Abstract
In the title compound, C10H10N4O4, the but-2-enal chain is almost planar, the largest deviation from the mean plane being 0.013 (1) Å, and this plane makes a dihedral angle of 9.95 (24)° with the benzene ring,. Of the two nitro groups, one is twisted with respect to the benzene ring, making a dihedral angle of 5.7 (1)°, whereas the other is nearly in the plane of the benzene ring, with a twist angle of only 0.7 (1)°. This difference is related to the occurence of an intramolecular N—H⋯O hydrogen bond with the O atom of the less twisted nitro group. The NH group is also involved in a weak interaction with the same O atom of a symmetry-related molecule, thus forming a pseudo inversion dimer.
Related literature
For general background, see: Okabe et al. (1993 ▶). For related structures, see: Bolte & Dill (1998 ▶); Ohba (1996 ▶). 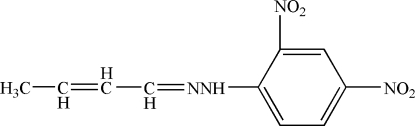
Experimental
Crystal data
C10H10N4O4
M r = 250.22
Monoclinic,

a = 4.6699 (8) Å
b = 13.188 (2) Å
c = 18.880 (3) Å
β = 92.565 (3)°
V = 1161.6 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 293 (2) K
0.27 × 0.23 × 0.23 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 1998 ▶) T min = 0.972, T max = 0.976
9152 measured reflections
2458 independent reflections
1326 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.111
S = 0.95
2458 reflections
164 parameters
H-atom parameters constrained
Δρmax = 0.16 e Å−3
Δρmin = −0.11 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 1998 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPIII (Burnett & Johnson, 1996 ▶), ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033254/dn2390sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033254/dn2390Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O1 | 0.86 | 2.02 | 2.6314 (18) | 127 |
| N2—H2⋯O1i | 0.86 | 2.52 | 3.331 (2) | 159 |
Symmetry code: (i)  .
.
Acknowledgments
The authors express their deep appreciation to the Outstanding Youth Fund for Henan Natural Scientific Research (grant No. 0512001100) and the Fund for Scientific and Technical Emphasis (grant No. 072102270006)
supplementary crystallographic information
Comment
2,4-Dinitrophenylhydrazine has applications in organic synthesis and some of its derivatives have been shown to be potentially DNA-damaging and mutagenic agents (Okabe et al., 1993). Some phenylhydrazone derivatives have been synthesized in our laboratory. As part of our work, we report the synthesis and crystal structure of the title compound(I).
In the title compound, the but-2-enal chain is planar with the largest deviation from the mean plane being 0.013 (1)Å at C1. This plane makes a dihedral angle of 9.95 (24)° with the benzene ring, so the whole molecule is roughly planar (Fig. 1). Of the the two nitro groups, one is twisted with respect to the benzene ring making a dihedral angle of 5.7 (1)° whereas the other is nearly in the plane of the benzene ring with a twist angle of only 0.7 (1)°. This difference is related to the occurence of an intramolecular hydrogen N-H···bond with the O atom of the less twisted nitro group (Table 1). The NH is also in weak intermolecular interaction with the same O atom of a symmetry related molecule building a pseudo dimer (Table 1, Fig. 2).
Bond lengths and bond angles are consistent with those of other dinitrophenylhydrazone derivatives(Ohba,1996; Bolte & Dill, 1998)
Experimental
2,4-Dinitrophenylhydrazine (1 mmol, 0.198 g) was dissolved in anhydrous methanol, H2SO4 (98% 0.5 ml) was added to this, the mixture was stirred for several minitutes at 351 K, but-2-enal (1 mmol 0.070 g) in methanol (8 ml) was added dropwise and the mixture was stirred at refluxing temperature for 2 h. The product was isolated and recrystallized in methanol, red single crystals of (I) was obtained after two weeks.
