |
| C25—H25A···Cl4ii | 0.98 | 2.89 | 3.832 (6) | 162 |
| C25—H25B···Cl29iv | 0.98 | 2.88 | 3.742 (7) | 148 |
| C25—H25C···Cl2iii | 0.98 | 2.97 | 3.901 (6) | 160 |
| C25—H25C···Cl3iii | 0.98 | 3.03 | 3.691 (6) | 126 |
| C26—H26B···Cl4 | 0.98 | 2.90 | 3.829 (7) | 158 |
| C27—H27C···Cl30v | 0.98 | 3.04 | 3.838 (8) | 140 |
| C28—H28A···Cl3 | 0.99 | 2.63 | 3.486 (7) | 145 |
Symmetry codes: (i) x, y−1, z; (ii) x, −y+1/2, z−1/2; (iii) x, −y+3/2, z−1/2; (iv) x, y, z−1; (v) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HK2528).
References
- Adé, A., Cerrada, E., Contel, M., Laguna, M., Merino, P. & Tejero, T. (2004). J. Organomet. Chem.689, 1788–1795.
- Arduengo, A. J. III, Goerlich, J. R. & Marshall, W. J. (1995). J. Am. Chem. Soc.117, 11027–11028.
- Asaji, T., Akiyama, E., Tajima, F., Eda, K., Hashimoto, M. & Furukawa, Y. (2004). Polyhedron, 23, 1605–1611.
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Brammer, L., Bruton, E. A. & Sherwood, P. (2001). Cryst. Growth Des.1, 277–290.
- Bruker (2001). SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2002). SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Costa, R. C. da, Hampel, F. & Gladysz, J. A. (2007). Polyhedron, 26, 581–588.
- Makotchenko, E. V., Baidina, I. A. & Naumov, D. Yu. (2006). J. Struct. Chem.47, 499–503.
- Sheldrick, G. M. (1997). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808031115/hk2528sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808031115/hk2528Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report