Abstract
The title compound, C20H23Cl2NO, was prepared by condensation of (R)-1-(2-chlorophenyl)-1-cyclopentylmethanamine with 1-(5-chloro-2-hydroxyphenyl)ethanone, resulting in the formation of a new chiral center. The structural analysis confirms the absolute configuration of the title compound and the formation of the (R,R) diastereoisomer. There is an intramolecular O—H⋯N hydrogen bond which stabilizes the conformation of the molecule. The molecules are linked to each other through weak C—H⋯π interactions.
Related literature
For general background, see: Ager et al. (1996 ▶); Berrisford et al. (1995 ▶); Cimarelli & Palmieri (1998 ▶); Cimarelli et al. (2001 ▶, 2002 ▶); Hayase et al. (1997 ▶); Nakano et al. (1997 ▶); Palmieri (1999 ▶, 2000 ▶); Soai & Niwa (1992 ▶); Watanabe et al. (1991 ▶); Xu & Pu (2004 ▶); Yang et al. (2005 ▶).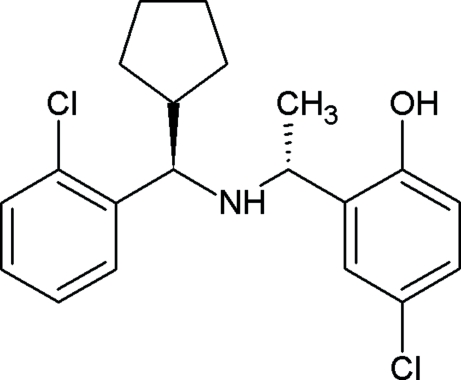
Experimental
Crystal data
C20H23Cl2NO
M r = 364.29
Orthorhombic,

a = 11.286 (2) Å
b = 11.539 (2) Å
c = 14.740 (3) Å
V = 1919.5 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.34 mm−1
T = 298 (2) K
0.42 × 0.29 × 0.18 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 1997 ▶) T min = 0.869, T max = 0.941
10145 measured reflections
3573 independent reflections
2761 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.113
S = 1.02
3573 reflections
219 parameters
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.22 e Å−3
Absolute structure: Flack (1983 ▶), 1629 Friedel pairs
Flack parameter: 0.03 (8)
Data collection: SMART (Bruker, 1997 ▶); cell refinement: SAINT (Bruker, 1997 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPIII (Burnett & Johnson, 1996 ▶) and ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808034624/dn2393sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808034624/dn2393Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯N1 | 0.82 | 1.92 | 2.639 (3) | 146 |
| C3—H3⋯Cg1i | 0.93 | 2.76 | 3.661 (3) | 164 |
Symmetry code: (i)  . Cg is the centroid of the C15–C20 benzene ring
. Cg is the centroid of the C15–C20 benzene ring
Acknowledgments
The authors are grateful to the Natural Science Foundation of Shandong Province China (grant No. G0231) and the Foundation of the Education Ministry of China for Returned Students (grant No. G0220) for financial support. The X-ray data were collected at Shandong Normal University, China.
supplementary crystallographic information
Comment
The synthesis of enantiopure amine alcohols with a variety of functionalities is an important subject of research because this class of compounds has found widespread application in biological systems showing pharmacological activity. These compounds are used as resolving agents, chiral bases and auxiliaries in asymmetric synthesis (Cimarelli et al.,2002), and most have been derived from new readily available natural products (Ager et al.,1996). Chiral aminophenols, which are similar to aminoalcohols, are important building blocks in organic synthesis, and have attracted increasing attention in recent years owing to their effects in asymmetric synthesis and asymmetric induction (Cimarelli et al.,2001; Palmieri, 1999, 2000; Xu & Pu, 2004; Berrisford et al., 1995; Cimarelli & Palmieri, 1998; Hayase et al., 1997; Nakano et al.,1997; Soai et al., 1992; Watanabe et al., 1991).
As part of our continuing studies of chiral aminophenols, we have established the molecular structure of the title compound which was intially synthesized to test its asymmetric catalytic activity. The compound has been prepared by conventional condensation of (R)-1-(2-chlorophenyl)-1-cyclopentylmethanamine with 1-(5-chloro-2-hydroxyphenyl)ethanone, resulting in the formation of a new chiral center as shown in Fig. 1.
The structural analyses confirms the absolute configuration of the title compound and the formation of the (R,R) diastereoisomer. There is an intramolecular O-H···N hydrogen bond which stabilizes the conformation of the molecule. The molecules are linked to each other through weak C-H···π interaction involving the C15-C20 benzene ring (Table 1).
