Abstract
In the title compound, C29H22O4, the chromene ring is almost planar with a small puckering [0.143 (2) Å]. The crystal structure is stabilized by C—H⋯O and C—H⋯π interactions. Edge-to-face (centroid–centroid distances of 3.894 and 3.673 Å) and face-to-face (centroid–centroid distance of 3.460 Å) π–π-ring electron interactions are also observed.
Related literature
For the biological and pharmacological properties of benzopyrans and their derivatives, see: Brooks (1998 ▶); Hatakeyama et al. (1988 ▶); Hyana & Saimoto (1987 ▶); Tang et al. (2007 ▶). For the importance of 4H-chromenes, see Liu et al. (2007 ▶); Wang, Fang et al. (2003 ▶); Wang, Zhang et al. (2003 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶); Desiraju (1989 ▶); Desiraju & Steiner (1999 ▶); Etter (1990 ▶).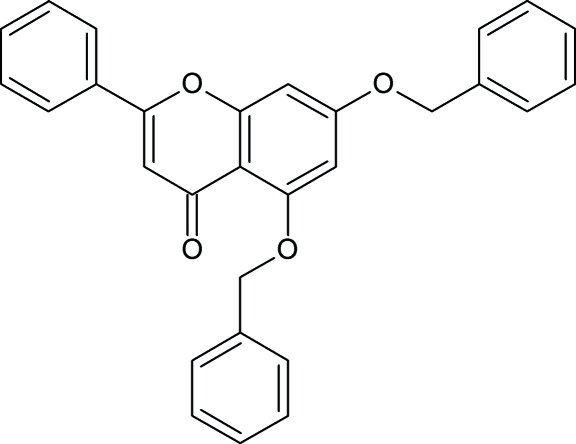
Experimental
Crystal data
C29H22O4
M r = 434.47
Triclinic,
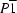
a = 9.496 (3) Å
b = 11.572 (3) Å
c = 11.767 (3) Å
α = 66.564 (4)°
β = 79.668 (5)°
γ = 73.836 (5)°
V = 1136.1 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 293 (2) K
0.45 × 0.33 × 0.23 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1998 ▶) T min = 0.963, T max = 0.981
13435 measured reflections
5302 independent reflections
3534 reflections with I > 2σ(I)
R int = 0.018
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.155
S = 1.05
5302 reflections
298 parameters
H-atom parameters constrained
Δρmax = 0.20 e Å−3
Δρmin = −0.23 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680803938X/fb2125sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803938X/fb2125Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16⋯O17i | 0.93 | 2.57 | 3.212 (3) | 127 |
| C30—H30⋯Cg1ii | 0.93 | 3.12 | 3.838 | 135 |
| C8—H8⋯Cg2iii | 0.93 | 3.29 | 4.066 | 142 |
| C27—H27B⋯Cg2iii | 0.97 | 3.18 | 4.083 | 156 |
Symmetry codes: (i) 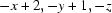 ; (ii)
; (ii) 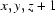 ; (iii)
; (iii) 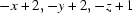 . Cg1 and Cg2 are the centroids of the C20–C25 and C28–C33 rings, respectively.
. Cg1 and Cg2 are the centroids of the C20–C25 and C28–C33 rings, respectively.
Acknowledgments
AN thanks Dr Naresh Kumar and Dr G. Vengatachalam, School of Chemistry, Bharathidasan University, Tiruchirappalli, and Organica Aromatics Pvt Ltd Bangalore, India, for providing laboratory facilities.
supplementary crystallographic information
Comment
Chromenes (benzopyrans) and their derivatives have numerous biological and pharmacological properties (Tang et al., 2007) such as antisterility (Brooks, 1998) and anticancer activity (Hyana & Saimoto, 1987). In addition, polyfunctionalized chromene units are present in numerous natural products (Hatakeyama et al., 1988). 4H-chromenes are important synthons for some natural products (Liu et al., 2007). As a part of our structural investigations on 4H-chromene derivatives and compounds containing the benzopyran fragment, the single-crystal X-ray diffraction study on the title compound was carried out.
