Abstract
In the title compound, C15H13Cl2NO2, the dihedral angle between the aromatic rings is 63.80 (12)°. The conformation may be stabilized by a weak N—H⋯O hydrogen bond. In the crystal structure, a short C—Cl⋯π interaction occurs, with a Cl⋯π separation of 3.5706 (13) Å.
Related literature
For general background, see: Hashem et al. (2007 ▶); Husain et al. (2005 ▶). For bond-length data, see: Allen et al. (1987 ▶).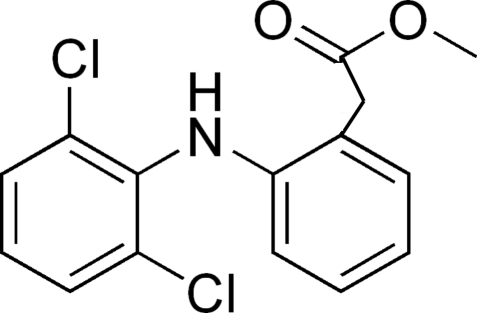
Experimental
Crystal data
C15H13Cl2NO2
M r = 310.16
Monoclinic,

a = 4.9319 (4) Å
b = 20.0288 (14) Å
c = 14.5542 (10) Å
β = 97.711 (1)°
V = 1424.66 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.46 mm−1
T = 173 (2) K
0.38 × 0.24 × 0.20 mm
Data collection
Bruker SMART APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.850, T max = 1.000 (expected range = 0.776–0.913)
8526 measured reflections
3423 independent reflections
2777 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.171
S = 1.04
3423 reflections
181 parameters
H-atom parameters constrained
Δρmax = 0.62 e Å−3
Δρmin = −0.52 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL, PARST (Nardelli, 1995 ▶) and PLATON (Spek, 2003 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808038336/pv2122sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808038336/pv2122Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O1 | 0.88 | 2.64 | 3.152 (2) | 118 |
Acknowledgments
The authors gratefully acknowledge funds from the Higher Education Commission, Islamabad, Pakistan.
supplementary crystallographic information
Comment
Esters are important intermediates in heterocyclic chemistry and have been used for the synthesis of various biologically active five-membered heterocyles such as butenolides, pyrrolones (Husain et al., 2005), oxadiazoles and triazoles (Hashem et al., 2007). In view of the versatility of these compounds, we have synthesized the title compound and report herein its crystal structure.
In the title compound (Fig. 1), the bond lengths and angles are within normal ranges (Allen et al., 1987). The planar ester group (O1/O2/C13/C14/C15) is oriented with respect to the plane of the benzene ring (C7–C12) at an angle 41.33 (2)°. There is a short intramolecular N—H···O hydrogen bond (Table 1) and a π-ring interaction of the type C—Cl···Cg with Cl1···Cg1 (centroid of C1–C6 ring) perpendicular distance 3.5706 (13) Å.
Experimental
A mixture of 2-(2,6-dichlorophenylamino)benzoic acid (2.81 g, 10 mmol) and absolute methanol (50 ml) in the presence of a few drops of sulfuric acid was refluxed for 5 h. The excess of the solvent was removed by distillation. The solid residue was filltered off, washed with water and recystallized from ethanol (30%) to give the title compound. Suitable single crystals of the title compound were obtained by slow evaporation of an ethanol solution at room temperature. (Yield, 88%; m.p. 331–332 K)
Refinement
H atoms were positioned geometrically, with O—H = 0.82 Å and C—H = 0.93, 0.97 and 0.96 Å for aryl, methylene and methyl H, respectively, and constrained to ride on their parent atoms with Uiso(H) = 1.5Ueq(methyl C) and 1.2Ueq(aryl and methylene C and O)
Figures
Fig. 1.
The molecular structure of the title compound, with the atom-numbering scheme; thermal ellipsoids have been plotted at 50% probability level.
Fig. 2.
The formation of the title compound.