Experimental
The title compound were prepared according to the procedure of Yang et al.(2005). (R)-1-(2-chlorophenyl)-1-cyclopentylmethanamine (0.9 mmol) and 1-(5-chloro-2-hydroxyphenyl)ethanone (0.9 mmol) were dissolved in methanol (10 ml) and reacted at room temperature for 48 h. After removal of the solvent, NaBH4 (4.5 mmol) was added to the solution in THF/ethanol (1:1 v/v, 20 ml) and stirred at 273 K until the solution became colourless. The solvent was then removed under reduced pressure. Water (10 ml)was added to the residue and 1 N HCl was added dropwise until hydrogen production ceased. The mixture was neutralized with aqueous Na2CO3 , then extracted with CHCl3, and the organic layer was dried over anhydrous sodium sulfate. The solvent was removed under reduced pressure. Further purification was carried out by thin-layer silica-gel chromatography (chloroform) to give a colourless solid (yield 83.5%). Crystals of (I) were grown from a n-hexane.
Refinement
All H atoms were included in calculated positions and treated as riding on their parent atoms, with N—H = 0.90 Å, O—H = 0.82 Å, aromatic C—H = 0.93 Å, methyl C—H =0.96 Å, methylene C—H =0.97 Å and methine C—H = 0.98 Å, and with Uiso(H) = 1.2Ueq(C,N,O)or 1.5Ueq(methyl C).
Figures
Fig. 1.
Molecular view of (I) with the atom-labeling scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are shown as small spheres of arbitrary radii. H bond is shown as dashed line.
Crystal data
| C20H23Cl2NO | F(000) = 768 |
| Mr = 364.29 | Dx = 1.261 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2608 reflections |
| a = 11.286 (2) Å | θ = 2.2–20.5° |
| b = 11.539 (2) Å | µ = 0.34 mm−1 |
| c = 14.740 (3) Å | T = 298 K |
| V = 1919.5 (6) Å3 | Block, colourless |
| Z = 4 | 0.42 × 0.29 × 0.18 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3573 independent reflections |
| Radiation source: fine-focus sealed tube | 2761 reflections with I > 2σ(I) |
| graphite | Rint = 0.032 |
| φ and ω scans | θmax = 25.5°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 1997) | h = −13→13 |
| Tmin = 0.869, Tmax = 0.941 | k = −13→11 |
| 10145 measured reflections | l = −17→15 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.048 | H-atom parameters constrained |
| wR(F2) = 0.113 | w = 1/[σ2(Fo2) + (0.0486P)2 + 0.3378P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 3573 reflections | Δρmax = 0.17 e Å−3 |
| 219 parameters | Δρmin = −0.22 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1629 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.03 (8) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.4448 (2) | 0.9053 (2) | 0.71545 (19) | 0.0527 (7) | |
| C2 | 0.5587 (2) | 0.9243 (2) | 0.68485 (16) | 0.0450 (6) | |
