Abstract
The title compound, C20H15Br2NO3, shows the furan ring to adopt a half-chair conformation and the two ring systems to be approximately perpendicular [dihedral angle = 71.0 (2)°]. In the crystal structure, intermolecular C—H⋯O contacts link the molecules.
Related literature
For general background, see: Hillard et al. (2008 ▶); Pinto et al., (1997 ▶); Dos Santos et al. (2001 ▶); Lima et al. (2004 ▶). For related structures and biological activity, see: da Silva Júnior et al. (2007 ▶, 2008 ▶); Lima et al. (2002 ▶). For the synthesis, see: da Silva Júnior et al. (2007 ▶, 2008 ▶). For geometric analysis, see: Cremer & Pople (1975 ▶).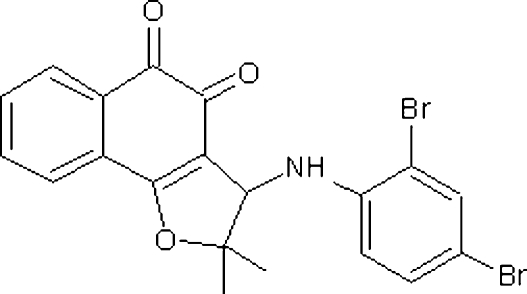
Experimental
Crystal data
C20H15Br2NO3
M r = 477.15
Triclinic,

a = 8.1430 (3) Å
b = 11.2584 (4) Å
c = 11.4742 (5) Å
α = 112.073 (2)°
β = 95.546 (2)°
γ = 108.696 (2)°
V = 894.70 (6) Å3
Z = 2
Mo Kα radiation
μ = 4.55 mm−1
T = 293 (2) K
0.31 × 0.28 × 0.16 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.272, T max = 0.490
9784 measured reflections
4099 independent reflections
3610 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.139
S = 1.09
4070 reflections
235 parameters
H-atom parameters constrained
Δρmax = 0.59 e Å−3
Δρmin = −1.49 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808034545/tk2311sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808034545/tk2311Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O2i | 0.98 | 2.64 | 3.347 (6) | 129 |
| C1′′—H1A⋯O2i | 0.96 | 2.67 | 3.389 (6) | 132 |
Symmetry code: (i)  .
.
Acknowledgments
This work received partial support from CNPq, CAPES, FAPEAL, FAPERJ, IM-INOFAR, CTPETRO and FINEP.
supplementary crystallographic information
Comment
The search for substances with pharmacological activity has grown exponentially in recent years and quinones play a major role as bioreductive drugs, reactive oxygen species (ROS) enhancers, and redox catalysts (Hillard et al., 2008). Substances such as lapachol (I), (Pinto et al., 1997; dos Santos et al., 2001; Lima et al., 2004), β-lapachone (II) (Hillard et al., 2008) and nor-β-lapachone (III) (da Silva Júnior et al., 2007), Scheme 1, are prototypes that can be used as starting points for the synthesis of new bioactive molecules. In this context, different compounds with anti-cancer activity (da Silva Júnior et al., 2007), trypanocides (da Silva Júnior et al., 2008), molluscicides (Lima et al., 2002) among others, were synthesized. The introduction of arylamino groups in the furan ring of III retained/enhanced the anticancer activity against six cancer cell lines with IC50 values below 1µg/ml (da Silva Júnior et al., 2007), as well as intensified trypanocidal activity (da Silva Júnior et al., 2008), demonstrating clearly that the arylamino group is important for pharmacological activity. In this paper, we report the molecular and crystal structure of (IV) that was easily obtained as described in the literature (da Silva Júnior et al., 2007; 2008).
The atoms of the naphthoquinonic ring of (IV), Fig. 1, are co-planar and the greatest deviation from their least-squares plane is exhibited by atom C5 [0.069 (4) Å]. The O1 atom lies in the mean least-square plane of the naphthoquinonic ring with a deviation of 0.051 (3) Å while atoms O2 and O3 are -0.129 (3) and 0.212 (4)Å out of that plane, respectively. The furane ring adopts a half chair conformation and the calculated puckering parameters are: q2 = 0.229 (4) Å and φ = -12 8.04 (1)° (Cremer & Pople, 1975). The dihedral angle between planes passing through atoms C1'-C6' of the aromatic ring and the naphthoquinonic ring is 71.0 (2) °. In the crystal packing, molecules interact through two intermolecular C—H···O contacts, Table 1.
