Abstract
In the title compound, C21H13N5O4, the dihedral angles formed between the planes of the phenyl and nitrophenyl rings and that of the heterotricyclic plane are 41.29 (7) and 61.35 (6)°, respectively. In the crystal, weak C—H⋯O interactions help to establish the packing.
Related literature
For background, see: Carneiro et al. (2005 ▶); Menegatti et al. (2006 ▶); Barreiro et al. (2006 ▶).
Experimental
Crystal data
C21H13N5O4
M r = 399.36
Monoclinic,
a = 9.677 (2) Å
b = 12.141 (3) Å
c = 17.438 (4) Å
β = 119.451 (16)°
V = 1784.0 (8) Å3
Z = 4
Cu Kα radiation
μ = 0.89 mm−1
T = 297 (2) K
0.3 × 0.1 × 0.08 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: none
3813 measured reflections
3259 independent reflections
2578 reflections with I > 2σ(I)
R int = 0.023
2 standard reflections frequency: 120 min intensity decay: 1%
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.198
S = 1.07
3259 reflections
273 parameters
H-atom parameters constrained
Δρmax = 0.48 e Å−3
Δρmin = −0.38 e Å−3
Data collection: CAD-4-PC (Enraf–Nonius, 1993 ▶); cell refinement: CAD-4-PC; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808037240/tk2318sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808037240/tk2318Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H7⋯O30i | 0.93 | 2.55 | 3.216 (3) | 129 |
| C18—H18⋯O30ii | 0.93 | 2.44 | 3.197 (3) | 139 |
Symmetry codes: (i) 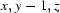 ; (ii)
; (ii) 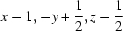 .
.
Acknowledgments
The authors gratefully acknowledge the financial support of Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq, Pró-Reitoria de Pesquisa e Pós Graduação-PRPPG/UFG.
supplementary crystallographic information
Comment
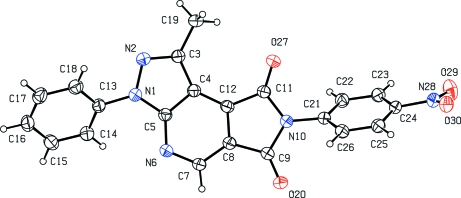
The title compound (I) (Carneiro et al., 2005; Menegatti et al., 2006; Barreiro et al., 2006) features a novel functionalized three-fused rings skeleton, Fig. 1. The bond distances within the pyridine ring are consistent with aromatic delocalization but the pyrazole ring presents a double bond, Table 1. The C8—C9 and C11—C12 bond lengths are elongated from the expected distances by 0.025 and 0.03 Å, respectively.
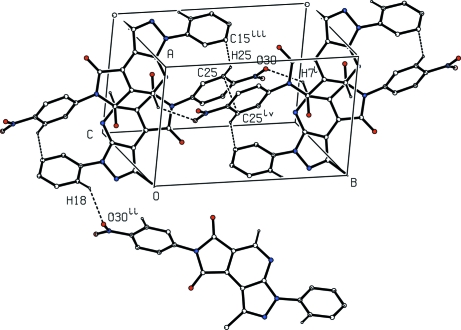
The pyrazolo[3,4-b]pyrrolo[3,4-d]pyridine moiety is planar (plane α, with r.m.s. deviation of about 0.01 Å). Relative to plane α, the phenyl rings C21/C26 and C13/C18 form dihedral angles of 41.29 (7)° and 61.35 (6)°, respectively. The latter ring is slanted from the plane α so that the C21 and C24 atoms are shifted by 0.118 (3) and 0.436 (4) Å from the plane α, respectively. This might be in part due to the participation of the oxygen atom O30 in two C-H···O contacts, C7—H7···O30i and C18—H18···O30ii; see Table 2 for details. The former interaction connects molecules into a linear chain along the b-axis and the latter connects parallel chains to form sheets in the plane of approximate indices (-1 0 2). The molecules in the linear chains interact with centrosymmetrically related chains via π···π stacking [closest distance for C25···C25iv: 3.380 (5) Å; (iv): 1 - x, 1 - y, 1 - z]. In addition, molecules participate in a C—H···π interaction: C25—H25···C15iii [2.74 Å, 3.539 (4) Å, 144°; (iii) 1 -x, -y, 1 -z] to consolidate the crystal packing, Fig. 2.
