Abstract
The two aromatic rings in the title compound, C10H9N3, are inclined at 15.2 (1)° to each other; this opens up the angle at the amino N atom to 130.4 (1)°. The amino N atom forms a hydrogen bond to the 4-N atom of an adjacent molecule to create a chain motif.
Related literature
For the structure of aminopyrazine, see: Chao et al. (1976 ▶). For the structure of 2-pyrazinyl-N-2-nitrophenylaniline; see: Parsons et al. (2006 ▶).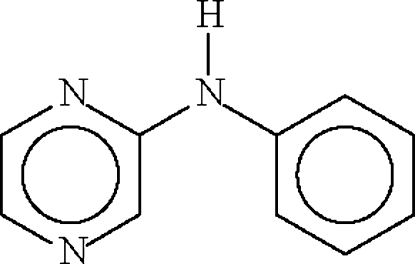
Experimental
Crystal data
C10H9N3
M r = 171.20
Monoclinic,

a = 11.0644 (3) Å
b = 7.8423 (3) Å
c = 10.8907 (3) Å
β = 116.439 (2)°
V = 846.15 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 100 (2) K
0.20 × 0.10 × 0.05 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: none
5664 measured reflections
1934 independent reflections
1463 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.101
S = 1.03
1934 reflections
122 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.23 e Å−3
Δρmin = −0.23 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808031942/pk2121sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808031942/pk2121Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯N2i | 0.89 (1) | 2.12 (1) | 2.977 (2) | 162 (1) |
Symmetry code: (i)  .
.
Acknowledgments
The authors thank the University of Malaya for supporting this study (grant Nos. FS 358/2008A and FA 067/2006A).
supplementary crystallographic information
Comment
There are few structural examples of pyrazine compounds having an amino substituent; these are limited to, for example, aminopyrazine (Chao et al., 1976) and pyrazinyl-N-2-nitrophenylaniline (Parsons et al., 2006). In the title compound (Scheme I, Fig. 1), the two aromatic rings are aligned at 15.2 (1)°; these open up the angle at the amino nitrogen to 130.4 (1) °. The amino nitrogen forms a hydrogen bond to the 4-nitrogen atom of an adjacent molecule to furnish a chain motif.
Experimental
Chloropyrazine (1 ml, 1.1 mmol) and aniline (1 ml, 1.1 mmol) were heated at 423–433 K for 3 h. The solid was dissolved in water. The compound was extracted with ether. The ether extract was dried over sodium sulfate; evaporation of the solvent gave a colorless crystals among some unidentified dark brown materials.
Refinement
Carbon-bound H-atoms were placed in calculated positions (C—H 0.95 Å) and were included in the refinement in the riding model approximation, with U(H) fixed at 1.2U(C). The amino H-atom was located in a difference Fourier map, and was refined with a distance restraint of N—H 0.88 (1) Å.
Figures
Fig. 1.
Thermal ellipsoid plot (Barbour, 2001) of C10H9N3 at the 70% probability level; hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C10H9N3 | F(000) = 360 |
| Mr = 171.20 | Dx = 1.344 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3723 reflections |
| a = 11.0644 (3) Å | θ = 3.3–26.4° |
| b = 7.8423 (3) Å | µ = 0.09 mm−1 |