Refinement
H atoms were placed in calculated position and treated as riding with C—H= 0.93Å (aromatic), 0.96Å(methyl) and N-H= 0.86\%A with Uiso(H)=1.2Ueq(C,N) or Uiso(H)=1.5Ueq(methyl).
Figures
Fig. 1.
Molecular view of (I) with the atom-labeling scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are presented as small spheres of arbitrary radii. Intramolecular H bond is shown as dashed line.
Fig. 2.
Partial packing view showing the formation of pseudo dimer through weak intermolecular N-H···O hydrogen bonds which are shown as dashed lines. H atoms not involved in hydrogen bondings have been omitted for clarity.[Symmetry code: (i) -x+1, -y+1, -z]
Crystal data
| C10H10N4O4 | F(000) = 520 |
| Mr = 250.22 | Dx = 1.432 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 1673 reflections |
| a = 4.6699 (8) Å | θ = 2.1–25.5° |
| b = 13.188 (2) Å | µ = 0.11 mm−1 |
| c = 18.880 (3) Å | T = 293 K |
| β = 92.565 (3)° | Block, red |
| V = 1161.6 (3) Å3 | 0.27 × 0.23 × 0.23 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 2458 independent reflections |
| Radiation source: sealed tube | 1326 reflections with I > 2σ(I) |
| graphite | Rint = 0.039 |
| φ and ω scans | θmax = 27.0°, θmin = 2.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 1998) | h = −5→5 |
| Tmin = 0.972, Tmax = 0.976 | k = −16→16 |
| 9152 measured reflections | l = −24→24 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.111 | H-atom parameters constrained |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0552P)2] where P = (Fo2 + 2Fc2)/3 |
| 2458 reflections | (Δ/σ)max < 0.001 |
| 164 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.11 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.0380 (3) | 0.35052 (10) | 0.11439 (8) | 0.0607 (4) | |
| N2 | 0.1292 (3) | 0.44685 (9) | 0.09740 (8) | 0.0563 (4) | |
| H2 | 0.2613 | 0.4552 | 0.0676 | 0.068* | |
| N3 | 0.2785 (3) | 0.65667 (11) | 0.05938 (8) | 0.0554 (4) | |
| N4 | −0.3915 (4) | 0.77803 (13) | 0.22849 (9) | 0.0673 (5) | |
| O1 | 0.4042 (3) | 0.58766 (9) | 0.02881 (7) | 0.0693 (4) | |
| O2 | 0.3266 (3) | 0.74610 (9) | 0.04720 (8) | 0.0761 (4) | |
| O3 | −0.5851 (3) | 0.75891 (12) | 0.26884 (9) | 0.0872 (5) | |
| O4 | −0.3125 (3) | 0.86398 (12) | 0.21507 (9) | 0.0909 (5) | |
| C1 | 0.1822 (6) | −0.01443 (14) | 0.09371 (14) | 0.1054 (9) | |
| H1A | 0.0157 | −0.0191 | 0.1216 | 0.158* | |
| H1B | 0.3428 | −0.0449 | 0.1191 | 0.158* | |
| H1C | 0.1469 | −0.0492 | 0.0495 | 0.158* | |