Experimental
Compound (I) was synthesized as described previously (Menegatti et al., 2006). A colourless needle for crystallography was obtained by slow evaporation of a MeOH solution of (I) at room temperature.
Refinement
All H atoms were positioned in idealized positions in the riding model approximation with C—H = 0.93 - 0.96 Å, and with Uiso(H) = 1.2-1.5 Ueq(C).
Figures
Fig. 1.
View of (I) with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level and H atoms are shown as small spheres of arbitrary radii.
Fig. 2.
Packing diagram of (I). Intermolecular interactions are shown as dashed lines. Only the H atoms involved in intermolecular contacts are shown.
Crystal data
| C21H13N5O4 | F000 = 824 |
| Mr = 399.36 | Dx = 1.487 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation λ = 1.5418 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 9.677 (2) Å | θ = 14.8–40.8º |
| b = 12.141 (3) Å | µ = 0.89 mm−1 |
| c = 17.438 (4) Å | T = 297 (2) K |
| β = 119.451 (16)º | Prism, colourless |
| V = 1784.0 (8) Å3 | 0.3 × 0.1 × 0.08 mm |
| Z = 4 |
Data collection
| Enraf–Nonius CAD-4 diffractometer | θmax = 67.9º |
| T = 297(2) K | θmin = 4.7º |
| Non–profiled ω/2ω scans | h = 0→11 |
| Absorption correction: none | k = −14→1 |
| 3813 measured reflections | l = −20→18 |
| 3259 independent reflections | 2 standard reflections |
| 2578 reflections with I > 2σ(I) | every 120 min |
| Rint = 0.023 | intensity decay: 1% |
Refinement
| Refinement on F2 | H-atom parameters constrained |
| Least-squares matrix: full | w = 1/[σ2(Fo2) + (0.152P)2 + 0.0892P] where P = (Fo2 + 2Fc2)/3 |
| R[F2 > 2σ(F2)] = 0.049 | (Δ/σ)max < 0.001 |
| wR(F2) = 0.198 | Δρmax = 0.48 e Å−3 |
| S = 1.07 | Δρmin = −0.38 e Å−3 |
| 3259 reflections | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 273 parameters | Extinction coefficient: 0.0114 (15) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C3 | −0.0408 (3) | 0.00360 (19) | 0.37884 (14) | 0.0446 (5) | |
| C4 | 0.1214 (3) | 0.00403 (18) | 0.44674 (14) | 0.0425 (5) | |
| C5 | 0.1708 (3) | −0.10707 (19) | 0.45566 (14) | 0.0431 (5) | |
| C7 | 0.4230 (3) | −0.07508 (19) | 0.56234 (16) | 0.0500 (6) | |
| H7 | 0.5254 | −0.0987 | 0.6016 | 0.06* | |
