Abstract
The title compound, C18H13ClO3, has an intramolecular O—H⋯O=C hydrogen bond between the carbonyl group and the hydroxy substituent on the naphthalene ring system. The angle between the C=O bond plane and the naphthalene ring system is relatively small [20.96 (8)°]. The angle between the benzene ring and the carbonyl group is rather large [35.65 (9)°] compared to that in an analogous compound [3.43 (11)°] having a methoxy group instead of a hydroxy substituent.
Related literature
For the structures of closely related compounds, see: Nakaema et al. (2007 ▶, 2008 ▶); Mitsui et al. (2008 ▶).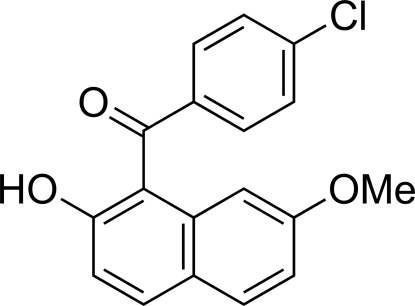
Experimental
Crystal data
C18H13ClO3
M r = 312.73
Orthorhombic,

a = 17.8030 (3) Å
b = 8.68121 (10) Å
c = 18.8683 (3) Å
V = 2916.14 (8) Å3
Z = 8
Cu Kα radiation
μ = 2.41 mm−1
T = 123 K
0.60 × 0.15 × 0.05 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: numerical (NUMABS; Higashi, 1999 ▶) T min = 0.485, T max = 0.886
49864 measured reflections
2669 independent reflections
2347 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.033
wR(F 2) = 0.096
S = 1.08
2669 reflections
205 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.17 e Å−3
Δρmin = −0.25 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 1998 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2004 ▶); program(s) used to solve structure: SIR2004 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPIII (Burnett & Johnson, 1996 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808039603/su2069sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808039603/su2069Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1 | 0.94 (2) | 1.71 (2) | 2.5573 (16) | 148 (2) |
Acknowledgments
This work was financially supported by SEIKI KOGYO CO., Ltd, Tokorozawa, Saitama, Japan.
supplementary crystallographic information
Comment
Recently, we have reported on the crystal structure of 1-(4-chlorobenzoyl)-2,7-dimethoxynaphthalene, (I) [Mitsui et al., 2008]. As a part of our ongoing studies on the synthesis and crystal structure analyses of aroylated naphthalene derivatives, we prepared and analysed the crystal structure of the title compound, (II). Compound (II) was prepared by regioselective demethylation reaction of compound (I) with aluminium trichloride.
The molecular structure of compound (II) is illustrated in Fig. 1. In analogous aroylated naphthalenes, for example compound (I) shown in Fig. 2, the C═O bond plane is almost perpendicular to the mean plane of the naphthalene ring (Nakaema et al., 2007, 2008; Mitsui et al., 2008). In contrast, the angle between the C═O bond and the naphthalene ring in compound (II) is considerably smaller, i.e. 20.96 (8)°. This is apparently caused by the intramolecular O—H···O═C hydrogen bond, which forms a six-membered ring including the carbonyl group and an edge of the naphthalene ring (Fig. 1 and Table 1).
In compound (I) the C═O bond and the benzene ring are almost coplanar with a dihedral angle of 3.43 (11)°. In compound (II) the mean plane of the benzene ring is twisted away from the C═O bond by 35.65 (9)°. This is presumably caused by the release of the rather large steric repulsion between the benzene ring and the naphthalene ring brought about by the small angle of the C═O bond plane and the naphthalene ring. The dihedral angle between the naphthalene ring (C1—C10) and the benzene ring (C12—C17) is 58.10 (6)°.
In the crystal structure the molecular packing of (II) is mainly stabilized by van der Waals interactions. The naphthalene rings interact with the phenyl rings [C5···C13 = 3.363 (2) Å] and the carbonyl groups [H6···O1 = 2.70 Å] along the a-axis. They also interact with the methyl groups [H3···C18 = 2.79 Å] and aroyl groups [H6···Cl1 = 2.88 Å] along the c-axis (Fig. 3). On the other hand, the naphthalene rings also interact with the methyl groups [C6···H18B = 2.81 Å, C7···H18B = 2.70 Å] and the phenyl rings [C6···H17 = 2.88 Å, C7···H17 = 2.79 Å] along the b-axis. The naphthalene rings are almost perpendicular to the phenyl rings of the adjacent molecules along the b-axis. In addition, the hydroxy groups interact with the phenyl rings [O2···H14 = 2.71 Å] along the b-axis (Fig. 4).