| C3 | 0.5716 (3) | 1.0086 (2) | 0.61779 (19) | 0.0583 (7) | |
| H3 | 0.6468 | 1.0242 | 0.5952 | 0.070* | |
| C4 | 0.4774 (3) | 1.0688 (3) | 0.5843 (2) | 0.0662 (9) | |
| H4 | 0.4891 | 1.1245 | 0.5395 | 0.079* | |
| C5 | 0.3652 (3) | 1.0476 (3) | 0.6164 (2) | 0.0702 (9) | |
| H5 | 0.3007 | 1.0886 | 0.5939 | 0.084* | |
| C6 | 0.3501 (3) | 0.9649 (3) | 0.6822 (2) | 0.0679 (9) | |
| H6 | 0.2746 | 0.9494 | 0.7043 | 0.082* | |
| C7 | 0.6676 (2) | 0.8606 (2) | 0.71942 (18) | 0.0494 (7) | |
| H7 | 0.6412 | 0.8068 | 0.7667 | 0.059* | |
| C8 | 0.7239 (3) | 0.7890 (3) | 0.6445 (2) | 0.0597 (8) | |
| H8 | 0.7496 | 0.8417 | 0.5963 | 0.072* | |
| C9 | 0.8298 (3) | 0.7153 (3) | 0.6731 (3) | 0.0842 (11) | |
| H9A | 0.9015 | 0.7616 | 0.6749 | 0.101* | |
| H9B | 0.8168 | 0.6814 | 0.7324 | 0.101* | |
| C10 | 0.8389 (4) | 0.6226 (4) | 0.6016 (3) | 0.1137 (16) | |
| H10A | 0.8922 | 0.6467 | 0.5536 | 0.136* | |
| H10B | 0.8687 | 0.5512 | 0.6278 | 0.136* | |
| C11 | 0.7165 (4) | 0.6053 (4) | 0.5653 (3) | 0.1091 (15) | |
| H11A | 0.7171 | 0.6091 | 0.4995 | 0.131* | |
| H11B | 0.6863 | 0.5301 | 0.5834 | 0.131* | |
| C12 | 0.6404 (3) | 0.6996 (3) | 0.6033 (2) | 0.0747 (10) | |
| H12A | 0.5877 | 0.6688 | 0.6493 | 0.090* | |
| H12B | 0.5930 | 0.7345 | 0.5557 | 0.090* | |
| C13 | 0.7159 (3) | 1.0138 (3) | 0.83381 (19) | 0.0558 (7) | |
| H13 | 0.6433 | 1.0529 | 0.8140 | 0.067* | |
| C14 | 0.8106 (4) | 1.1051 (3) | 0.8523 (2) | 0.0835 (11) | |
| H14A | 0.8828 | 1.0677 | 0.8705 | 0.125* | |
| H14B | 0.7842 | 1.1557 | 0.8999 | 0.125* | |
| H14C | 0.8245 | 1.1494 | 0.7983 | 0.125* | |
| C15 | 0.6889 (3) | 0.9446 (3) | 0.91861 (19) | 0.0537 (7) | |
| C16 | 0.6008 (3) | 0.9804 (3) | 0.9767 (2) | 0.0606 (8) | |
| H16 | 0.5558 | 1.0452 | 0.9620 | 0.073* | |
| C17 | 0.5781 (3) | 0.9215 (4) | 1.0566 (2) | 0.0757 (10) | |
| C18 | 0.6416 (5) | 0.8248 (4) | 1.0777 (3) | 0.0969 (15) | |
| H18 | 0.6254 | 0.7844 | 1.1308 | 0.116* | |
| C19 | 0.7279 (4) | 0.7875 (3) | 1.0216 (3) | 0.0909 (14) | |
| H19 | 0.7700 | 0.7207 | 1.0362 | 0.109* | |
| C20 | 0.7551 (3) | 0.8472 (3) | 0.9425 (2) | 0.0669 (9) | |
| Cl1 | 0.41737 (8) | 0.79949 (9) | 0.79694 (7) | 0.0876 (3) | |
| Cl2 | 0.47184 (9) | 0.97355 (14) | 1.13076 (7) | 0.1212 (5) | |
| N1 | 0.75723 (19) | 0.9378 (2) | 0.76056 (16) | 0.0590 (6) | |
| O1 | 0.8465 (2) | 0.8094 (2) | 0.89205 (18) | 0.0974 (9) | |
| H1A | 0.8471 | 0.8437 | 0.8433 | 0.146* | |
| H1 | 0.7853 | 0.9784 | 0.7146 | 0.117* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0501 (16) | 0.0547 (17) | 0.0533 (16) | −0.0074 (14) | 0.0004 (14) | 0.0055 (13) |
| C2 | 0.0518 (15) | 0.0445 (14) | 0.0386 (14) | −0.0076 (13) | −0.0041 (12) | −0.0018 (12) |