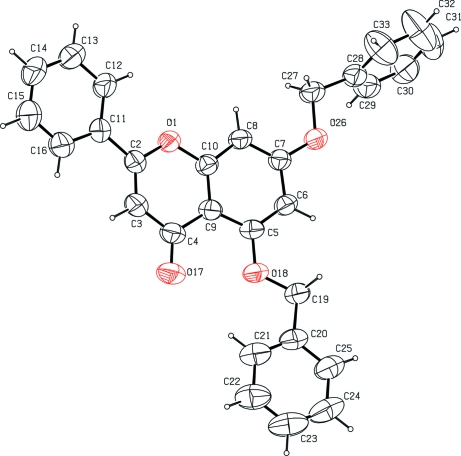
The chromene ring is almost planar similarly as those found in the related chromene derivatives (Wang, Zhang et al., 2003; Wang, Fang et al., 2003). The total puckering amplitude of the chromene ring is 0.143 (2) Å in the title structure. The interplanar angle between the chromene ring and the 2-phenyl ring is 6.8 (2)° thereby indicating the almost coplanar arrangement (Fig. 1). The benzyl group at C5 is slightly distorted from coplanarity with the chromene ring whereas the benzyl group at C7 is clearly non-coplanar as discerned from the respective interplanar angles of 7.6 (1)° and 70.01 (7)°.
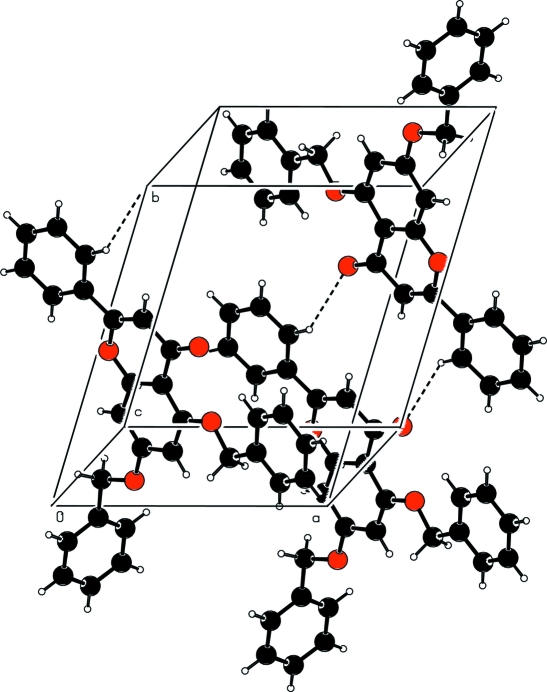
The crystal structure is stabilized by the interplay of C–H···O and C–H···π interactions (Fig. 2, Table 1; Desiraju, 1989; Desiraju & Steiner, 1999). The C12–H12···O1 interaction is involved in a motif of a graph set S(5) (Bernstein et al., 1995; Etter, 1990). In another S(5) motif, C21–H21···O18 interaction is involved. The C8–H8···Cg2ii and C27–H27···Cg2ii (Cg2 is the centroid of the ring C28\C29···C33) interactions take part in the motif of the graph set R12(7) where the entire Cg2 ring C28\C29···C33 is considered as a single acceptor atom.
There are two edge-to-face π···π interactions between Cg3 (O1\C2\C3\C4\C9\C10) and Cg4 (C5\C6\C7\C8\C9\C10) [2-x, 2-y, -z] at 3.894 Å with α = 4.44, β = 26.68, γ = 31.06° and perpendicular distances being 3.336 and 3.480 Å, Cg4 and Cg1 (C20\C21\C22\C23\C24\C25) [1-x, 2-y, -z] at 3.673 Å with α = 6.87, β = 20.69, γ = 16.31° and perpendicular distances being 3.525 and 3.436 Å. There is a face to face π···π interaction between two symmetery related Cg4 (2-x, 2-y, -z) rings at 3.460 Å with α = 0.00, β = 10.24, γ = 10.24° and perpendicular distances being 3.405 Å (α is the dihedral angle between the planes I and J where I is the plane of centroid 1 and J is the plane of centroid 2, β is the angle between the vector Cg(I)→Cg(J) and the normal to plane I, γ is the angle between the vector Cg(I)→ Cg(J) and the normal to plane J, the two perpendicular distances denote the perpendicular distances of Cg(I) on ring J and Cg(J) on ring I).