Experimental
To a chloroform (25mL) solution of the nor-lapachol (2-hydroxy-3-(2-methylprop-1-enyl)naphthalene-1,4-dione, 228 mg, 1 mmol), bromine (2 mL, 38 mmol) was added. The bromo intermediate, 3-bromo-2,2-dimethyl-2,3-dihydro-naphtho[1,2-b]furan-4,5-dione, precipitated immediately as an orange solid. Over this mixture, an excess of 2,4-dibromobenzenamine was added and the mixture was left under agitation overnight. After the addition of water (50 ml), the organic phase was separated and washed with 10% HCl (3 x 50 ml), dried over sodium sulfate, filtered, and concentrated under reduced pressure. The arylaminoderivative,3-(2',4'-dibromophenylamine)-2H,3H-2,2- dimethylnaphtho[1,2 - b]furan-4,5-dione species, was purified by column chromatography over silica-gel, using as eluent a gradient mixture of hexane/ethyl acetate (9/1 to 7/3) with increasing polarity and obtained as a red solid (330 mg, 0.70 mmol, 70% yield). 1H NMR (300 MHz, CDCl3) δ: 8.14 (1H, dd, J = 6.7, 1.3 Hz), 7.76–7.63 (3H, m), 7.56 (1H, d, J = 2.0 Hz), 7.30 (1H, dd, J = 8.8, 2.0 Hz), 6.53 (1H, d, J = 8.8 Hz), 4.83 (1H, d, J = 7.5 Hz), 4.48 (NH, d,J = 7.5 Hz), 1.66 (3H, s), 1.54 (3H, s). 13C NMR (75 MHz, CDCl3) δ: 180.6 (C=O), 175.0 (C=O), 169.7 (Cq), 143.0 (Cq), 134.6 (CH), 132.6 (CH),131.1 (CH), 131.0 (Cq), 129.5 (CH), 127.1 (Cq), 125.1 (CH), 114.4 (Cq),112.6(CH), 110.2 (Cq), 108.9 (Cq), 96.0 (Cq), 134.5(CH), 61.2 (CH), 27.5 (CH3), 21.6 (CH3).
Refinement
H atoms were located on stereochemical grounds and refined with fixed geometry, each riding on a carrier atom, with N—H = 0.86 Å and C—H = 0.93 - 0.98 Å, and with U(H) set to 1.2–1.5 times Ueq(N, C). The maximum and minimum residual electron density peaks were located 0.59 and -1.49 Å, respectively, from the Br1 atom.
Figures
Fig. 1.
Projection of a molecule of (IV), showing the atom labelling with 50% probability displacement ellipsoids.