Crystal data
| C15H13Cl2NO2 | F000 = 640 |
| Mr = 310.16 | Dx = 1.446 Mg m−3 |
| Monoclinic, P21/n | Melting point: 331 K |
| Hall symbol: -P 2yn | Mo Kα radiation λ = 0.71073 Å |
| a = 4.9319 (4) Å | Cell parameters from 9949 reflections |
| b = 20.0288 (14) Å | θ = 2.4–28.3º |
| c = 14.5542 (10) Å | µ = 0.46 mm−1 |
| β = 97.711 (1)º | T = 173 (2) K |
| V = 1424.66 (18) Å3 | Block, pale yellow |
| Z = 4 | 0.38 × 0.24 × 0.20 mm |
Data collection
| Bruker SMART APEXII CCD diffractometer | 3423 independent reflections |
| Radiation source: fine-focus sealed tube | 2777 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.020 |
| T = 173(2) K | θmax = 28.3º |
| ω and φ scans | θmin = 2.5º |
| Absorption correction: multi-scan(SADABS; Bruker, 2005) | h = −6→6 |
| Tmin = 0.850, Tmax = 1.000 | k = −26→26 |
| 8526 measured reflections | l = −19→15 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.052 | H-atom parameters constrained |
| wR(F2) = 0.171 | w = 1/[σ2(Fo2) + (0.1042P)2 + 0.5254P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 3423 reflections | Δρmax = 0.62 e Å−3 |
| 181 parameters | Δρmin = −0.52 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.8063 (4) | 0.18670 (11) | 0.42183 (15) | 0.0459 (5) | |
| C2 | 0.9559 (6) | 0.23352 (14) | 0.47685 (18) | 0.0592 (7) | |
| H2A | 0.9401 | 0.2361 | 0.5411 | 0.071* | |
| C3 | 1.1298 (6) | 0.27691 (13) | 0.4384 (2) | 0.0646 (7) | |
| H3A | 1.2365 | 0.3084 | 0.4764 | 0.077* | |
| C4 | 1.1462 (6) | 0.27396 (11) | 0.3451 (2) | 0.0572 (6) | |
| H4A | 1.2586 | 0.3046 | 0.3178 | 0.069* | |
| C5 | 0.9985 (5) | 0.22618 (10) | 0.29100 (16) | 0.0454 (5) | |
| C6 | 0.8295 (4) | 0.17922 (9) | 0.32730 (13) | 0.0386 (4) | |
| C7 | 0.6955 (4) | 0.06202 (9) | 0.29122 (13) | 0.0360 (4) | |
| C8 | 0.8988 (5) | 0.03457 (12) | 0.35515 (15) | 0.0468 (5) | |
| H8A | 1.0348 | 0.0624 | 0.3879 | 0.056* | |
| C9 | 0.9027 (6) | −0.03407 (14) | 0.37114 (18) | 0.0614 (7) | |
| H9A | 1.0410 | −0.0529 | 0.4153 | 0.074* | |
| C10 | 0.7088 (7) | −0.07451 (13) | 0.3237 (2) | 0.0676 (8) | |
| H10A | 0.7142 | −0.1213 | 0.3347 | 0.081* | |
| C11 | 0.5040 (5) | −0.04753 (12) | 0.25957 (19) | 0.0549 (6) | |
| H11A | 0.3696 | −0.0759 | 0.2271 | 0.066* | |
| C12 | 0.4948 (4) | 0.02126 (10) | 0.24258 (14) | 0.0388 (4) | |
| C13 | 0.2660 (4) | 0.04998 (12) | 0.17557 (15) | 0.0434 (5) | |
| H13A | 0.1975 | 0.0907 | 0.2033 | 0.052* | |
| H13B | 0.1140 | 0.0173 | 0.1670 | 0.052* | |
| C14 | 0.3438 (4) | 0.06753 (10) | 0.08154 (14) | 0.0374 (4) | |
| C15 | 0.1597 (4) | 0.09199 (12) | −0.07671 (14) | 0.0433 (5) | |