| C3 | 0.0571 (17) | 0.0609 (18) | 0.0567 (17) | −0.0135 (15) | −0.0047 (15) | 0.0086 (15) |
| C4 | 0.088 (2) | 0.0532 (19) | 0.0577 (19) | −0.0095 (18) | −0.0138 (18) | 0.0100 (15) |
| C5 | 0.074 (2) | 0.063 (2) | 0.074 (2) | 0.0047 (18) | −0.0227 (19) | 0.0004 (19) |
| C6 | 0.0472 (15) | 0.078 (2) | 0.079 (2) | −0.0036 (16) | −0.0035 (15) | 0.0018 (19) |
| C7 | 0.0468 (15) | 0.0528 (16) | 0.0485 (16) | −0.0063 (13) | −0.0026 (13) | 0.0001 (13) |
| C8 | 0.0586 (17) | 0.0647 (19) | 0.0558 (18) | −0.0008 (15) | −0.0021 (15) | −0.0062 (15) |
| C9 | 0.0568 (19) | 0.093 (3) | 0.102 (3) | 0.013 (2) | −0.0081 (18) | −0.028 (2) |
| C10 | 0.088 (3) | 0.124 (3) | 0.129 (4) | 0.024 (3) | −0.010 (3) | −0.059 (3) |
| C11 | 0.098 (3) | 0.097 (3) | 0.132 (4) | 0.014 (3) | −0.008 (3) | −0.052 (3) |
| C12 | 0.068 (2) | 0.077 (2) | 0.079 (2) | 0.0014 (19) | −0.0103 (18) | −0.0250 (19) |
| C13 | 0.0533 (16) | 0.0598 (17) | 0.0544 (17) | −0.0024 (15) | −0.0074 (14) | −0.0045 (14) |
| C14 | 0.099 (3) | 0.082 (2) | 0.069 (2) | −0.028 (2) | −0.001 (2) | −0.0123 (19) |
| C15 | 0.0525 (16) | 0.0569 (17) | 0.0516 (16) | −0.0085 (15) | −0.0162 (14) | −0.0034 (14) |
| C16 | 0.0534 (17) | 0.070 (2) | 0.0587 (19) | −0.0094 (16) | −0.0122 (15) | 0.0018 (16) |
| C17 | 0.070 (2) | 0.095 (3) | 0.062 (2) | −0.035 (2) | −0.0145 (18) | 0.000 (2) |
| C18 | 0.145 (4) | 0.086 (3) | 0.060 (2) | −0.051 (3) | −0.037 (3) | 0.013 (2) |
| C19 | 0.143 (4) | 0.059 (2) | 0.071 (3) | −0.007 (2) | −0.057 (3) | 0.001 (2) |
| C20 | 0.081 (2) | 0.059 (2) | 0.060 (2) | 0.0069 (18) | −0.0332 (18) | −0.0136 (16) |
| Cl1 | 0.0671 (5) | 0.1041 (7) | 0.0916 (6) | −0.0106 (5) | 0.0140 (5) | 0.0452 (5) |
| Cl2 | 0.0834 (6) | 0.2043 (14) | 0.0759 (6) | −0.0444 (8) | 0.0151 (5) | −0.0046 (8) |
| N1 | 0.0475 (13) | 0.0749 (17) | 0.0547 (14) | −0.0100 (13) | −0.0024 (11) | −0.0098 (13) |
| O1 | 0.1023 (19) | 0.102 (2) | 0.0875 (18) | 0.0454 (17) | −0.0382 (15) | −0.0257 (16) |
Geometric parameters (Å, °)
| C1—C6 | 1.363 (4) | C11—H11A | 0.9700 |
| C1—C2 | 1.379 (4) | C11—H11B | 0.9700 |
| C1—Cl1 | 1.740 (3) | C12—H12A | 0.9700 |
| C2—C3 | 1.394 (4) | C12—H12B | 0.9700 |
| C2—C7 | 1.520 (4) | C13—N1 | 1.468 (3) |
| C3—C4 | 1.363 (4) | C13—C15 | 1.515 (4) |
| C3—H3 | 0.9300 | C13—C14 | 1.525 (4) |
| C4—C5 | 1.373 (5) | C13—H13 | 0.9800 |
| C4—H4 | 0.9300 | C14—H14A | 0.9600 |
| C5—C6 | 1.371 (5) | C14—H14B | 0.9600 |
| C5—H5 | 0.9300 | C14—H14C | 0.9600 |
| C6—H6 | 0.9300 | C15—C16 | 1.375 (4) |
| C7—N1 | 1.477 (3) | C15—C20 | 1.394 (4) |
| C7—C8 | 1.518 (4) | C16—C17 | 1.384 (4) |
| C7—H7 | 0.9800 | C16—H16 | 0.9300 |
| C8—C12 | 1.524 (4) | C17—C18 | 1.363 (6) |
| C8—C9 | 1.526 (4) | C17—Cl2 | 1.730 (4) |