Experimental
A suspension of chrysin (3.93 mmol, 1.00 g) and potassium carbonate (11.81 mmol, 1.64 g) in dimethyl formamide (10 ml) were added into a round bottom flask. The reaction mixture was heated to 383 K for 2–3 h. The reaction mixture was then cooled to 353 K and benzyl chloride (15.74 mmol, 1.99 g) was slowly added to the reaction mixture with the help of a dropping funnel. The reaction mixture was maintained for 8–9 h at 353 K and monitored by a high pressure liquid chromatography (HPLC). After completion of the reaction, the content was quenched with water and stirred for 30–45 min at 303 K. The obtained crude solid was filtered and washed with plenty of water followed by methanol and dried under vacuum at 343 K. The crude product was dissolved in 20 ml of 1:1 (volume) mixture of dichloromethane and n-hexane. The clear solution was kept for a week without stirring. Diffraction quality prism shaped crystals of average size 0.3 mm were obtained which were filtered and washed with n-hexane and dried under vacuum at 343 K. Yield: 90%
Refinement
All the H-atoms were observed in the difference electron density map. However, they were situated into idealized positions with C–H = 0.93 and 0.97 Å for aryl and methylene H, respectively, and constrained to ride on their parent atoms with Uiso(H)=1.2Ueq(C).
Figures
Fig. 1.
The title molecule showing the displacement ellipsoids depicted at the 50% probability level for all non-H atoms. The hydrogen atoms are drawn as spheres of arbitrary radius.
Fig. 2.
The molecular packing viewed down the a-axis. Dashed lines represent weak C–H···O interactions.
Crystal data
| C29H22O4 | Z = 2 |
| Mr = 434.47 | F000 = 456 |
| Triclinic, P1 | Dx = 1.270 Mg m−3 |
| Hall symbol: -P 1 | Melting point = 439–441 K |
| a = 9.496 (3) Å | Mo Kα radiation λ = 0.71073 Å |
| b = 11.572 (3) Å | Cell parameters from 589 reflections |
| c = 11.767 (3) Å | θ = 2.5–27.5º |
| α = 66.564 (4)º | µ = 0.08 mm−1 |
| β = 79.668 (5)º | T = 293 (2) K |
| γ = 73.836 (5)º | Prism, colourless |
| V = 1136.1 (5) Å3 | 0.45 × 0.33 × 0.23 mm |
Data collection
| Bruker SMART APEX CCD diffractometer | 5302 independent reflections |
| Radiation source: fine-focus sealed tube | 3534 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.018 |
| Detector resolution: 0.3 pixels mm-1 | θmax = 28.0º |
| T = 293(2) K | θmin = 1.9º |
| ω scans | h = −12→12 |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1998) | k = −15→15 |
| Tmin = 0.963, Tmax = 0.981 | l = −15→15 |
| 13435 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | H-atom parameters constrained |
| wR(F2) = 0.155 | w = 1/[σ2(Fo2) + (0.0655P)2 + 0.1818P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 5302 reflections | Δρmax = 0.20 e Å−3 |