| C8 | 0.3874 (3) | 0.03680 (18) | 0.55955 (14) | 0.0452 (5) | |
| C9 | 0.4917 (3) | 0.13014 (18) | 0.61003 (15) | 0.0455 (5) | |
| C11 | 0.2390 (3) | 0.19832 (18) | 0.51522 (14) | 0.0442 (5) | |
| C12 | 0.2381 (3) | 0.07694 (18) | 0.50241 (14) | 0.0427 (5) | |
| C13 | 0.0263 (3) | −0.28159 (19) | 0.37880 (16) | 0.0462 (5) | |
| C14 | 0.0917 (3) | −0.3569 (2) | 0.44682 (16) | 0.0514 (6) | |
| H14 | 0.1508 | −0.3332 | 0.5049 | 0.062* | |
| C15 | 0.0681 (3) | −0.4684 (2) | 0.42733 (19) | 0.0584 (7) | |
| H15 | 0.1132 | −0.5198 | 0.4726 | 0.07* | |
| C16 | −0.0219 (3) | −0.5039 (2) | 0.3414 (2) | 0.0606 (7) | |
| H16 | −0.0394 | −0.5787 | 0.3289 | 0.073* | |
| C17 | −0.0852 (3) | −0.4279 (2) | 0.27450 (19) | 0.0616 (7) | |
| H17 | −0.1454 | −0.4516 | 0.2165 | 0.074* | |
| C18 | −0.0604 (3) | −0.3157 (2) | 0.29235 (16) | 0.0542 (6) | |
| H18 | −0.1016 | −0.2645 | 0.2468 | 0.065* | |
| C19 | −0.1519 (3) | 0.0984 (2) | 0.34157 (17) | 0.0549 (6) | |
| H19A | −0.2592 | 0.0727 | 0.3172 | 0.082* | |
| H19B | −0.1383 | 0.1329 | 0.2962 | 0.082* | |
| H19C | −0.13 | 0.1509 | 0.3874 | 0.082* | |
| C21 | 0.4456 (3) | 0.33472 (18) | 0.60655 (14) | 0.0426 (5) | |
| C22 | 0.3722 (3) | 0.3988 (2) | 0.64181 (16) | 0.0489 (5) | |
| H22 | 0.2948 | 0.3685 | 0.6523 | 0.059* | |
| C23 | 0.4150 (3) | 0.5083 (2) | 0.66135 (16) | 0.0507 (6) | |
| H23 | 0.3682 | 0.5525 | 0.6859 | 0.061* | |
| C24 | 0.5284 (3) | 0.55016 (18) | 0.64357 (14) | 0.0461 (5) | |
| C25 | 0.6055 (3) | 0.4867 (2) | 0.61069 (15) | 0.0496 (6) | |
| H25 | 0.6842 | 0.5169 | 0.6012 | 0.06* | |
| C26 | 0.5631 (3) | 0.37726 (19) | 0.59215 (15) | 0.0480 (5) | |
| H26 | 0.6134 | 0.3325 | 0.5701 | 0.058* | |
| O20 | 0.6301 (2) | 0.12980 (14) | 0.66441 (12) | 0.0587 (5) | |
| O27 | 0.1327 (2) | 0.26369 (14) | 0.47771 (13) | 0.0592 (5) | |
| O30 | 0.6424 (2) | 0.70822 (16) | 0.62526 (14) | 0.0675 (6) | |
| N1 | 0.0428 (2) | −0.16562 (16) | 0.39668 (12) | 0.0469 (5) | |
| N2 | −0.0858 (2) | −0.09733 (17) | 0.34974 (13) | 0.0497 (5) | |
| N6 | 0.3155 (2) | −0.14946 (16) | 0.51075 (13) | 0.0489 (5) | |
| N10 | 0.3932 (2) | 0.22413 (15) | 0.58107 (12) | 0.0453 (5) | |
| N28 | 0.5686 (3) | 0.66741 (18) | 0.65876 (13) | 0.0561 (6) | |