Experimental
To a solution of 1-(4-chlorobenzoyl)-2,7-dimethoxynaphthalene (33 mg, 0.10 mmol) in CH2Cl2 (1.0 ml) was added AlCl3 (67 mg, 0.50 mmol). The reaction mixture was refluxed for 30 min giving a dark red solution, which was then poured into H2O (5 ml) and CHCl3 (3 ml). The aqueous layer was extracted with CHCl3 (3 × 5 ml). The combined organic layers were washed with brine (3 × 10 ml), and dried over MgSO4 overnight. The solvent was removed in vacuo and the crude material was purified by recrystallization from hexane to give compound (II) as yellow platelets (m.p. 391–391.5 K, yield 23 mg, 75%).
Spectroscopic Data: 1H NMR (300 MHz, CDCl3) δ 11.35 (s, 1H), 7.85 (d, 1H), 7.63 (d, 1H), 7.58 (d, 2H), 7.40 (d, 2H), 7.07 (d, 1H), 6.91 (dd, 1H), 6.58 (d, 1H), 3.37 (s, 3H); 13C NMR (75 MHz, CDCl3) δ 199.1, 162.6, 158.2, 138.8, 138.7, 136.5, 133.8, 130.7, 130.2, 128.9, 123.7, 116.4, 115.8, 113.4, 106.5, 54.5; IR (KBr): 3434, 1623, 1583, 1513, 1214, 843.
Anal. Calcd for C18H13ClO3: C 69.13, H 4.19. Found: C 69.11, H 4.09.
Refinement
All the H-atoms could be located in difference Fourier maps. The OH hydrogen atom was freely refined: O2—H2 = 0.94 (2) Å. The C-bound H-atoms were subsequently refined as riding atoms, with C—H = 0.95 (aromatic) and 0.98 (methyl) Å, and Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of compound (II), showing 50% probability displacement ellipsoids. The intramolecular hydrogen bond is shown as a dashed line.
Fig. 2.
The molecular structure of compound (I) [Mitsui et al., 2008], showing 50% probability displacement ellipsoids.
Fig. 3.
A partial crystal packing diagram of compound (II), viewed down the b-axis (the intermolecular C—H···O and C—H···π interactions are shown as dashed lines).
Fig. 4.
A partial crystal packing diagram of compound (II), viewed down the c-axis (the intermolecular C—H···O and C—H···π interactions are shown as dashed lines).
Crystal data
| C18H13ClO3 | Dx = 1.425 Mg m−3 |
| Mr = 312.73 | Melting point = 391.0–391.5 K |
| Orthorhombic, Pbca | Cu Kα radiation λ = 1.54187 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 42602 reflections |
| a = 17.8030 (3) Å | θ = 3.4–68.2º |
| b = 8.68121 (10) Å | µ = 2.41 mm−1 |
| c = 18.8683 (3) Å | T = 123 K |
| V = 2916.14 (8) Å3 | Plate, yellow |
| Z = 8 | 0.60 × 0.15 × 0.05 mm |
| F000 = 1296 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 2669 independent reflections |
| Radiation source: rotating anode | 2347 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.033 |
| Detector resolution: 10.00 pixels mm-1 | θmax = 68.2º |
| T = 123 K | θmin = 4.7º |
| ω scans | h = −21→21 |