| 298 parameters | Δρmin = −0.23 e Å−3 |
| 88 constraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 1.10569 (12) | 0.71663 (11) | 0.18466 (11) | 0.0536 (3) | |
| C2 | 1.13210 (18) | 0.63546 (16) | 0.12145 (16) | 0.0511 (4) | |
| C3 | 1.0413 (2) | 0.65365 (18) | 0.03827 (18) | 0.0619 (5) | |
| H3 | 1.0625 | 0.5959 | −0.0026 | 0.074* | |
| C4 | 0.91202 (19) | 0.75798 (18) | 0.00843 (17) | 0.0585 (5) | |
| C5 | 0.78763 (16) | 0.96910 (16) | 0.04245 (15) | 0.0487 (4) | |
| C6 | 0.77971 (17) | 1.05076 (16) | 0.10297 (15) | 0.0502 (4) | |
| H6 | 0.7084 | 1.1279 | 0.0852 | 0.060* | |
| C7 | 0.87887 (17) | 1.01797 (16) | 0.19122 (15) | 0.0483 (4) | |
| C8 | 0.98609 (17) | 0.90462 (16) | 0.21876 (15) | 0.0503 (4) | |
| H8 | 1.0521 | 0.8825 | 0.2777 | 0.060* | |
| C9 | 0.89526 (17) | 0.85064 (16) | 0.06822 (15) | 0.0482 (4) | |
| C10 | 0.99182 (16) | 0.82506 (15) | 0.15545 (15) | 0.0469 (4) | |
| C11 | 1.26652 (19) | 0.53363 (16) | 0.15672 (17) | 0.0547 (4) | |
| C12 | 1.3574 (2) | 0.53208 (19) | 0.23695 (19) | 0.0690 (5) | |
| H12 | 1.3321 | 0.5957 | 0.2711 | 0.083* | |
| C13 | 1.4850 (3) | 0.4377 (2) | 0.2670 (2) | 0.0883 (7) | |
| H13 | 1.5451 | 0.4385 | 0.3209 | 0.106* | |
| C14 | 1.5238 (3) | 0.3431 (2) | 0.2185 (3) | 0.0958 (8) | |
| H14 | 1.6105 | 0.2799 | 0.2381 | 0.115* | |
| C15 | 1.4336 (3) | 0.3424 (2) | 0.1410 (3) | 0.1085 (10) | |
| H15 | 1.4585 | 0.2773 | 0.1087 | 0.130* | |
| C16 | 1.3067 (2) | 0.4365 (2) | 0.1098 (2) | 0.0867 (7) | |
| H16 | 1.2470 | 0.4346 | 0.0562 | 0.104* | |
| O17 | 0.82679 (16) | 0.76574 (15) | −0.06285 (15) | 0.0858 (5) | |
| O18 | 0.69794 (12) | 0.99537 (12) | −0.04619 (11) | 0.0609 (3) | |
| C19 | 0.59954 (19) | 1.11810 (18) | −0.08836 (17) | 0.0587 (5) | |
| H19A | 0.6522 | 1.1861 | −0.1123 | 0.070* | |
| H19B | 0.5245 | 1.1272 | −0.0227 | 0.070* | |
| C20 | 0.52982 (18) | 1.12834 (19) | −0.19821 (16) | 0.0588 (5) | |
| C21 | 0.5695 (2) | 1.0332 (2) | −0.24758 (18) | 0.0691 (5) | |
| H21 | 0.6430 | 0.9601 | −0.2137 | 0.083* | |
| C22 | 0.5008 (3) | 1.0456 (3) | −0.3475 (2) | 0.0871 (7) | |
| H22 | 0.5285 | 0.9806 | −0.3801 | 0.105* | |
| C23 | 0.3932 (3) | 1.1517 (3) | −0.3984 (2) | 0.0995 (8) | |
| H23 | 0.3478 | 1.1597 | −0.4657 | 0.119* | |
| C24 | 0.3522 (3) | 1.2469 (3) | −0.3498 (2) | 0.0956 (8) | |
| H24 | 0.2784 | 1.3196 | −0.3843 | 0.115* | |
| C25 | 0.4196 (2) | 1.2359 (2) | −0.2496 (2) | 0.0767 (6) | |
| H25 | 0.3907 | 1.3009 | −0.2170 | 0.092* | |
| O26 | 0.86159 (12) | 1.10706 (11) | 0.24369 (11) | 0.0595 (3) | |
| C27 | 0.9671 (2) | 1.08276 (19) | 0.32789 (18) | 0.0655 (5) | |
| H27A | 1.0648 | 1.0779 | 0.2855 | 0.079* | |
| H27B | 0.9663 | 1.0009 | 0.3963 | 0.079* | |
| C28 | 0.9288 (2) | 1.19031 (17) | 0.37608 (16) | 0.0578 (5) | |
| C29 | 0.8056 (2) | 1.2047 (2) | 0.45477 (18) | 0.0678 (5) | |