Crystal data
| C20H15Br2NO3 | Z = 2 |
| Mr = 477.15 | F000 = 472 |
| Triclinic, P1 | Dx = 1.771 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 8.1430 (3) Å | Cell parameters from 7882 reflections |
| b = 11.2584 (4) Å | θ = 1.0–27.5º |
| c = 11.4742 (5) Å | µ = 4.55 mm−1 |
| α = 112.073 (2)º | T = 293 (2) K |
| β = 95.546 (2)º | Plate, colorless |
| γ = 108.696 (2)º | 0.31 × 0.28 × 0.16 mm |
| V = 894.70 (6) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 4099 independent reflections |
| Radiation source: Enraf Nonius FR590 | 3610 reflections with I > 2σ(I) |
| Monochromator: horizonally mounted graphite crystal | Rint = 0.032 |
| Detector resolution: 9 pixels mm-1 | θmax = 27.5º |
| CCD rotation images, thick slices scans | θmin = 2.7º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −10→9 |
| Tmin = 0.272, Tmax = 0.490 | k = −14→14 |
| 9784 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.048 | H-atom parameters constrained |
| wR(F2) = 0.139 | w = 1/[σ2(Fo2) + (0.0642P)2 + 1.7472P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 4070 reflections | Δρmax = 0.59 e Å−3 |
| 235 parameters | Δρmin = −1.49 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.13493 (6) | 0.15969 (6) | 0.15092 (5) | 0.06302 (18) | |
| Br2 | 0.61904 (6) | 0.13670 (4) | −0.16856 (4) | 0.04728 (15) | |
| O1 | 0.5871 (4) | 0.4358 (3) | 0.6646 (3) | 0.0405 (6) | |
| O2 | 0.2827 (4) | −0.0404 (3) | 0.4343 (3) | 0.0475 (7) | |
| O3 | 0.0912 (5) | −0.0405 (3) | 0.6162 (4) | 0.0647 (10) | |
| N1 | 0.4572 (4) | 0.2348 (4) | 0.3578 (3) | 0.0388 (7) | |
| H1 | 0.3585 | 0.2460 | 0.3654 | 0.047* | |
| C2 | 0.6940 (5) | 0.3902 (4) | 0.5704 (4) | 0.0376 (8) | |
| C3 | 0.5652 (5) | 0.2412 (4) | 0.4710 (4) | 0.0343 (7) | |
| H3 | 0.6291 | 0.1787 | 0.4452 | 0.041* | |
| C3A | 0.4399 (5) | 0.2026 (4) | 0.5504 (3) | 0.0335 (7) | |
| C4 | 0.3082 (5) | 0.0695 (4) | 0.5251 (4) | 0.0355 (7) | |
| C5 | 0.1911 (5) | 0.0697 (4) | 0.6241 (4) | 0.0396 (8) | |
| C5A | 0.2076 (5) | 0.2057 (4) | 0.7271 (4) | 0.0356 (7) | |
| C6 | 0.0916 (5) | 0.2100 (4) | 0.8081 (4) | 0.0430 (9) | |
| H6 | 0.0005 | 0.1284 | 0.7967 | 0.052* | |
| C7 | 0.1114 (6) | 0.3356 (5) | 0.9059 (4) | 0.0487 (10) | |
| H7 | 0.0348 | 0.3379 | 0.9610 | 0.058* | |
| C8 | 0.2437 (7) | 0.4573 (5) | 0.9222 (5) | 0.0503 (10) | |
| H8 | 0.2560 | 0.5413 | 0.9882 | 0.060* | |
| C9 | 0.3589 (6) | 0.4550 (4) | 0.8403 (4) | 0.0449 (9) | |
| H9 | 0.4480 | 0.5374 | 0.8514 | 0.054* | |