| Absorption correction: numerical(NUMABS; Higashi, 1999) | k = −10→10 |
| Tmin = 0.485, Tmax = 0.886 | l = −22→22 |
| 49864 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: dfimap |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.033 | w = 1/[σ2(Fo2) + (0.0544P)2 + 0.6658P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.096 | (Δ/σ)max = 0.001 |
| S = 1.08 | Δρmax = 0.17 e Å−3 |
| 2669 reflections | Δρmin = −0.25 e Å−3 |
| 205 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00062 (11) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | −0.05110 (2) | 0.23154 (6) | 0.500766 (19) | 0.05295 (17) | |
| O1 | 0.04658 (5) | 0.08073 (14) | 0.17181 (6) | 0.0439 (3) | |
| O2 | 0.12423 (7) | 0.21814 (15) | 0.07692 (6) | 0.0485 (3) | |
| H2 | 0.0840 (13) | 0.163 (3) | 0.0974 (11) | 0.070 (7)* | |
| O3 | 0.29872 (6) | 0.03670 (13) | 0.41875 (6) | 0.0474 (3) | |
| C1 | 0.16824 (8) | 0.17285 (16) | 0.19660 (7) | 0.0299 (3) | |
| C2 | 0.17846 (9) | 0.22425 (17) | 0.12659 (8) | 0.0373 (3) | |
| C3 | 0.24694 (10) | 0.28979 (18) | 0.10428 (8) | 0.0432 (4) | |
| H3 | 0.2518 | 0.3289 | 0.0575 | 0.052* | |
| C4 | 0.30589 (9) | 0.29694 (18) | 0.14974 (9) | 0.0415 (4) | |
| H4 | 0.3514 | 0.3439 | 0.1347 | 0.050* | |
| C5 | 0.36473 (8) | 0.23357 (18) | 0.26380 (10) | 0.0419 (4) | |
| H5 | 0.4104 | 0.2779 | 0.2476 | 0.050* | |
| C6 | 0.36219 (8) | 0.16956 (18) | 0.32922 (9) | 0.0433 (4) | |
| H6 | 0.4052 | 0.1714 | 0.3590 | 0.052* | |
| C7 | 0.29510 (8) | 0.10024 (17) | 0.35260 (8) | 0.0368 (3) | |
| C8 | 0.23210 (7) | 0.09796 (16) | 0.31021 (7) | 0.0312 (3) | |
| H8 | 0.1880 | 0.0473 | 0.3262 | 0.037* | |
| C9 | 0.23251 (7) | 0.17038 (15) | 0.24311 (7) | 0.0293 (3) | |
| C10 | 0.30113 (7) | 0.23607 (16) | 0.21894 (9) | 0.0341 (3) | |
| C11 | 0.09175 (7) | 0.12732 (16) | 0.21759 (8) | 0.0325 (3) | |
| C12 | 0.06218 (7) | 0.14707 (16) | 0.29105 (8) | 0.0304 (3) | |
| C13 | 0.00695 (8) | 0.04620 (17) | 0.31572 (8) | 0.0354 (3) | |
| H13 | −0.0079 | −0.0389 | 0.2873 | 0.042* | |
| C14 | −0.02637 (8) | 0.06910 (18) | 0.38113 (8) | 0.0389 (4) | |
| H14 | −0.0630 | −0.0011 | 0.3984 | 0.047* | |
| C15 | −0.00543 (8) | 0.19623 (18) | 0.42098 (8) | 0.0364 (3) | |
| C16 | 0.04883 (8) | 0.29823 (17) | 0.39772 (8) | 0.0346 (3) | |
| H16 | 0.0626 | 0.3846 | 0.4258 | 0.041* | |
| C17 | 0.08296 (8) | 0.27253 (16) | 0.33264 (8) | 0.0320 (3) | |
| H17 | 0.1208 | 0.3411 | 0.3163 | 0.038* | |
| C18 | 0.23232 (12) | −0.0353 (2) | 0.44536 (9) | 0.0560 (5) | |
| H18A | 0.2422 | −0.0770 | 0.4927 | 0.067* | |
| H18B | 0.2174 | −0.1192 | 0.4135 | 0.067* | |