| C8—H8 | 0.9800 | C18—C19 | 1.348 (6) |
| C9—C10 | 1.504 (5) | C18—H18 | 0.9300 |
| C9—H9A | 0.9700 | C19—C20 | 1.389 (5) |
| C9—H9B | 0.9700 | C19—H19 | 0.9300 |
| C10—C11 | 1.495 (6) | C20—O1 | 1.344 (4) |
| C10—H10A | 0.9700 | N1—H1 | 0.8826 |
| C10—H10B | 0.9700 | O1—H1A | 0.8200 |
| C11—C12 | 1.495 (5) | ||
| C6—C1—C2 | 122.2 (3) | C10—C11—H11A | 110.2 |
| C6—C1—Cl1 | 117.6 (2) | C12—C11—H11B | 110.2 |
| C2—C1—Cl1 | 120.2 (2) | C10—C11—H11B | 110.2 |
| C1—C2—C3 | 116.1 (3) | H11A—C11—H11B | 108.5 |
| C1—C2—C7 | 124.5 (2) | C11—C12—C8 | 106.7 (3) |
| C3—C2—C7 | 119.4 (2) | C11—C12—H12A | 110.4 |
| C4—C3—C2 | 122.1 (3) | C8—C12—H12A | 110.4 |
| C4—C3—H3 | 118.9 | C11—C12—H12B | 110.4 |
| C2—C3—H3 | 118.9 | C8—C12—H12B | 110.4 |
| C3—C4—C5 | 120.2 (3) | H12A—C12—H12B | 108.6 |
| C3—C4—H4 | 119.9 | N1—C13—C15 | 110.8 (2) |
| C5—C4—H4 | 119.9 | N1—C13—C14 | 108.8 (2) |
| C6—C5—C4 | 118.9 (3) | C15—C13—C14 | 110.9 (2) |
| C6—C5—H5 | 120.6 | N1—C13—H13 | 108.7 |
| C4—C5—H5 | 120.6 | C15—C13—H13 | 108.7 |
| C1—C6—C5 | 120.5 (3) | C14—C13—H13 | 108.7 |
| C1—C6—H6 | 119.7 | C13—C14—H14A | 109.5 |
| C5—C6—H6 | 119.7 | C13—C14—H14B | 109.5 |
| N1—C7—C8 | 109.9 (2) | H14A—C14—H14B | 109.5 |
| N1—C7—C2 | 113.6 (2) | C13—C14—H14C | 109.5 |
| C8—C7—C2 | 111.0 (2) | H14A—C14—H14C | 109.5 |
| N1—C7—H7 | 107.4 | H14B—C14—H14C | 109.5 |
| C8—C7—H7 | 107.4 | C16—C15—C20 | 118.2 (3) |
| C2—C7—H7 | 107.4 | C16—C15—C13 | 120.0 (3) |
| C7—C8—C12 | 113.6 (2) | C20—C15—C13 | 121.7 (3) |
| C7—C8—C9 | 115.5 (3) | C15—C16—C17 | 121.1 (3) |
| C12—C8—C9 | 102.5 (3) | C15—C16—H16 | 119.4 |
| C7—C8—H8 | 108.3 | C17—C16—H16 | 119.4 |
| C12—C8—H8 | 108.3 | C18—C17—C16 | 119.9 (4) |
| C9—C8—H8 | 108.3 | C18—C17—Cl2 | 120.3 (3) |
| C10—C9—C8 | 104.9 (3) | C16—C17—Cl2 | 119.8 (3) |
| C10—C9—H9A | 110.8 | C19—C18—C17 | 120.2 (4) |
| C8—C9—H9A | 110.8 | C19—C18—H18 | 119.9 |
| C10—C9—H9B | 110.8 | C17—C18—H18 | 119.9 |
| C8—C9—H9B | 110.8 | C18—C19—C20 | 121.1 (4) |
| H9A—C9—H9B | 108.8 | C18—C19—H19 | 119.5 |
| C11—C10—C9 | 106.4 (3) | C20—C19—H19 | 119.5 |
| C11—C10—H10A | 110.4 | O1—C20—C19 | 118.2 (3) |
| C9—C10—H10A | 110.4 | O1—C20—C15 | 122.2 (3) |
| C11—C10—H10B | 110.4 | C19—C20—C15 | 119.5 (4) |
| C9—C10—H10B | 110.4 | C13—N1—C7 | 116.4 (2) |
| H10A—C10—H10B | 108.6 | C13—N1—H1 | 111.2 |
| C12—C11—C10 | 107.4 (3) | C7—N1—H1 | 104.5 |
| C12—C11—H11A | 110.2 | C20—O1—H1A | 109.5 |
| C6—C1—C2—C3 | 0.2 (4) | C7—C8—C12—C11 | −154.8 (3) |
| Cl1—C1—C2—C3 | 178.3 (2) | C9—C8—C12—C11 | −29.4 (4) |
| C6—C1—C2—C7 | 179.9 (3) | N1—C13—C15—C16 | −148.4 (2) |
| Cl1—C1—C2—C7 | −1.9 (4) | C14—C13—C15—C16 | 90.6 (3) |
| C1—C2—C3—C4 | 0.0 (4) | N1—C13—C15—C20 | 34.8 (4) |
| C7—C2—C3—C4 | −179.8 (3) | C14—C13—C15—C20 | −86.2 (3) |