| C9A | 0.3410 (5) | 0.3304 (4) | 0.7427 (4) | 0.0347 (7) | |
| C9B | 0.4547 (5) | 0.3204 (4) | 0.6524 (4) | 0.0339 (7) | |
| C1' | 0.4990 (5) | 0.2127 (4) | 0.2403 (4) | 0.0336 (7) | |
| C2' | 0.3688 (5) | 0.1795 (4) | 0.1324 (4) | 0.0349 (7) | |
| C3' | 0.4010 (5) | 0.1562 (4) | 0.0115 (4) | 0.0384 (8) | |
| H3' | 0.3101 | 0.1327 | −0.0586 | 0.046* | |
| C4' | 0.5725 (5) | 0.1686 (4) | −0.0031 (4) | 0.0370 (8) | |
| C5' | 0.7048 (6) | 0.2006 (5) | 0.1004 (4) | 0.0428 (9) | |
| H5' | 0.8189 | 0.2081 | 0.0898 | 0.051* | |
| C6' | 0.6691 (5) | 0.2220 (4) | 0.2212 (4) | 0.0406 (8) | |
| H6' | 0.7597 | 0.2429 | 0.2904 | 0.049* | |
| C1'' | 0.8530 (6) | 0.3854 (5) | 0.6478 (5) | 0.0509 (10) | |
| H1A | 0.8108 | 0.3159 | 0.6791 | 0.076* | |
| H1B | 0.9175 | 0.4744 | 0.7201 | 0.076* | |
| H1C | 0.9308 | 0.3628 | 0.5930 | 0.076* | |
| C2'' | 0.7497 (6) | 0.4983 (4) | 0.5182 (5) | 0.0466 (9) | |
| H2A | 0.6454 | 0.4971 | 0.4703 | 0.070* | |
| H2B | 0.8268 | 0.4777 | 0.4621 | 0.070* | |
| H2C | 0.8124 | 0.5886 | 0.5891 | 0.070* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0428 (3) | 0.0937 (4) | 0.0536 (3) | 0.0282 (3) | 0.0146 (2) | 0.0306 (3) |
| Br2 | 0.0576 (3) | 0.0534 (3) | 0.0380 (2) | 0.0228 (2) | 0.02256 (19) | 0.02340 (19) |
| O1 | 0.0463 (15) | 0.0328 (12) | 0.0396 (14) | 0.0101 (11) | 0.0189 (12) | 0.0153 (11) |
| O2 | 0.0541 (17) | 0.0357 (14) | 0.0432 (16) | 0.0129 (12) | 0.0138 (13) | 0.0105 (12) |
| O3 | 0.075 (2) | 0.0385 (16) | 0.074 (2) | 0.0084 (15) | 0.0367 (19) | 0.0246 (16) |
| N1 | 0.0419 (17) | 0.0533 (19) | 0.0326 (16) | 0.0262 (15) | 0.0153 (13) | 0.0222 (14) |
| C2 | 0.0393 (19) | 0.0401 (19) | 0.0381 (19) | 0.0139 (15) | 0.0151 (16) | 0.0217 (16) |
| C3 | 0.0386 (18) | 0.0405 (18) | 0.0338 (18) | 0.0199 (15) | 0.0151 (15) | 0.0209 (15) |
| C3A | 0.0394 (18) | 0.0368 (17) | 0.0285 (16) | 0.0143 (15) | 0.0105 (14) | 0.0184 (14) |
| C4 | 0.0398 (19) | 0.0353 (17) | 0.0323 (18) | 0.0144 (15) | 0.0083 (14) | 0.0157 (14) |
| C5 | 0.0404 (19) | 0.0386 (19) | 0.041 (2) | 0.0121 (15) | 0.0120 (16) | 0.0207 (16) |
| C5A | 0.0364 (18) | 0.0389 (18) | 0.0357 (18) | 0.0140 (15) | 0.0106 (15) | 0.0204 (15) |
| C6 | 0.0378 (19) | 0.050 (2) | 0.048 (2) | 0.0166 (17) | 0.0189 (17) | 0.0268 (18) |
| C7 | 0.051 (2) | 0.062 (3) | 0.045 (2) | 0.028 (2) | 0.0240 (19) | 0.027 (2) |
| C8 | 0.061 (3) | 0.049 (2) | 0.042 (2) | 0.025 (2) | 0.022 (2) | 0.0159 (18) |
| C9 | 0.054 (2) | 0.0382 (19) | 0.041 (2) | 0.0149 (17) | 0.0172 (18) | 0.0162 (17) |