| c = 10.8907 (3) Å | T = 100 K |
| β = 116.439 (2)° | Prism, colourless |
| V = 846.15 (5) Å3 | 0.20 × 0.10 × 0.05 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 1463 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.033 |
| graphite | θmax = 27.5°, θmin = 3.3° |
| ω scans | h = −14→14 |
| 5664 measured reflections | k = −10→10 |
| 1934 independent reflections | l = −14→14 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.101 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0421P)2 + 0.247P] where P = (Fo2 + 2Fc2)/3 |
| 1934 reflections | (Δ/σ)max = 0.001 |
| 122 parameters | Δρmax = 0.23 e Å−3 |
| 1 restraint | Δρmin = −0.23 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.36080 (11) | 0.49391 (15) | 0.56127 (12) | 0.0188 (3) | |
| H1 | 0.4485 (9) | 0.479 (2) | 0.6153 (13) | 0.024 (4)* | |
| N2 | 0.36741 (11) | 0.89784 (15) | 0.72193 (11) | 0.0205 (3) | |
| N3 | 0.18404 (11) | 0.69151 (16) | 0.51024 (12) | 0.0221 (3) | |
| C1 | 0.29407 (14) | 0.36077 (18) | 0.46992 (13) | 0.0181 (3) | |
| C2 | 0.37632 (14) | 0.23785 (18) | 0.45198 (14) | 0.0201 (3) | |
| H2 | 0.4717 | 0.2479 | 0.5007 | 0.024* | |
| C3 | 0.32046 (15) | 0.1019 (2) | 0.36410 (15) | 0.0246 (3) | |
| H3 | 0.3775 | 0.0188 | 0.3534 | 0.029* | |
| C4 | 0.18124 (15) | 0.0862 (2) | 0.29138 (15) | 0.0255 (3) | |
| H4 | 0.1427 | −0.0058 | 0.2293 | 0.031* | |
| C5 | 0.09930 (14) | 0.20604 (19) | 0.31037 (14) | 0.0235 (3) | |
| H5 | 0.0040 | 0.1948 | 0.2617 | 0.028* | |
| C6 | 0.15425 (14) | 0.34292 (18) | 0.39962 (14) | 0.0204 (3) | |
| H6 | 0.0969 | 0.4236 | 0.4125 | 0.025* | |
| C7 | 0.31216 (13) | 0.64466 (17) | 0.58462 (13) | 0.0174 (3) | |
| C8 | 0.40342 (13) | 0.74982 (17) | 0.69058 (14) | 0.0184 (3) | |
| H8 | 0.4941 | 0.7127 | 0.7412 | 0.022* | |
| C9 | 0.23812 (14) | 0.94629 (19) | 0.64641 (14) | 0.0232 (3) | |
| H9 | 0.2078 | 1.0522 | 0.6649 | 0.028* | |
| C10 | 0.14961 (14) | 0.84350 (19) | 0.54274 (15) | 0.0245 (3) | |
| H10 | 0.0594 | 0.8820 | 0.4914 | 0.029* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0140 (6) | 0.0182 (6) | 0.0186 (6) | 0.0013 (5) | 0.0023 (5) | −0.0019 (5) |
| N2 | 0.0201 (6) | 0.0197 (6) | 0.0210 (6) | −0.0006 (5) | 0.0085 (5) | −0.0008 (5) |
| N3 | 0.0182 (6) | 0.0214 (7) | 0.0220 (6) | 0.0020 (5) | 0.0048 (5) | −0.0008 (5) |
| C1 | 0.0201 (7) | 0.0171 (7) | 0.0147 (6) | −0.0012 (5) | 0.0057 (5) | 0.0009 (5) |
| C2 | 0.0176 (7) | 0.0223 (8) | 0.0194 (7) | −0.0005 (6) | 0.0074 (6) | 0.0000 (6) |
| C3 | 0.0285 (8) | 0.0227 (8) | 0.0257 (8) | −0.0008 (6) | 0.0150 (6) | −0.0045 (6) |
| C4 | 0.0286 (8) | 0.0236 (8) | 0.0233 (7) | −0.0072 (6) | 0.0108 (6) | −0.0075 (6) |
| C5 | 0.0197 (7) | 0.0251 (8) | 0.0214 (7) | −0.0047 (6) | 0.0051 (6) | −0.0003 (6) |
| C6 | 0.0189 (7) | 0.0197 (7) | 0.0194 (7) | −0.0005 (6) | 0.0056 (6) | 0.0008 (6) |
| C7 | 0.0180 (7) | 0.0175 (7) | 0.0164 (7) | −0.0003 (5) | 0.0073 (5) | 0.0019 (5) |