| H18C | 0.1918 | 0.0407 | 0.4481 | 0.067* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0399 (3) | 0.0813 (4) | 0.0376 (2) | −0.0016 (2) | 0.00670 (15) | 0.00181 (19) |
| O1 | 0.0292 (6) | 0.0561 (7) | 0.0463 (6) | −0.0002 (5) | −0.0077 (4) | −0.0137 (5) |
| O2 | 0.0513 (7) | 0.0577 (8) | 0.0364 (6) | 0.0083 (6) | −0.0084 (5) | −0.0005 (5) |
| O3 | 0.0542 (7) | 0.0414 (6) | 0.0466 (6) | 0.0105 (5) | −0.0200 (5) | −0.0052 (5) |
| C1 | 0.0286 (7) | 0.0260 (7) | 0.0351 (7) | 0.0025 (5) | −0.0004 (5) | −0.0043 (5) |
| C2 | 0.0416 (8) | 0.0317 (8) | 0.0385 (8) | 0.0069 (6) | −0.0016 (6) | −0.0048 (6) |
| C3 | 0.0554 (9) | 0.0328 (9) | 0.0413 (8) | 0.0008 (7) | 0.0116 (8) | 0.0006 (7) |
| C4 | 0.0395 (8) | 0.0313 (8) | 0.0536 (10) | −0.0038 (6) | 0.0153 (7) | −0.0075 (7) |
| C5 | 0.0245 (7) | 0.0363 (8) | 0.0649 (10) | −0.0006 (6) | 0.0027 (7) | −0.0195 (7) |
| C6 | 0.0268 (7) | 0.0396 (9) | 0.0635 (11) | 0.0073 (6) | −0.0130 (7) | −0.0200 (8) |
| C7 | 0.0368 (8) | 0.0295 (8) | 0.0441 (8) | 0.0104 (6) | −0.0112 (6) | −0.0111 (6) |
| C8 | 0.0274 (7) | 0.0263 (7) | 0.0398 (7) | 0.0028 (5) | −0.0033 (5) | −0.0053 (6) |
| C9 | 0.0253 (7) | 0.0237 (7) | 0.0388 (7) | 0.0025 (5) | 0.0001 (5) | −0.0074 (5) |
| C10 | 0.0276 (7) | 0.0267 (7) | 0.0482 (9) | 0.0006 (5) | 0.0051 (6) | −0.0102 (6) |
| C11 | 0.0264 (7) | 0.0290 (7) | 0.0422 (8) | 0.0029 (6) | −0.0056 (6) | −0.0043 (6) |
| C12 | 0.0213 (6) | 0.0298 (7) | 0.0402 (7) | 0.0025 (5) | −0.0033 (5) | −0.0007 (6) |
| C13 | 0.0263 (7) | 0.0305 (8) | 0.0493 (8) | 0.0003 (6) | −0.0043 (6) | −0.0018 (6) |
| C14 | 0.0274 (7) | 0.0392 (9) | 0.0502 (9) | −0.0027 (6) | −0.0010 (6) | 0.0097 (7) |
| C15 | 0.0281 (7) | 0.0448 (9) | 0.0363 (7) | 0.0057 (6) | 0.0007 (6) | 0.0059 (6) |
| C16 | 0.0312 (7) | 0.0342 (8) | 0.0383 (8) | 0.0026 (6) | −0.0017 (6) | −0.0028 (6) |
| C17 | 0.0258 (7) | 0.0298 (7) | 0.0405 (8) | 0.0001 (5) | −0.0008 (6) | 0.0000 (6) |
| C18 | 0.0896 (14) | 0.0381 (9) | 0.0402 (9) | −0.0063 (9) | −0.0156 (9) | 0.0008 (7) |
Geometric parameters (Å, °)
| Cl1—C15 | 1.7381 (15) | C7—C8 | 1.3778 (19) |
| O1—C11 | 1.2476 (17) | C8—C9 | 1.414 (2) |
| O2—C2 | 1.3466 (19) | C8—H8 | 0.9500 |
| O2—H2 | 0.94 (2) | C9—C10 | 1.4233 (19) |
| O3—C7 | 1.3661 (19) | C11—C12 | 1.493 (2) |
| O3—C18 | 1.429 (2) | C12—C17 | 1.392 (2) |
| C1—C2 | 1.406 (2) | C12—C13 | 1.397 (2) |
| C1—C9 | 1.4422 (19) | C13—C14 | 1.384 (2) |
| C1—C11 | 1.4722 (19) | C13—H13 | 0.9500 |
| C2—C3 | 1.410 (2) | C14—C15 | 1.387 (2) |
| C3—C4 | 1.357 (2) | C14—H14 | 0.9500 |
| C3—H3 | 0.9500 | C15—C16 | 1.382 (2) |
| C4—C10 | 1.411 (2) | C16—C17 | 1.388 (2) |
| C4—H4 | 0.9500 | C16—H16 | 0.9500 |
| C5—C6 | 1.354 (3) | C17—H17 | 0.9500 |
| C5—C10 | 1.414 (2) | C18—H18A | 0.9800 |