| H29 | 0.7427 | 1.1492 | 0.4750 | 0.081* | |
| C30 | 0.7731 (3) | 1.2992 (2) | 0.5043 (2) | 0.0807 (6) | |
| H30 | 0.6885 | 1.3082 | 0.5569 | 0.097* | |
| C31 | 0.8657 (4) | 1.3795 (2) | 0.4758 (2) | 0.1003 (9) | |
| H31 | 0.8448 | 1.4434 | 0.5095 | 0.120* | |
| C32 | 0.9893 (4) | 1.3666 (3) | 0.3978 (3) | 0.1228 (12) | |
| H32 | 1.0527 | 1.4214 | 0.3787 | 0.147* | |
| C33 | 1.0199 (3) | 1.2724 (3) | 0.3475 (2) | 0.0965 (8) | |
| H33 | 1.1034 | 1.2647 | 0.2936 | 0.116* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0515 (6) | 0.0537 (7) | 0.0603 (7) | 0.0017 (5) | −0.0168 (5) | −0.0296 (6) |
| C2 | 0.0536 (9) | 0.0488 (9) | 0.0564 (10) | −0.0114 (7) | −0.0058 (8) | −0.0247 (8) |
| C3 | 0.0644 (11) | 0.0605 (11) | 0.0732 (12) | −0.0072 (9) | −0.0160 (9) | −0.0375 (10) |
| C4 | 0.0571 (10) | 0.0673 (11) | 0.0617 (11) | −0.0133 (9) | −0.0150 (8) | −0.0309 (9) |
| C5 | 0.0392 (8) | 0.0589 (10) | 0.0505 (9) | −0.0107 (7) | −0.0094 (7) | −0.0207 (8) |
| C6 | 0.0413 (8) | 0.0527 (9) | 0.0542 (10) | −0.0030 (7) | −0.0119 (7) | −0.0189 (8) |
| C7 | 0.0452 (8) | 0.0530 (9) | 0.0508 (9) | −0.0063 (7) | −0.0084 (7) | −0.0246 (8) |
| C8 | 0.0461 (9) | 0.0573 (10) | 0.0523 (9) | −0.0024 (7) | −0.0154 (7) | −0.0265 (8) |
| C9 | 0.0433 (8) | 0.0562 (10) | 0.0500 (9) | −0.0111 (7) | −0.0063 (7) | −0.0236 (8) |
| C10 | 0.0417 (8) | 0.0505 (9) | 0.0489 (9) | −0.0060 (7) | −0.0066 (7) | −0.0203 (7) |
| C11 | 0.0560 (10) | 0.0471 (9) | 0.0635 (11) | −0.0069 (8) | −0.0085 (8) | −0.0246 (8) |
| C12 | 0.0745 (12) | 0.0592 (11) | 0.0775 (13) | 0.0086 (9) | −0.0264 (10) | −0.0371 (10) |
| C13 | 0.0869 (15) | 0.0769 (14) | 0.1058 (18) | 0.0158 (12) | −0.0460 (14) | −0.0452 (14) |
| C14 | 0.0860 (16) | 0.0741 (15) | 0.129 (2) | 0.0242 (12) | −0.0414 (15) | −0.0530 (15) |
| C15 | 0.1037 (18) | 0.0862 (17) | 0.159 (3) | 0.0276 (14) | −0.0466 (18) | −0.0866 (18) |
| C16 | 0.0804 (14) | 0.0795 (15) | 0.1213 (19) | 0.0102 (12) | −0.0371 (13) | −0.0655 (14) |
| O17 | 0.0821 (10) | 0.0934 (11) | 0.1075 (11) | 0.0004 (8) | −0.0444 (9) | −0.0614 (9) |
| O18 | 0.0536 (7) | 0.0666 (8) | 0.0682 (8) | −0.0024 (6) | −0.0269 (6) | −0.0290 (6) |
| C19 | 0.0521 (10) | 0.0597 (11) | 0.0642 (11) | −0.0109 (8) | −0.0187 (8) | −0.0178 (9) |
| C20 | 0.0453 (9) | 0.0740 (12) | 0.0528 (10) | −0.0185 (9) | −0.0099 (8) | −0.0130 (9) |
| C21 | 0.0544 (11) | 0.0946 (15) | 0.0583 (11) | −0.0131 (10) | −0.0102 (9) | −0.0284 (11) |
| C22 | 0.0783 (14) | 0.127 (2) | 0.0646 (13) | −0.0213 (14) | −0.0128 (11) | −0.0418 (14) |
| C23 | 0.0856 (17) | 0.147 (3) | 0.0628 (14) | −0.0260 (17) | −0.0273 (12) | −0.0263 (16) |
| C24 | 0.0728 (15) | 0.108 (2) | 0.0810 (16) | −0.0075 (13) | −0.0355 (12) | −0.0054 (15) |