| C2—C3—C4—C5 | 0.0 (5) | C20—C15—C16—C17 | −0.1 (4) |
| C3—C4—C5—C6 | −0.2 (5) | C13—C15—C16—C17 | −177.1 (3) |
| C2—C1—C6—C5 | −0.3 (5) | C15—C16—C17—C18 | −1.6 (5) |
| Cl1—C1—C6—C5 | −178.5 (2) | C15—C16—C17—Cl2 | 176.6 (2) |
| C4—C5—C6—C1 | 0.3 (5) | C16—C17—C18—C19 | 1.2 (5) |
| C1—C2—C7—N1 | −119.6 (3) | Cl2—C17—C18—C19 | −177.0 (3) |
| C3—C2—C7—N1 | 60.2 (3) | C17—C18—C19—C20 | 1.0 (6) |
| C1—C2—C7—C8 | 116.1 (3) | C18—C19—C20—O1 | 176.3 (3) |
| C3—C2—C7—C8 | −64.2 (3) | C18—C19—C20—C15 | −2.7 (5) |
| N1—C7—C8—C12 | 175.2 (3) | C16—C15—C20—O1 | −176.8 (3) |
| C2—C7—C8—C12 | −58.3 (3) | C13—C15—C20—O1 | 0.1 (4) |
| N1—C7—C8—C9 | 57.2 (3) | C16—C15—C20—C19 | 2.2 (4) |
| C2—C7—C8—C9 | −176.4 (3) | C13—C15—C20—C19 | 179.1 (3) |
| C7—C8—C9—C10 | 158.9 (3) | C15—C13—N1—C7 | 70.8 (3) |
| C12—C8—C9—C10 | 34.8 (4) | C14—C13—N1—C7 | −166.9 (3) |
| C8—C9—C10—C11 | −27.7 (5) | C8—C7—N1—C13 | 179.4 (2) |
| C9—C10—C11—C12 | 9.1 (5) | C2—C7—N1—C13 | 54.4 (3) |
| C10—C11—C12—C8 | 13.0 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···N1 | 0.82 | 1.92 | 2.639 (3) | 146 |
| C3—H3···Cg1i | 0.93 | 2.76 | 3.661 (3) | 164 |
Symmetry codes: (i) −x+3/2, −y+2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2393).
References
- Ager, D. J., Prakash, I. & Schaad, D. R. (1996). Chem. Rev.96, 835–875. [DOI] [PubMed]
- Berrisford, D. J., Bolm, C. & Sharpless, K. B. (1995). Angew. Chem. Int. Ed. Engl.34, 1059–1070.
- Bruker (1997). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Cimarelli, C., Mazzanti, A., Palmieri, G. & Volpini, E. (2001). J. Org. Chem.66, 4759–4765. [DOI] [PubMed]
- Cimarelli, C. & Palmieri, G. (1998). Tetrahedron, 54, 15711–15720.
- Cimarelli, C., Palmieri, G. & Volpini, E. (2002). Tetrahedron Asymmetry, 13, 2011–2018.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Hayase, T., Sugiyama, T., Suzuki, M., Shibata, T. & Soai, K. (1997). J. Fluorine Chem.84, 1–5.
- Nakano, H., Kumagai, N., Matsuzaki, H., Kabuto, C. & Hongo, V. (1997). Tetrahedron Asymmetry, 8, 1391–1401.
- Palmieri, G. (1999). Eur. J. Org. Chem. pp. 805–811.
- Palmieri, G. (2000). Tetrahedron Asymmetry, 11, 3361–3373.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Soai, K. & Niwa, S. (1992). Chem. Rev.92, 833–856.
- Watanabe, V., Araki, V. & Butsugan, V. (1991). J. Org. Chem.56, 2218–2224.
- Xu, M. H. & Pu, L. (2004). Org. Lett 4, 4555-4557. [DOI] [PubMed]
- Yang, X.-F., Zhang, G.-Y., Zhang, Y., Zhao, J.-Y. & Wang, X.-B. (2005). Acta Cryst. C61, o262–o264. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808034624/dn2393sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808034624/dn2393Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