| H15A | −0.0226 | 0.0958 | −0.1125 | 0.065* | |
| H15B | 0.2626 | 0.0566 | −0.1032 | 0.065* | |
| H15C | 0.2570 | 0.1345 | −0.0789 | 0.065* | |
| Cl1 | 0.57692 (13) | 0.13766 (4) | 0.47192 (4) | 0.0602 (2) | |
| Cl2 | 1.01835 (17) | 0.22543 (3) | 0.17271 (4) | 0.0654 (2) | |
| N1 | 0.6851 (4) | 0.13126 (8) | 0.27114 (12) | 0.0425 (4) | |
| H1A | 0.5820 | 0.1447 | 0.2205 | 0.051* | |
| O1 | 0.5844 (3) | 0.07442 (10) | 0.06748 (12) | 0.0591 (5) | |
| O2 | 0.1341 (3) | 0.07623 (9) | 0.01586 (12) | 0.0550 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0417 (11) | 0.0525 (12) | 0.0422 (11) | 0.0138 (9) | 0.0005 (8) | −0.0030 (9) |
| C2 | 0.0595 (15) | 0.0663 (15) | 0.0479 (12) | 0.0187 (12) | −0.0069 (10) | −0.0198 (11) |
| C3 | 0.0656 (16) | 0.0500 (13) | 0.0718 (17) | 0.0071 (11) | −0.0138 (13) | −0.0208 (12) |
| C4 | 0.0564 (14) | 0.0363 (10) | 0.0754 (17) | −0.0006 (9) | −0.0040 (12) | −0.0007 (10) |
| C5 | 0.0512 (12) | 0.0358 (9) | 0.0473 (11) | 0.0055 (8) | −0.0006 (9) | 0.0021 (8) |
| C6 | 0.0373 (10) | 0.0373 (9) | 0.0385 (10) | 0.0064 (7) | −0.0048 (7) | −0.0008 (7) |
| C7 | 0.0370 (9) | 0.0388 (9) | 0.0331 (9) | 0.0030 (7) | 0.0075 (7) | 0.0035 (7) |
| C8 | 0.0460 (11) | 0.0554 (12) | 0.0386 (10) | 0.0100 (9) | 0.0045 (8) | 0.0072 (9) |
| C9 | 0.0735 (17) | 0.0627 (15) | 0.0506 (13) | 0.0289 (13) | 0.0176 (12) | 0.0193 (11) |
| C10 | 0.097 (2) | 0.0419 (12) | 0.0690 (17) | 0.0124 (13) | 0.0313 (16) | 0.0124 (11) |
| C11 | 0.0661 (15) | 0.0420 (11) | 0.0606 (14) | −0.0056 (10) | 0.0230 (12) | 0.0000 (10) |
| C12 | 0.0384 (10) | 0.0412 (10) | 0.0385 (10) | −0.0019 (7) | 0.0111 (8) | 0.0003 (7) |
| C13 | 0.0311 (9) | 0.0565 (12) | 0.0430 (11) | −0.0068 (8) | 0.0073 (8) | −0.0036 (9) |
| C14 | 0.0286 (9) | 0.0407 (9) | 0.0436 (10) | −0.0040 (7) | 0.0075 (7) | −0.0060 (8) |
| C15 | 0.0329 (10) | 0.0601 (12) | 0.0359 (10) | −0.0045 (8) | 0.0005 (7) | 0.0023 (8) |
| Cl1 | 0.0537 (4) | 0.0771 (4) | 0.0521 (4) | 0.0141 (3) | 0.0159 (3) | 0.0074 (3) |
| Cl2 | 0.0911 (5) | 0.0551 (4) | 0.0509 (4) | −0.0072 (3) | 0.0123 (3) | 0.0118 (2) |
| N1 | 0.0489 (10) | 0.0383 (8) | 0.0363 (8) | −0.0016 (7) | −0.0084 (7) | 0.0037 (6) |
| O1 | 0.0273 (7) | 0.1015 (14) | 0.0486 (9) | −0.0057 (7) | 0.0057 (6) | 0.0102 (9) |
| O2 | 0.0413 (8) | 0.0703 (11) | 0.0532 (10) | −0.0023 (7) | 0.0060 (7) | −0.0026 (8) |
Geometric parameters (Å, °)
| C1—C2 | 1.381 (3) | C9—C10 | 1.367 (5) |
| C1—C6 | 1.404 (3) | C9—H9A | 0.9500 |
| C1—Cl1 | 1.731 (3) | C10—C11 | 1.390 (4) |
| C2—C3 | 1.390 (4) | C10—H10A | 0.9500 |