| C25 | 0.0621 (12) | 0.0810 (14) | 0.0764 (14) | −0.0089 (10) | −0.0246 (10) | −0.0151 (11) |
| O26 | 0.0566 (7) | 0.0597 (7) | 0.0690 (8) | 0.0068 (6) | −0.0251 (6) | −0.0357 (6) |
| C27 | 0.0673 (11) | 0.0692 (12) | 0.0670 (12) | 0.0059 (9) | −0.0303 (9) | −0.0360 (10) |
| C28 | 0.0663 (11) | 0.0582 (11) | 0.0517 (10) | −0.0029 (9) | −0.0181 (9) | −0.0249 (9) |
| C29 | 0.0647 (12) | 0.0765 (13) | 0.0680 (12) | −0.0101 (10) | −0.0132 (10) | −0.0333 (11) |
| C30 | 0.0873 (15) | 0.0879 (16) | 0.0690 (14) | 0.0021 (13) | −0.0108 (11) | −0.0439 (12) |
| C31 | 0.167 (3) | 0.0714 (15) | 0.0735 (16) | −0.0217 (17) | −0.0086 (17) | −0.0416 (13) |
| C32 | 0.192 (3) | 0.120 (2) | 0.098 (2) | −0.093 (2) | 0.033 (2) | −0.0632 (19) |
| C33 | 0.119 (2) | 0.116 (2) | 0.0828 (16) | −0.0573 (17) | 0.0248 (14) | −0.0581 (15) |
Geometric parameters (Å, °)
| O1—C2 | 1.3623 (19) | C19—C20 | 1.505 (2) |
| O1—C10 | 1.3772 (19) | C19—H19A | 0.9700 |
| C2—C3 | 1.336 (2) | C19—H19B | 0.9700 |
| C2—C11 | 1.468 (2) | C20—C21 | 1.375 (3) |
| C3—C4 | 1.442 (2) | C20—C25 | 1.384 (3) |
| C3—H3 | 0.9300 | C21—C22 | 1.386 (3) |
| C4—O17 | 1.2287 (19) | C21—H21 | 0.9300 |
| C4—C9 | 1.461 (2) | C22—C23 | 1.359 (3) |
| C5—O18 | 1.3521 (18) | C22—H22 | 0.9300 |
| C5—C6 | 1.372 (2) | C23—C24 | 1.369 (4) |
| C5—C9 | 1.420 (2) | C23—H23 | 0.9300 |
| C6—C7 | 1.395 (2) | C24—C25 | 1.386 (3) |
| C6—H6 | 0.9300 | C24—H24 | 0.9300 |
| C7—O26 | 1.3590 (19) | C25—H25 | 0.9300 |
| C7—C8 | 1.378 (2) | O26—C27 | 1.4291 (19) |
| C8—C10 | 1.382 (2) | C27—C28 | 1.496 (2) |
| C8—H8 | 0.9300 | C27—H27A | 0.9700 |
| C9—C10 | 1.388 (2) | C27—H27B | 0.9700 |
| C11—C16 | 1.376 (2) | C28—C33 | 1.365 (3) |
| C11—C12 | 1.381 (2) | C28—C29 | 1.371 (3) |
| C12—C13 | 1.376 (3) | C29—C30 | 1.373 (3) |
| C12—H12 | 0.9300 | C29—H29 | 0.9300 |
| C13—C14 | 1.364 (3) | C30—C31 | 1.361 (4) |
| C13—H13 | 0.9300 | C30—H30 | 0.9300 |
| C14—C15 | 1.362 (3) | C31—C32 | 1.367 (4) |
| C14—H14 | 0.9300 | C31—H31 | 0.9300 |
| C15—C16 | 1.373 (3) | C32—C33 | 1.378 (3) |
| C15—H15 | 0.9300 | C32—H32 | 0.9300 |
| C16—H16 | 0.9300 | C33—H33 | 0.9300 |
| O18—C19 | 1.415 (2) | ||
| C2—O1—C10 | 119.99 (12) | O18—C19—H19A | 110.1 |
| C3—C2—O1 | 120.68 (15) | C20—C19—H19A | 110.1 |
| C3—C2—C11 | 127.46 (16) | O18—C19—H19B | 110.1 |
| O1—C2—C11 | 111.85 (14) | C20—C19—H19B | 110.1 |
| C2—C3—C4 | 123.57 (16) | H19A—C19—H19B | 108.4 |
| C2—C3—H3 | 118.2 | C21—C20—C25 | 118.81 (18) |
| C4—C3—H3 | 118.2 | C21—C20—C19 | 122.32 (17) |
| O17—C4—C3 | 121.05 (16) | C25—C20—C19 | 118.86 (19) |
| O17—C4—C9 | 124.67 (17) | C20—C21—C22 | 120.4 (2) |
| C3—C4—C9 | 114.27 (14) | C20—C21—H21 | 119.8 |
| O18—C5—C6 | 123.46 (15) | C22—C21—H21 | 119.8 |
| O18—C5—C9 | 115.42 (14) | C23—C22—C21 | 120.6 (2) |
| C6—C5—C9 | 121.11 (14) | C23—C22—H22 | 119.7 |