| O29 | 0.5258 (3) | 0.71978 (17) | 0.70287 (16) | 0.0830 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C3 | 0.0414 (11) | 0.0460 (11) | 0.0430 (11) | 0.0022 (9) | 0.0181 (9) | 0.0009 (9) |
| C4 | 0.0394 (11) | 0.0436 (11) | 0.0430 (11) | 0.0017 (9) | 0.0190 (9) | 0.0022 (9) |
| C5 | 0.0401 (11) | 0.0427 (11) | 0.0459 (11) | 0.0016 (9) | 0.0207 (9) | 0.0017 (9) |
| C7 | 0.0398 (11) | 0.0408 (11) | 0.0580 (13) | 0.0049 (9) | 0.0152 (10) | 0.0057 (9) |
| C8 | 0.0418 (11) | 0.0406 (11) | 0.0479 (11) | 0.0017 (9) | 0.0180 (10) | 0.0039 (9) |
| C9 | 0.0436 (12) | 0.0393 (12) | 0.0490 (12) | 0.0048 (9) | 0.0192 (10) | 0.0033 (9) |
| C11 | 0.0407 (11) | 0.0425 (12) | 0.0464 (11) | 0.0035 (9) | 0.0191 (9) | 0.0036 (9) |
| C12 | 0.0410 (11) | 0.0411 (11) | 0.0444 (11) | 0.0046 (9) | 0.0196 (9) | 0.0045 (8) |
| C13 | 0.0411 (11) | 0.0424 (12) | 0.0567 (12) | −0.0020 (9) | 0.0252 (10) | −0.0056 (9) |
| C14 | 0.0511 (13) | 0.0476 (13) | 0.0544 (13) | −0.0011 (10) | 0.0250 (11) | −0.0023 (10) |
| C15 | 0.0586 (15) | 0.0462 (13) | 0.0755 (17) | 0.0040 (11) | 0.0368 (13) | 0.0043 (12) |
| C16 | 0.0550 (14) | 0.0457 (13) | 0.0872 (19) | −0.0057 (11) | 0.0397 (14) | −0.0145 (12) |
| C17 | 0.0524 (14) | 0.0661 (17) | 0.0650 (15) | −0.0068 (12) | 0.0280 (12) | −0.0226 (13) |
| C18 | 0.0492 (13) | 0.0567 (14) | 0.0548 (13) | −0.0003 (11) | 0.0241 (11) | −0.0034 (11) |
| C19 | 0.0426 (12) | 0.0534 (14) | 0.0545 (13) | 0.0090 (10) | 0.0130 (11) | 0.0022 (10) |
| C21 | 0.0389 (11) | 0.0386 (11) | 0.0436 (10) | 0.0011 (8) | 0.0152 (9) | 0.0014 (8) |
| C22 | 0.0469 (12) | 0.0464 (12) | 0.0550 (12) | −0.0016 (10) | 0.0264 (11) | −0.0028 (10) |
| C23 | 0.0475 (12) | 0.0483 (12) | 0.0530 (12) | 0.0023 (10) | 0.0222 (10) | −0.0079 (10) |
| C24 | 0.0420 (11) | 0.0390 (11) | 0.0441 (11) | −0.0020 (9) | 0.0109 (9) | −0.0046 (9) |
| C25 | 0.0441 (12) | 0.0463 (12) | 0.0581 (13) | −0.0018 (9) | 0.0248 (11) | 0.0008 (10) |
| C26 | 0.0454 (12) | 0.0457 (12) | 0.0528 (12) | 0.0029 (10) | 0.0241 (10) | −0.0026 (10) |
| O20 | 0.0414 (9) | 0.0510 (10) | 0.0623 (11) | 0.0035 (7) | 0.0089 (8) | 0.0036 (7) |
| O27 | 0.0457 (9) | 0.0434 (9) | 0.0709 (11) | 0.0100 (7) | 0.0152 (8) | 0.0002 (8) |
| O30 | 0.0607 (11) | 0.0490 (10) | 0.0811 (13) | −0.0093 (9) | 0.0258 (10) | 0.0001 (9) |