| C9A | 0.0393 (18) | 0.0355 (17) | 0.0302 (17) | 0.0124 (14) | 0.0101 (14) | 0.0165 (14) |
| C9B | 0.0392 (18) | 0.0356 (17) | 0.0329 (17) | 0.0141 (14) | 0.0123 (14) | 0.0203 (14) |
| C1' | 0.0389 (18) | 0.0373 (17) | 0.0314 (17) | 0.0177 (15) | 0.0121 (14) | 0.0185 (14) |
| C2' | 0.0329 (17) | 0.0388 (18) | 0.0382 (19) | 0.0162 (14) | 0.0122 (14) | 0.0192 (15) |
| C3' | 0.043 (2) | 0.043 (2) | 0.0331 (18) | 0.0185 (16) | 0.0097 (15) | 0.0195 (16) |
| C4' | 0.046 (2) | 0.0383 (18) | 0.0341 (18) | 0.0186 (16) | 0.0189 (16) | 0.0199 (15) |
| C5' | 0.041 (2) | 0.055 (2) | 0.044 (2) | 0.0241 (18) | 0.0193 (17) | 0.0271 (19) |
| C6' | 0.0374 (19) | 0.055 (2) | 0.0371 (19) | 0.0220 (17) | 0.0111 (15) | 0.0235 (17) |
| C1'' | 0.042 (2) | 0.054 (2) | 0.055 (3) | 0.0127 (18) | 0.0059 (19) | 0.029 (2) |
| C2'' | 0.052 (2) | 0.044 (2) | 0.056 (3) | 0.0180 (18) | 0.027 (2) | 0.0305 (19) |
Geometric parameters (Å, °)
| Br1—C2' | 1.887 (4) | C7—C8 | 1.377 (7) |
| Br2—C4' | 1.895 (4) | C7—H7 | 0.9300 |
| O1—C9B | 1.344 (4) | C8—C9 | 1.389 (6) |
| O1—C2 | 1.497 (5) | C8—H8 | 0.9300 |
| O2—C4 | 1.219 (5) | C9—C9A | 1.379 (6) |
| O3—C5 | 1.210 (5) | C9—H9 | 0.9300 |
| N1—C1' | 1.374 (5) | C9A—C9B | 1.451 (5) |
| N1—C3 | 1.461 (5) | C1'—C2' | 1.394 (5) |
| N1—H1 | 0.8600 | C1'—C6' | 1.400 (5) |
| C2—C2'' | 1.514 (5) | C2'—C3' | 1.379 (5) |
| C2—C1'' | 1.525 (6) | C3'—C4' | 1.392 (5) |
| C2—C3 | 1.556 (5) | C3'—H3' | 0.9300 |
| C3—C3A | 1.501 (5) | C4'—C5' | 1.372 (6) |
| C3—H3 | 0.9800 | C5'—C6' | 1.391 (6) |
| C3A—C9B | 1.360 (5) | C5'—H5' | 0.9300 |
| C3A—C4 | 1.436 (5) | C6'—H6' | 0.9300 |
| C4—C5 | 1.552 (5) | C1''—H1A | 0.9600 |
| C5—C5A | 1.496 (5) | C1''—H1B | 0.9600 |
| C5A—C6 | 1.387 (5) | C1''—H1C | 0.9600 |
| C5A—C9A | 1.406 (5) | C2''—H2A | 0.9600 |
| C6—C7 | 1.382 (6) | C2''—H2B | 0.9600 |
| C6—H6 | 0.9300 | C2''—H2C | 0.9600 |
| C9B—O1—C2 | 107.1 (3) | C9A—C9—H9 | 120.0 |
| C1'—N1—C3 | 125.9 (3) | C8—C9—H9 | 120.0 |
| C1'—N1—H1 | 117.0 | C9—C9A—C5A | 119.9 (4) |
| C3—N1—H1 | 117.0 | C9—C9A—C9B | 122.9 (3) |
| O1—C2—C2'' | 106.1 (3) | C5A—C9A—C9B | 117.2 (3) |
| O1—C2—C1'' | 106.1 (3) | O1—C9B—C3A | 114.0 (3) |
| C2''—C2—C1'' | 112.7 (3) | O1—C9B—C9A | 119.7 (3) |
| O1—C2—C3 | 104.0 (3) | C3A—C9B—C9A | 126.2 (3) |
| C2''—C2—C3 | 116.4 (3) | N1—C1'—C2' | 119.9 (3) |
| C1''—C2—C3 | 110.5 (3) | N1—C1'—C6' | 123.5 (3) |
| N1—C3—C3A | 106.9 (3) | C2'—C1'—C6' | 116.6 (3) |
| N1—C3—C2 | 114.7 (3) | C3'—C2'—C1' | 123.2 (3) |
| C3A—C3—C2 | 100.7 (3) | C3'—C2'—Br1 | 118.3 (3) |