| C2—H2A | 0.9500 | C11—C12 | 1.399 (3) |
| C3—C4 | 1.371 (4) | C11—H11A | 0.9500 |
| C3—H3A | 0.9500 | C12—C13 | 1.503 (3) |
| C4—C5 | 1.383 (3) | C13—C14 | 1.511 (3) |
| C4—H4A | 0.9500 | C13—H13A | 0.9900 |
| C5—C6 | 1.406 (3) | C13—H13B | 0.9900 |
| C5—Cl2 | 1.738 (2) | C14—O1 | 1.239 (2) |
| C6—N1 | 1.393 (3) | C14—O2 | 1.322 (3) |
| C7—C8 | 1.387 (3) | C15—O2 | 1.406 (3) |
| C7—C12 | 1.400 (3) | C15—H15A | 0.9800 |
| C7—N1 | 1.417 (2) | C15—H15B | 0.9800 |
| C8—C9 | 1.394 (4) | C15—H15C | 0.9800 |
| C8—H8A | 0.9500 | N1—H1A | 0.8800 |
| C2—C1—C6 | 122.2 (2) | C9—C10—H10A | 119.8 |
| C2—C1—Cl1 | 118.01 (19) | C11—C10—H10A | 119.8 |
| C6—C1—Cl1 | 119.72 (17) | C10—C11—C12 | 120.3 (2) |
| C1—C2—C3 | 120.1 (2) | C10—C11—H11A | 119.8 |
| C1—C2—H2A | 120.0 | C12—C11—H11A | 119.8 |
| C3—C2—H2A | 120.0 | C11—C12—C7 | 118.7 (2) |
| C4—C3—C2 | 119.5 (2) | C11—C12—C13 | 119.7 (2) |
| C4—C3—H3A | 120.2 | C7—C12—C13 | 121.54 (18) |
| C2—C3—H3A | 120.2 | C12—C13—C14 | 114.64 (16) |
| C3—C4—C5 | 119.8 (3) | C12—C13—H13A | 108.6 |
| C3—C4—H4A | 120.1 | C14—C13—H13A | 108.6 |
| C5—C4—H4A | 120.1 | C12—C13—H13B | 108.6 |
| C4—C5—C6 | 122.8 (2) | C14—C13—H13B | 108.6 |
| C4—C5—Cl2 | 118.4 (2) | H13A—C13—H13B | 107.6 |
| C6—C5—Cl2 | 118.79 (16) | O1—C14—O2 | 122.7 (2) |
| N1—C6—C1 | 123.1 (2) | O1—C14—C13 | 122.73 (18) |
| N1—C6—C5 | 121.52 (19) | O2—C14—C13 | 114.61 (16) |
| C1—C6—C5 | 115.29 (19) | O2—C15—H15A | 109.5 |
| C8—C7—C12 | 120.44 (19) | O2—C15—H15B | 109.5 |
| C8—C7—N1 | 121.95 (19) | H15A—C15—H15B | 109.5 |
| C12—C7—N1 | 117.59 (17) | O2—C15—H15C | 109.5 |
| C7—C8—C9 | 119.7 (2) | H15A—C15—H15C | 109.5 |
| C7—C8—H8A | 120.1 | H15B—C15—H15C | 109.5 |
| C9—C8—H8A | 120.1 | C6—N1—C7 | 123.53 (16) |
| C10—C9—C8 | 120.5 (2) | C6—N1—H1A | 118.2 |
| C10—C9—H9A | 119.8 | C7—N1—H1A | 118.2 |
| C8—C9—H9A | 119.8 | C14—O2—C15 | 124.07 (17) |
| C9—C10—C11 | 120.3 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O1 | 0.88 | 2.64 | 3.152 (2) | 118 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2122).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Hashem, A. I., Youssef, A. S. A., Kandeel, K. A. & Abou-Elmagd, W. S. I. (2007). Eur. J. Med. Chem.42, 934–939. [DOI] [PubMed]
- Husain, A., Khan, M. S. Y., Hasan, S. M. & Alam, M. M. (2005). Eur. J. Med. Chem.40, 1394–1404. [DOI] [PubMed]
- Nardelli, M. (1995). J. Appl. Cryst.28, 659.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808038336/pv2122sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808038336/pv2122Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