| N1 | 0.0415 (10) | 0.0421 (10) | 0.0497 (10) | −0.0009 (8) | 0.0167 (8) | −0.0029 (8) |
| N2 | 0.0411 (10) | 0.0498 (11) | 0.0498 (10) | 0.0019 (8) | 0.0159 (9) | −0.0013 (8) |
| N6 | 0.0420 (10) | 0.0395 (9) | 0.0568 (11) | 0.0039 (8) | 0.0178 (9) | 0.0014 (8) |
| N10 | 0.0408 (10) | 0.0379 (10) | 0.0493 (10) | 0.0013 (8) | 0.0161 (8) | 0.0012 (7) |
| N28 | 0.0528 (12) | 0.0460 (11) | 0.0535 (11) | −0.0040 (9) | 0.0139 (10) | −0.0058 (9) |
| O29 | 0.1122 (19) | 0.0534 (11) | 0.0905 (15) | −0.0043 (12) | 0.0553 (15) | −0.0207 (10) |
Geometric parameters (Å, °)
| C3—N2 | 1.316 (3) | C16—C17 | 1.373 (4) |
| C3—C4 | 1.428 (3) | C16—H16 | 0.93 |
| C3—C19 | 1.489 (3) | C17—C18 | 1.392 (4) |
| C4—C12 | 1.387 (3) | C17—H17 | 0.93 |
| C4—C5 | 1.414 (3) | C18—H18 | 0.93 |
| C5—N6 | 1.351 (3) | C19—H19A | 0.96 |
| C5—N1 | 1.358 (3) | C19—H19B | 0.96 |
| C7—N6 | 1.337 (3) | C19—H19C | 0.96 |
| C7—C8 | 1.396 (3) | C21—C26 | 1.380 (3) |
| C7—H7 | 0.93 | C21—C22 | 1.385 (3) |
| C8—C12 | 1.379 (3) | C21—N10 | 1.427 (3) |
| C8—C9 | 1.484 (3) | C22—C23 | 1.384 (3) |
| C9—O20 | 1.202 (3) | C22—H22 | 0.93 |
| C9—N10 | 1.411 (3) | C23—C24 | 1.376 (4) |
| C11—O27 | 1.206 (3) | C23—H23 | 0.93 |
| C11—N10 | 1.400 (3) | C24—C25 | 1.378 (3) |
| C11—C12 | 1.490 (3) | C24—N28 | 1.465 (3) |
| C13—C18 | 1.380 (3) | C25—C26 | 1.382 (3) |
| C13—C14 | 1.380 (3) | C25—H25 | 0.93 |
| C13—N1 | 1.434 (3) | C26—H26 | 0.93 |
| C14—C15 | 1.387 (3) | O30—N28 | 1.228 (3) |
| C14—H14 | 0.93 | N1—N2 | 1.380 (3) |
| C15—C16 | 1.381 (4) | N28—O29 | 1.216 (3) |
| C15—H15 | 0.93 | ||
| N2—C3—C4 | 110.1 (2) | C13—C18—C17 | 118.9 (2) |
| N2—C3—C19 | 121.4 (2) | C13—C18—H18 | 120.6 |
| C4—C3—C19 | 128.5 (2) | C17—C18—H18 | 120.6 |
| C12—C4—C5 | 114.68 (19) | C3—C19—H19A | 109.5 |
| C12—C4—C3 | 140.1 (2) | C3—C19—H19B | 109.5 |
| C5—C4—C3 | 105.19 (19) | H19A—C19—H19B | 109.5 |
| N6—C5—N1 | 125.4 (2) | C3—C19—H19C | 109.5 |
| N6—C5—C4 | 128.1 (2) | H19A—C19—H19C | 109.5 |
| N1—C5—C4 | 106.44 (19) | H19B—C19—H19C | 109.5 |
| N6—C7—C8 | 122.5 (2) | C26—C21—C22 | 121.3 (2) |
| N6—C7—H7 | 118.8 | C26—C21—N10 | 119.6 (2) |
| C8—C7—H7 | 118.8 | C22—C21—N10 | 119.1 (2) |
| C12—C8—C7 | 121.5 (2) | C23—C22—C21 | 119.5 (2) |
| C12—C8—C9 | 108.9 (2) | C23—C22—H22 | 120.2 |