| C8 | 0.0155 (6) | 0.0193 (7) | 0.0187 (7) | 0.0006 (5) | 0.0060 (5) | 0.0019 (5) |
| C9 | 0.0215 (7) | 0.0211 (7) | 0.0250 (7) | 0.0039 (6) | 0.0087 (6) | −0.0015 (6) |
| C10 | 0.0191 (7) | 0.0237 (8) | 0.0266 (8) | 0.0058 (6) | 0.0065 (6) | −0.0001 (6) |
Geometric parameters (Å, °)
| N1—C7 | 1.3689 (17) | C3—H3 | 0.9500 |
| N1—C1 | 1.4039 (17) | C4—C5 | 1.384 (2) |
| N1—H1 | 0.891 (9) | C4—H4 | 0.9500 |
| N2—C8 | 1.3207 (18) | C5—C6 | 1.393 (2) |
| N2—C9 | 1.3488 (17) | C5—H5 | 0.9500 |
| N3—C7 | 1.3335 (17) | C6—H6 | 0.9500 |
| N3—C10 | 1.3458 (19) | C7—C8 | 1.4120 (19) |
| C1—C6 | 1.3944 (18) | C8—H8 | 0.9500 |
| C1—C2 | 1.3978 (19) | C9—C10 | 1.378 (2) |
| C2—C3 | 1.381 (2) | C9—H9 | 0.9500 |
| C2—H2 | 0.9500 | C10—H10 | 0.9500 |
| C3—C4 | 1.389 (2) | ||
| C7—N1—C1 | 130.38 (12) | C4—C5—H5 | 119.5 |
| C7—N1—H1 | 113.3 (10) | C6—C5—H5 | 119.5 |
| C1—N1—H1 | 116.3 (10) | C5—C6—C1 | 119.56 (13) |
| C8—N2—C9 | 116.75 (12) | C5—C6—H6 | 120.2 |
| C7—N3—C10 | 115.67 (12) | C1—C6—H6 | 120.2 |
| C6—C1—C2 | 119.09 (13) | N3—C7—N1 | 121.64 (12) |
| C6—C1—N1 | 124.65 (13) | N3—C7—C8 | 121.03 (12) |
| C2—C1—N1 | 116.25 (12) | N1—C7—C8 | 117.32 (12) |
| C3—C2—C1 | 120.72 (13) | N2—C8—C7 | 122.44 (12) |
| C3—C2—H2 | 119.6 | N2—C8—H8 | 118.8 |
| C1—C2—H2 | 119.6 | C7—C8—H8 | 118.8 |
| C2—C3—C4 | 120.26 (14) | N2—C9—C10 | 120.58 (13) |
| C2—C3—H3 | 119.9 | N2—C9—H9 | 119.7 |
| C4—C3—H3 | 119.9 | C10—C9—H9 | 119.7 |
| C5—C4—C3 | 119.26 (14) | N3—C10—C9 | 123.53 (13) |
| C5—C4—H4 | 120.4 | N3—C10—H10 | 118.2 |
| C3—C4—H4 | 120.4 | C9—C10—H10 | 118.2 |
| C4—C5—C6 | 121.08 (13) | ||
| C7—N1—C1—C6 | −12.7 (2) | C10—N3—C7—N1 | −179.30 (12) |
| C7—N1—C1—C2 | 168.41 (13) | C10—N3—C7—C8 | 0.36 (19) |
| C6—C1—C2—C3 | 1.1 (2) | C1—N1—C7—N3 | −4.2 (2) |
| N1—C1—C2—C3 | −179.89 (12) | C1—N1—C7—C8 | 176.09 (13) |
| C1—C2—C3—C4 | 0.5 (2) | C9—N2—C8—C7 | −0.73 (19) |
| C2—C3—C4—C5 | −1.5 (2) | N3—C7—C8—N2 | 0.3 (2) |
| C3—C4—C5—C6 | 0.9 (2) | N1—C7—C8—N2 | −179.99 (12) |
| C4—C5—C6—C1 | 0.7 (2) | C8—N2—C9—C10 | 0.4 (2) |
| C2—C1—C6—C5 | −1.7 (2) | C7—N3—C10—C9 | −0.7 (2) |
| N1—C1—C6—C5 | 179.40 (13) | N2—C9—C10—N3 | 0.3 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N2i | 0.89 (1) | 2.12 (1) | 2.977 (2) | 162 (1) |
Symmetry codes: (i) −x+1, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PK2121).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chao, M., Schempp, E. & Rosenstein, R. D. (1976). Acta Cryst. B32, 288–290.
- Parsons, S., Wharton, S., McNab, H., Parkin, A. & Johnstone, R. (2006). Private communication (Deposition No. 610410). CCDC, Union Road, Cambridge, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2008). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808031942/pk2121sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808031942/pk2121Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