| C2 | 0.2468 (5) | 0.09525 (15) | 0.07954 (12) | 0.0770 (6) | |
| H22 | 0.4003 | 0.1089 | 0.0513 | 0.092* | |
| C3 | 0.1060 (5) | 0.17380 (13) | 0.10341 (10) | 0.0651 (5) | |
| H3 | −0.0512 | 0.1615 | 0.1307 | 0.078* | |
| C4 | 0.1811 (4) | 0.27691 (12) | 0.08969 (10) | 0.0585 (5) | |
| H4 | 0.3370 | 0.2906 | 0.0623 | 0.070* | |
| C5 | 0.0079 (4) | 0.52752 (12) | 0.12821 (9) | 0.0497 (4) | |
| C6 | 0.0733 (4) | 0.63020 (12) | 0.11150 (9) | 0.0506 (4) | |
| C7 | −0.0580 (4) | 0.71065 (13) | 0.14479 (10) | 0.0539 (5) | |
| H7 | −0.0131 | 0.7771 | 0.1330 | 0.065* | |
| C8 | −0.2534 (4) | 0.69186 (13) | 0.19495 (10) | 0.0556 (5) | |
| C9 | −0.3232 (4) | 0.59241 (15) | 0.21334 (10) | 0.0603 (5) | |
| H9 | −0.4562 | 0.5806 | 0.2476 | 0.072* | |
| C10 | −0.1960 (4) | 0.51299 (13) | 0.18104 (10) | 0.0582 (5) | |
| H10 | −0.2439 | 0.4473 | 0.1939 | 0.070* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0727 (10) | 0.0384 (8) | 0.0716 (10) | −0.0054 (7) | 0.0084 (8) | 0.0014 (7) |
| N2 | 0.0651 (9) | 0.0405 (8) | 0.0639 (9) | −0.0042 (7) | 0.0116 (7) | 0.0008 (7) |
| N3 | 0.0681 (10) | 0.0380 (8) | 0.0602 (9) | 0.0009 (7) | 0.0032 (8) | 0.0008 (7) |
| N4 | 0.0677 (11) | 0.0668 (12) | 0.0670 (11) | 0.0122 (9) | −0.0015 (9) | −0.0095 (9) |
| O1 | 0.0902 (10) | 0.0448 (7) | 0.0747 (9) | 0.0027 (7) | 0.0259 (7) | −0.0002 (6) |
| O2 | 0.1022 (11) | 0.0386 (7) | 0.0894 (10) | −0.0035 (7) | 0.0259 (8) | 0.0066 (6) |
| O3 | 0.0782 (10) | 0.0913 (11) | 0.0937 (11) | 0.0151 (8) | 0.0195 (9) | −0.0164 (8) |
| O4 | 0.1150 (13) | 0.0545 (9) | 0.1047 (12) | 0.0163 (9) | 0.0220 (10) | −0.0058 (8) |
| C1 | 0.162 (2) | 0.0418 (11) | 0.110 (2) | 0.0019 (14) | −0.0186 (17) | 0.0004 (12) |
| C2 | 0.1028 (17) | 0.0469 (11) | 0.0809 (14) | 0.0004 (11) | 0.0013 (12) | −0.0004 (10) |
| C3 | 0.0838 (14) | 0.0428 (10) | 0.0687 (12) | −0.0059 (9) | 0.0057 (10) | 0.0026 (9) |
| C4 | 0.0724 (12) | 0.0427 (10) | 0.0606 (11) | −0.0040 (9) | 0.0060 (9) | 0.0011 (8) |
| C5 | 0.0527 (10) | 0.0394 (9) | 0.0564 (11) | −0.0010 (8) | −0.0037 (8) | −0.0010 (7) |
| C6 | 0.0528 (10) | 0.0445 (9) | 0.0543 (10) | −0.0006 (8) | −0.0001 (9) | 0.0012 (8) |
| C7 | 0.0572 (11) | 0.0406 (9) | 0.0632 (11) | 0.0014 (8) | −0.0062 (9) | −0.0002 (8) |
| C8 | 0.0548 (11) | 0.0519 (11) | 0.0596 (11) | 0.0098 (9) | −0.0014 (9) | −0.0042 (9) |
| C9 | 0.0545 (11) | 0.0661 (12) | 0.0605 (12) | 0.0025 (9) | 0.0051 (9) | −0.0002 (9) |
| C10 | 0.0622 (11) | 0.0474 (10) | 0.0649 (12) | −0.0042 (9) | 0.0039 (9) | 0.0056 (9) |