| C7—C8—C9 | 129.6 (2) | C21—C22—H22 | 120.2 |
| O20—C9—N10 | 125.4 (2) | C24—C23—C22 | 118.3 (2) |
| O20—C9—C8 | 129.4 (2) | C24—C23—H23 | 120.8 |
| N10—C9—C8 | 105.23 (18) | C22—C23—H23 | 120.8 |
| O27—C11—N10 | 125.4 (2) | C23—C24—C25 | 122.8 (2) |
| O27—C11—C12 | 129.0 (2) | C23—C24—N28 | 119.2 (2) |
| N10—C11—C12 | 105.65 (18) | C25—C24—N28 | 117.9 (2) |
| C8—C12—C4 | 118.9 (2) | C24—C25—C26 | 118.4 (2) |
| C8—C12—C11 | 108.3 (2) | C24—C25—H25 | 120.8 |
| C4—C12—C11 | 132.8 (2) | C26—C25—H25 | 120.8 |
| C18—C13—C14 | 121.1 (2) | C21—C26—C25 | 119.6 (2) |
| C18—C13—N1 | 118.3 (2) | C21—C26—H26 | 120.2 |
| C14—C13—N1 | 120.6 (2) | C25—C26—H26 | 120.2 |
| C13—C14—C15 | 119.0 (2) | C5—N1—N2 | 110.82 (18) |
| C13—C14—H14 | 120.5 | C5—N1—C13 | 129.86 (19) |
| C15—C14—H14 | 120.5 | N2—N1—C13 | 119.32 (18) |
| C16—C15—C14 | 120.7 (2) | C3—N2—N1 | 107.44 (18) |
| C16—C15—H15 | 119.6 | C7—N6—C5 | 114.4 (2) |
| C14—C15—H15 | 119.6 | C11—N10—C9 | 111.87 (18) |
| C17—C16—C15 | 119.4 (2) | C11—N10—C21 | 122.56 (18) |
| C17—C16—H16 | 120.3 | C9—N10—C21 | 125.15 (19) |
| C15—C16—H16 | 120.3 | O29—N28—O30 | 123.2 (2) |
| C16—C17—C18 | 120.8 (3) | O29—N28—C24 | 118.7 (2) |
| C16—C17—H17 | 119.6 | O30—N28—C24 | 118.1 (2) |
| C18—C17—H17 | 119.6 | ||
| N2—C3—C4—C12 | 179.4 (3) | C22—C23—C24—N28 | 176.4 (2) |
| C19—C3—C4—C12 | −0.2 (5) | C23—C24—C25—C26 | 2.2 (4) |
| N2—C3—C4—C5 | 0.3 (3) | N28—C24—C25—C26 | −177.0 (2) |
| C19—C3—C4—C5 | −179.4 (2) | C22—C21—C26—C25 | −2.0 (3) |
| C12—C4—C5—N6 | −0.3 (3) | N10—C21—C26—C25 | 175.2 (2) |
| C3—C4—C5—N6 | 179.1 (2) | C24—C25—C26—C21 | 0.2 (3) |
| C12—C4—C5—N1 | −179.93 (19) | N6—C5—N1—N2 | −179.1 (2) |
| C3—C4—C5—N1 | −0.5 (2) | C4—C5—N1—N2 | 0.6 (2) |
| N6—C7—C8—C12 | −0.4 (4) | N6—C5—N1—C13 | 1.1 (4) |
| N6—C7—C8—C9 | −178.5 (2) | C4—C5—N1—C13 | −179.2 (2) |
| C12—C8—C9—O20 | −178.1 (3) | C18—C13—N1—C5 | −140.1 (2) |
| C7—C8—C9—O20 | 0.2 (4) | C14—C13—N1—C5 | 41.9 (4) |
| C12—C8—C9—N10 | 2.0 (3) | C18—C13—N1—N2 | 40.1 (3) |
| C7—C8—C9—N10 | −179.7 (2) | C14—C13—N1—N2 | −137.9 (2) |
| C7—C8—C12—C4 | 0.0 (3) | C4—C3—N2—N1 | 0.1 (3) |
| C9—C8—C12—C4 | 178.5 (2) | C19—C3—N2—N1 | 179.8 (2) |
| C7—C8—C12—C11 | −179.7 (2) | C5—N1—N2—C3 | −0.4 (3) |