| C5—C6—C7 | 119.92 (15) | C21—C22—H22 | 119.7 |
| C5—C6—H6 | 120.0 | C22—C23—C24 | 119.6 (2) |
| C7—C6—H6 | 120.0 | C22—C23—H23 | 120.2 |
| O26—C7—C8 | 124.22 (14) | C24—C23—H23 | 120.2 |
| O26—C7—C6 | 114.59 (14) | C23—C24—C25 | 120.6 (2) |
| C8—C7—C6 | 121.18 (15) | C23—C24—H24 | 119.7 |
| C7—C8—C10 | 117.46 (14) | C25—C24—H24 | 119.7 |
| C7—C8—H8 | 121.3 | C20—C25—C24 | 120.0 (2) |
| C10—C8—H8 | 121.3 | C20—C25—H25 | 120.0 |
| C10—C9—C5 | 115.96 (15) | C24—C25—H25 | 120.0 |
| C10—C9—C4 | 119.12 (15) | C7—O26—C27 | 116.84 (13) |
| C5—C9—C4 | 124.91 (14) | O26—C27—C28 | 108.76 (14) |
| O1—C10—C8 | 113.76 (13) | O26—C27—H27A | 109.9 |
| O1—C10—C9 | 121.86 (14) | C28—C27—H27A | 109.9 |
| C8—C10—C9 | 124.36 (15) | O26—C27—H27B | 109.9 |
| C16—C11—C12 | 117.70 (17) | C28—C27—H27B | 109.9 |
| C16—C11—C2 | 120.75 (16) | H27A—C27—H27B | 108.3 |
| C12—C11—C2 | 121.55 (15) | C33—C28—C29 | 118.51 (19) |
| C13—C12—C11 | 120.97 (18) | C33—C28—C27 | 120.59 (19) |
| C13—C12—H12 | 119.5 | C29—C28—C27 | 120.83 (19) |
| C11—C12—H12 | 119.5 | C28—C29—C30 | 121.4 (2) |
| C14—C13—C12 | 120.5 (2) | C28—C29—H29 | 119.3 |
| C14—C13—H13 | 119.8 | C30—C29—H29 | 119.3 |
| C12—C13—H13 | 119.8 | C31—C30—C29 | 119.4 (2) |
| C15—C14—C13 | 119.0 (2) | C31—C30—H30 | 120.3 |
| C15—C14—H14 | 120.5 | C29—C30—H30 | 120.3 |
| C13—C14—H14 | 120.5 | C30—C31—C32 | 120.1 (2) |
| C14—C15—C16 | 121.0 (2) | C30—C31—H31 | 119.9 |
| C14—C15—H15 | 119.5 | C32—C31—H31 | 119.9 |
| C16—C15—H15 | 119.5 | C31—C32—C33 | 119.9 (3) |
| C15—C16—C11 | 120.8 (2) | C31—C32—H32 | 120.0 |
| C15—C16—H16 | 119.6 | C33—C32—H32 | 120.0 |
| C11—C16—H16 | 119.6 | C28—C33—C32 | 120.7 (2) |
| C5—O18—C19 | 119.10 (13) | C28—C33—H33 | 119.7 |
| O18—C19—C20 | 107.99 (15) | C32—C33—H33 | 119.7 |
| C10—O1—C2—C3 | −5.7 (2) | C2—C11—C12—C13 | −178.5 (2) |
| C10—O1—C2—C11 | 174.00 (14) | C11—C12—C13—C14 | −0.4 (4) |
| O1—C2—C3—C4 | 0.4 (3) | C12—C13—C14—C15 | −0.7 (4) |
| C11—C2—C3—C4 | −179.18 (17) | C13—C14—C15—C16 | 1.1 (5) |
| C2—C3—C4—O17 | −175.26 (19) | C14—C15—C16—C11 | −0.4 (5) |
| C2—C3—C4—C9 | 5.6 (3) | C12—C11—C16—C15 | −0.7 (4) |
| O18—C5—C6—C7 | −178.38 (15) | C2—C11—C16—C15 | 178.9 (2) |
| C9—C5—C6—C7 | 0.2 (3) | C6—C5—O18—C19 | 6.3 (2) |
| C5—C6—C7—O26 | 179.15 (14) | C9—C5—O18—C19 | −172.41 (14) |
| C5—C6—C7—C8 | 0.3 (3) | C5—O18—C19—C20 | 171.97 (14) |
| O26—C7—C8—C10 | −178.51 (15) | O18—C19—C20—C21 | −3.9 (2) |
| C6—C7—C8—C10 | 0.2 (3) | O18—C19—C20—C25 | 174.59 (16) |
| O18—C5—C9—C10 | 177.49 (14) | C25—C20—C21—C22 | 0.4 (3) |
| C6—C5—C9—C10 | −1.2 (2) | C19—C20—C21—C22 | 178.92 (18) |
| O18—C5—C9—C4 | −1.2 (2) | C20—C21—C22—C23 | 0.0 (3) |
| C6—C5—C9—C4 | −179.92 (16) | C21—C22—C23—C24 | −0.3 (4) |
| O17—C4—C9—C10 | 174.32 (18) | C22—C23—C24—C25 | 0.2 (4) |