Geometric parameters (Å, °)
| N1—C4 | 1.278 (2) | C2—C3 | 1.317 (3) |
| N1—N2 | 1.3821 (17) | C2—H22 | 0.9300 |
| N2—C5 | 1.349 (2) | C3—C4 | 1.431 (2) |
| N2—H2 | 0.8600 | C3—H3 | 0.9300 |
| N3—O2 | 1.2245 (17) | C4—H4 | 0.9300 |
| N3—O1 | 1.2396 (17) | C5—C10 | 1.422 (3) |
| N3—C6 | 1.446 (2) | C5—C6 | 1.427 (2) |
| N4—O4 | 1.2221 (19) | C6—C7 | 1.390 (2) |
| N4—O3 | 1.234 (2) | C7—C8 | 1.367 (3) |
| N4—C8 | 1.465 (2) | C7—H7 | 0.9300 |
| C1—C2 | 1.504 (3) | C8—C9 | 1.399 (3) |
| C1—H1A | 0.9600 | C9—C10 | 1.362 (2) |
| C1—H1B | 0.9600 | C9—H9 | 0.9300 |
| C1—H1C | 0.9600 | C10—H10 | 0.9300 |
| C4—N1—N2 | 116.23 (15) | N1—C4—C3 | 121.30 (18) |
| C5—N2—N1 | 119.02 (14) | N1—C4—H4 | 119.4 |
| C5—N2—H2 | 120.5 | C3—C4—H4 | 119.4 |
| N1—N2—H2 | 120.5 | N2—C5—C10 | 120.21 (15) |
| O2—N3—O1 | 121.65 (15) | N2—C5—C6 | 123.70 (16) |
| O2—N3—C6 | 119.54 (15) | C10—C5—C6 | 116.08 (16) |
| O1—N3—C6 | 118.80 (14) | C7—C6—C5 | 121.43 (16) |
| O4—N4—O3 | 123.62 (17) | C7—C6—N3 | 116.26 (15) |
| O4—N4—C8 | 119.16 (19) | C5—C6—N3 | 122.30 (15) |
| O3—N4—C8 | 117.21 (17) | C8—C7—C6 | 119.78 (16) |
| C2—C1—H1A | 109.5 | C8—C7—H7 | 120.1 |
| C2—C1—H1B | 109.5 | C6—C7—H7 | 120.1 |
| H1A—C1—H1B | 109.5 | C7—C8—C9 | 120.80 (17) |
| C2—C1—H1C | 109.5 | C7—C8—N4 | 118.64 (17) |
| H1A—C1—H1C | 109.5 | C9—C8—N4 | 120.55 (18) |
| H1B—C1—H1C | 109.5 | C10—C9—C8 | 119.93 (18) |
| C3—C2—C1 | 126.1 (2) | C10—C9—H9 | 120.0 |
| C3—C2—H22 | 117.0 | C8—C9—H9 | 120.0 |
| C1—C2—H22 | 117.0 | C9—C10—C5 | 121.97 (16) |
| C2—C3—C4 | 123.7 (2) | C9—C10—H10 | 119.0 |
| C2—C3—H3 | 118.1 | C5—C10—H10 | 119.0 |
| C4—C3—H3 | 118.1 | ||
| C5—N2—N1—C4 | −172.68 (16) | N1—C4—C3—C2 | −179.7 (2) |
| N2—N1—C4—C3 | −179.67 (15) | C4—C3—C2—C1 | 178.41 (19) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1 | 0.86 | 2.02 | 2.6314 (18) | 127 |
| N2—H2···O1i | 0.86 | 2.52 | 3.331 (2) | 159 |
Symmetry codes: (i) −x+1, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2390).
References
- Bolte, M. & Dill, M. (1998). Acta Cryst. C54, IUC9800065.
- Bruker (1998). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Ohba, S. (1996). Acta Cryst. C52, 2118–2119.
- Okabe, N., Nakamura, T. & Fukuda, H. (1993). Acta Cryst. C49, 1678–1680.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808033254/dn2390sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808033254/dn2390Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