| N1—C3—H3 | 111.3 | C1'—C2'—Br1 | 118.5 (3) |
| C3A—C3—H3 | 111.3 | C2'—C3'—C4' | 118.5 (3) |
| C2—C3—H3 | 111.3 | C2'—C3'—H3' | 120.8 |
| C9B—C3A—C4 | 121.3 (3) | C4'—C3'—H3' | 120.8 |
| C9B—C3A—C3 | 108.9 (3) | C5'—C4'—C3' | 120.3 (3) |
| C4—C3A—C3 | 129.6 (3) | C5'—C4'—Br2 | 120.6 (3) |
| O2—C4—C3A | 125.6 (4) | C3'—C4'—Br2 | 119.0 (3) |
| O2—C4—C5 | 118.8 (3) | C4'—C5'—C6' | 120.3 (4) |
| C3A—C4—C5 | 115.6 (3) | C4'—C5'—H5' | 119.8 |
| O3—C5—C5A | 122.3 (4) | C6'—C5'—H5' | 119.8 |
| O3—C5—C4 | 118.7 (4) | C5'—C6'—C1' | 121.0 (4) |
| C5A—C5—C4 | 119.0 (3) | C5'—C6'—H6' | 119.5 |
| C6—C5A—C9A | 119.5 (4) | C1'—C6'—H6' | 119.5 |
| C6—C5A—C5 | 120.3 (3) | C2—C1''—H1A | 109.5 |
| C9A—C5A—C5 | 120.3 (3) | C2—C1''—H1B | 109.5 |
| C7—C6—C5A | 119.9 (4) | H1A—C1''—H1B | 109.5 |
| C7—C6—H6 | 120.0 | C2—C1''—H1C | 109.5 |
| C5A—C6—H6 | 120.0 | H1A—C1''—H1C | 109.5 |
| C8—C7—C6 | 120.5 (4) | H1B—C1''—H1C | 109.5 |
| C8—C7—H7 | 119.7 | C2—C2''—H2A | 109.5 |
| C6—C7—H7 | 119.7 | C2—C2''—H2B | 109.5 |
| C7—C8—C9 | 120.2 (4) | H2A—C2''—H2B | 109.5 |
| C7—C8—H8 | 119.9 | C2—C2''—H2C | 109.5 |
| C9—C8—H8 | 119.9 | H2A—C2''—H2C | 109.5 |
| C9A—C9—C8 | 119.9 (4) | H2B—C2''—H2C | 109.5 |
| C9B—O1—C2—C2'' | 143.2 (3) | C8—C9—C9A—C5A | −0.8 (6) |
| C9B—O1—C2—C1'' | −96.7 (3) | C8—C9—C9A—C9B | 179.1 (4) |
| C9B—O1—C2—C3 | 19.9 (4) | C6—C5A—C9A—C9 | 2.0 (6) |
| C1'—N1—C3—C3A | −154.9 (4) | C5—C5A—C9A—C9 | −178.1 (4) |
| C1'—N1—C3—C2 | 94.4 (4) | C6—C5A—C9A—C9B | −177.9 (4) |
| O1—C2—C3—N1 | 92.0 (3) | C5—C5A—C9A—C9B | 2.1 (5) |
| C2''—C2—C3—N1 | −24.3 (5) | C2—O1—C9B—C3A | −8.8 (4) |
| C1''—C2—C3—N1 | −154.5 (3) | C2—O1—C9B—C9A | 172.6 (3) |
| O1—C2—C3—C3A | −22.4 (3) | C4—C3A—C9B—O1 | 178.5 (3) |
| C2''—C2—C3—C3A | −138.7 (3) | C3—C3A—C9B—O1 | −6.9 (4) |
| C1''—C2—C3—C3A | 91.1 (4) | C4—C3A—C9B—C9A | −3.0 (6) |
| N1—C3—C3A—C9B | −101.7 (4) | C3—C3A—C9B—C9A | 171.6 (3) |
| C2—C3—C3A—C9B | 18.4 (4) | C9—C9A—C9B—O1 | 1.7 (6) |
| N1—C3—C3A—C4 | 72.3 (5) | C5A—C9A—C9B—O1 | −178.5 (3) |
| C2—C3—C3A—C4 | −167.6 (4) | C9—C9A—C9B—C3A | −176.7 (4) |
| C9B—C3A—C4—O2 | 178.0 (4) | C5A—C9A—C9B—C3A | 3.1 (6) |
| C3—C3A—C4—O2 | 4.7 (7) | C3—N1—C1'—C2' | 166.3 (3) |
| C9B—C3A—C4—C5 | −2.1 (5) | C3—N1—C1'—C6' | −14.0 (6) |
| C3—C3A—C4—C5 | −175.4 (3) | N1—C1'—C2'—C3' | 179.8 (3) |
| O2—C4—C5—O3 | 7.7 (6) | C6'—C1'—C2'—C3' | 0.1 (5) |
| C3A—C4—C5—O3 | −172.1 (4) | N1—C1'—C2'—Br1 | −2.3 (5) |
| O2—C4—C5—C5A | −173.3 (4) | C6'—C1'—C2'—Br1 | 178.0 (3) |