| C3—C4—C9—C10 | −6.5 (2) | C21—C20—C25—C24 | −0.5 (3) |
| O17—C4—C9—C5 | −7.0 (3) | C19—C20—C25—C24 | −179.12 (19) |
| C3—C4—C9—C5 | 172.14 (16) | C23—C24—C25—C20 | 0.3 (4) |
| C2—O1—C10—C8 | −174.37 (14) | C8—C7—O26—C27 | 3.3 (3) |
| C2—O1—C10—C9 | 4.4 (2) | C6—C7—O26—C27 | −175.52 (15) |
| C7—C8—C10—O1 | 177.45 (14) | C7—O26—C27—C28 | −179.56 (15) |
| C7—C8—C10—C9 | −1.3 (3) | O26—C27—C28—C33 | −114.1 (2) |
| C5—C9—C10—O1 | −176.87 (14) | O26—C27—C28—C29 | 69.2 (2) |
| C4—C9—C10—O1 | 1.9 (2) | C33—C28—C29—C30 | 0.1 (3) |
| C5—C9—C10—C8 | 1.8 (2) | C27—C28—C29—C30 | 176.85 (18) |
| C4—C9—C10—C8 | −179.40 (16) | C28—C29—C30—C31 | −0.7 (3) |
| C3—C2—C11—C16 | −5.0 (3) | C29—C30—C31—C32 | 0.5 (4) |
| O1—C2—C11—C16 | 175.35 (18) | C30—C31—C32—C33 | 0.3 (5) |
| C3—C2—C11—C12 | 174.54 (19) | C29—C28—C33—C32 | 0.7 (4) |
| O1—C2—C11—C12 | −5.1 (2) | C27—C28—C33—C32 | −176.1 (2) |
| C16—C11—C12—C13 | 1.1 (3) | C31—C32—C33—C28 | −0.9 (5) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···O1 | 0.93 | 2.37 | 2.701 (2) | 101 |
| C21—H21···O18 | 0.93 | 2.33 | 2.685 (2) | 102 |
| C16—H16···O17i | 0.93 | 2.57 | 3.212 (3) | 127 |
| C30—H30···Cg1ii | 0.93 | 3.12 | 3.838 | 135 |
| C8—H8···Cg2iii | 0.93 | 3.30 | 4.066 | 142 |
| C27—H27B···Cg2iii | 0.97 | 3.18 | 4.083 | 156 |
Symmetry codes: (i) −x+2, −y+1, −z; (ii) x, y, z+1; (iii) −x+2, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FB2125).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Brooks, G. T. (1998). Pestic. Sci.22, 41–50.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Desiraju, G. R. (1989). Crystal Engineering: The Design of Organic Solids, pp. 125–167. Amsterdam: Elsevier.
- Desiraju, G. R. & Steiner, T. (1999). The Weak Hydrogen Bond in Structural Chemistry and Biology, pp. 11–40. New York: Oxford University Press.
- Etter, M. C. (1990). Acc. Chem. Res.23, 120–126.
- Hatakeyama, S., Ochi, N., Numata, H. & Takano, S. (1988). J. Chem. Soc. Chem. Commun. pp. 1202–1204.
- Hyana, T. & Saimoto, H. (1987). Jpn Patent JP 621 812 768.
- Liu, C.-B., Chen, Y.-H., Zhou, X.-Y., Ding, L. & Wen, H.-L. (2007). Acta Cryst. E63, o90–o91.
- Sheldrick, G. M. (1998). SADABS University of Gottingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Tang, Q.-G., Wu, W.-Y., He, W., Sun, H.-S. & Guo, C. (2007). Acta Cryst. E63, o1437–o1438.
- Wang, J.-F., Fang, M.-J., Huang, H.-Q., Li, G.-L., Su, W.-J. & Zhao, Y.-F. (2003). Acta Cryst. E59, o1517–o1518.
- Wang, J.-F., Zhang, Y.-J., Fang, M.-J., Huang, Y.-J., Wei, Z.-B., Zheng, Z.-H., Su, W.-J. & Zhao, Y.-F. (2003). Acta Cryst. E59, o1244–o1245.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680803938X/fb2125sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803938X/fb2125Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report