| C9—C8—C12—C11 | −1.2 (2) | C13—N1—N2—C3 | 179.4 (2) |
| C5—C4—C12—C8 | 0.3 (3) | C8—C7—N6—C5 | 0.4 (4) |
| C3—C4—C12—C8 | −178.8 (3) | N1—C5—N6—C7 | 179.5 (2) |
| C5—C4—C12—C11 | 179.9 (2) | C4—C5—N6—C7 | −0.1 (4) |
| C3—C4—C12—C11 | 0.9 (5) | O27—C11—N10—C9 | −178.1 (2) |
| O27—C11—C12—C8 | 179.4 (2) | C12—C11—N10—C9 | 1.3 (2) |
| N10—C11—C12—C8 | 0.0 (2) | O27—C11—N10—C21 | −5.2 (4) |
| O27—C11—C12—C4 | −0.3 (4) | C12—C11—N10—C21 | 174.2 (2) |
| N10—C11—C12—C4 | −179.6 (2) | O20—C9—N10—C11 | 178.1 (2) |
| C18—C13—C14—C15 | −0.5 (4) | C8—C9—N10—C11 | −2.0 (3) |
| N1—C13—C14—C15 | 177.4 (2) | O20—C9—N10—C21 | 5.4 (4) |
| C13—C14—C15—C16 | −1.1 (4) | C8—C9—N10—C21 | −174.7 (2) |
| C14—C15—C16—C17 | 1.5 (4) | C26—C21—N10—C11 | −115.1 (2) |
| C15—C16—C17—C18 | −0.3 (4) | C22—C21—N10—C11 | 62.1 (3) |
| C14—C13—C18—C17 | 1.8 (4) | C26—C21—N10—C9 | 56.9 (3) |
| N1—C13—C18—C17 | −176.2 (2) | C22—C21—N10—C9 | −125.9 (2) |
| C16—C17—C18—C13 | −1.4 (4) | C23—C24—N28—O29 | 14.8 (3) |
| C26—C21—C22—C23 | 1.4 (4) | C25—C24—N28—O29 | −166.1 (2) |
| N10—C21—C22—C23 | −175.8 (2) | C23—C24—N28—O30 | −164.4 (2) |
| C21—C22—C23—C24 | 1.0 (4) | C25—C24—N28—O30 | 14.8 (3) |
| C22—C23—C24—C25 | −2.8 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7···O30i | 0.93 | 2.55 | 3.216 (3) | 129 |
| C18—H18···O30ii | 0.93 | 2.44 | 3.197 (3) | 139 |
Symmetry codes: (i) x, y−1, z; (ii) x−1, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2318).
References
- Barreiro, E. J. L., Fraga, C. A. M., Sudo, G. Z., Sudo, R. T. & Menegatti, R. (2006). WO Patent 092 032 A3, September 8, 2006.
- Carneiro, E. O., De Oliveira, V., Menegatti, R., Fraga, C. A. M. & Barreiro, E. J. L. (2005). Rev. Electron. Farm.2, 44–47.
- Enraf–Nonius (1993). CAD-4-PC Enraf–Nonius, Delft, The Netherlands.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Harms, K. & Wocadlo, S. (1995). XCAD4-PC University of Marburg, Germany.
- Menegatti, R., Silva, G. M. S., Zapata-Sudo, G., Raimundo, J. M., Sudo, R. T., Barreiro, E. J. & Fraga, C. A. M. (2006). Bioorg. Med. Chem.14, 632–640. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808037240/tk2318sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808037240/tk2318Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report