| C3A—C4—C5—C5A | 6.8 (5) | C1'—C2'—C3'—C4' | −1.1 (6) |
| O3—C5—C5A—C6 | −8.0 (6) | Br1—C2'—C3'—C4' | −179.0 (3) |
| C4—C5—C5A—C6 | 173.1 (4) | C2'—C3'—C4'—C5' | 1.3 (6) |
| O3—C5—C5A—C9A | 172.0 (4) | C2'—C3'—C4'—Br2 | 179.9 (3) |
| C4—C5—C5A—C9A | −6.9 (5) | C3'—C4'—C5'—C6' | −0.4 (6) |
| C9A—C5A—C6—C7 | −2.1 (6) | Br2—C4'—C5'—C6' | −179.1 (3) |
| C5—C5A—C6—C7 | 177.9 (4) | C4'—C5'—C6'—C1' | −0.6 (6) |
| C5A—C6—C7—C8 | 1.2 (7) | N1—C1'—C6'—C5' | −179.0 (4) |
| C6—C7—C8—C9 | 0.0 (7) | C2'—C1'—C6'—C5' | 0.7 (6) |
| C7—C8—C9—C9A | −0.1 (7) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O2i | 0.98 | 2.64 | 3.347 (6) | 129 |
| C1''—H1A···O2i | 0.96 | 2.67 | 3.389 (6) | 132 |
Symmetry codes: (i) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2311).
References
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc.97, 1354–1358.
- Dos Santos, A. F., Ferraz, P. A. L., De Abreu, F. C., Chiari, E., Goulart, M. O. F. & Sant’Ana, A. E. G. (2001). Planta Med.67, 92–93. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Hillard, E. A., de Abreu, F. C., Ferreira, D. C. M., Jaouen, G., Goulart, M. O. F. & Amatore, C. (2008). Chem. Commun. pp. 2612–2628. [DOI] [PubMed]
- Lima, N. M. F., Correia, C. S., Ferraz, P. A. L., Pinto, A. V., Pinto, M. C. R. F., Sant’Ana, A. E. G. & Goulart, M. O. F. (2002). J. Braz. Chem. Soc.13, 822–829.
- Lima, N. M. F., Correia, C. S., Leon, L. L., Machado, G. M. C., Madeira, M. F., Santana, A. E. G. & Goulart, M. O. F. (2004). Mem. Inst. Oswaldo Cruz, 99, 757–761. [DOI] [PubMed]
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet Vol. 276, 307–326, New York: Academic Press.
- Pinto, A. V., Neves-Pinto, C., Pinto, M. C. F. R., Santa-Rita, R. M., Pezzella, C. & De Castro, S. L. (1997). Arzneim. Forsch.47, 74–79. [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Silva Júnior, E. N. da, de Souza, M. C. B. V., Fernades, M. C., Menna-Barreto, R. F. S., Pinto, A. V., Pinto, M. C. F. R., Lopes, F. A., De Simone, C. A., Andrade, C. K. Z., Ferreira, V. F. & De Castro, S. L. (2008). Bioorg. Med. Chem.16, 5030–5038. [DOI] [PubMed]
- Silva Júnior, E. N. da, de Souza, M. C. B. V., Pinto, A. V., Pinto, M. C. F. R., Goulart, M. O. F., Pessoa, C., Costa-Lotufo, L., Montenegro, R. C., Moraes, M. O. & Ferreira, V. F. (2007). Bioorg. Med. Chem.15, 7035–7041. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808034545/tk2311sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808034545/tk2311Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