| C5—H5 | 0.9500 | C18—H18B | 0.9800 |
| C6—C7 | 1.408 (2) | C18—H18C | 0.9800 |
| C6—H6 | 0.9500 | ||
| C2—O2—H2 | 106.3 (13) | C4—C10—C9 | 119.88 (14) |
| C7—O3—C18 | 117.31 (12) | C5—C10—C9 | 119.30 (15) |
| C2—C1—C9 | 118.28 (13) | O1—C11—C1 | 119.80 (13) |
| C2—C1—C11 | 117.23 (13) | O1—C11—C12 | 116.93 (12) |
| C9—C1—C11 | 124.48 (12) | C1—C11—C12 | 123.04 (12) |
| O2—C2—C1 | 123.27 (15) | C17—C12—C13 | 119.32 (13) |
| O2—C2—C3 | 115.35 (14) | C17—C12—C11 | 121.30 (13) |
| C1—C2—C3 | 121.36 (14) | C13—C12—C11 | 119.07 (13) |
| C4—C3—C2 | 119.91 (15) | C14—C13—C12 | 120.60 (14) |
| C4—C3—H3 | 120.0 | C14—C13—H13 | 119.7 |
| C2—C3—H3 | 120.0 | C12—C13—H13 | 119.7 |
| C3—C4—C10 | 121.42 (14) | C13—C14—C15 | 118.87 (14) |
| C3—C4—H4 | 119.3 | C13—C14—H14 | 120.6 |
| C10—C4—H4 | 119.3 | C15—C14—H14 | 120.6 |
| C6—C5—C10 | 121.68 (15) | C16—C15—C14 | 121.71 (14) |
| C6—C5—H5 | 119.2 | C16—C15—Cl1 | 119.28 (12) |
| C10—C5—H5 | 119.2 | C14—C15—Cl1 | 118.97 (12) |
| C5—C6—C7 | 119.28 (14) | C15—C16—C17 | 118.95 (14) |
| C5—C6—H6 | 120.4 | C15—C16—H16 | 120.5 |
| C7—C6—H6 | 120.4 | C17—C16—H16 | 120.5 |
| O3—C7—C8 | 124.26 (14) | C16—C17—C12 | 120.53 (13) |
| O3—C7—C6 | 114.75 (13) | C16—C17—H17 | 119.7 |
| C8—C7—C6 | 120.98 (15) | C12—C17—H17 | 119.7 |
| C7—C8—C9 | 120.61 (13) | O3—C18—H18A | 109.5 |
| C7—C8—H8 | 119.7 | O3—C18—H18B | 109.5 |
| C9—C8—H8 | 119.7 | H18A—C18—H18B | 109.5 |
| C8—C9—C10 | 118.02 (13) | O3—C18—H18C | 109.5 |
| C8—C9—C1 | 123.19 (12) | H18A—C18—H18C | 109.5 |
| C10—C9—C1 | 118.68 (13) | H18B—C18—H18C | 109.5 |
| C4—C10—C5 | 120.77 (14) | ||
| C9—C1—C2—O2 | 173.72 (13) | C6—C5—C10—C9 | 0.3 (2) |
| C11—C1—C2—O2 | −7.2 (2) | C8—C9—C10—C4 | 174.23 (12) |
| C9—C1—C2—C3 | −7.8 (2) | C1—C9—C10—C4 | −2.23 (19) |
| C11—C1—C2—C3 | 171.28 (13) | C8—C9—C10—C5 | −3.14 (19) |
| O2—C2—C3—C4 | −177.98 (14) | C1—C9—C10—C5 | −179.59 (12) |
| C1—C2—C3—C4 | 3.4 (2) | C2—C1—C11—O1 | 26.3 (2) |
| C2—C3—C4—C10 | 1.8 (2) | C9—C1—C11—O1 | −154.68 (14) |
| C10—C5—C6—C7 | 1.6 (2) | C2—C1—C11—C12 | −148.00 (14) |
| C18—O3—C7—C8 | −0.5 (2) | C9—C1—C11—C12 | 31.0 (2) |
| C18—O3—C7—C6 | 179.99 (13) | O1—C11—C12—C17 | −139.55 (14) |
| C5—C6—C7—O3 | 178.89 (13) | C1—C11—C12—C17 | 34.9 (2) |
| C5—C6—C7—C8 | −0.6 (2) | O1—C11—C12—C13 | 33.95 (19) |
| O3—C7—C8—C9 | 178.21 (12) | C1—C11—C12—C13 | −151.57 (13) |
| C6—C7—C8—C9 | −2.4 (2) | C17—C12—C13—C14 | −0.7 (2) |
| C7—C8—C9—C10 | 4.17 (19) | C11—C12—C13—C14 | −174.37 (13) |
| C7—C8—C9—C1 | −179.55 (13) | C12—C13—C14—C15 | 1.7 (2) |
| C2—C1—C9—C8 | −169.18 (13) | C13—C14—C15—C16 | −1.4 (2) |
| C11—C1—C9—C8 | 11.8 (2) | C13—C14—C15—Cl1 | 176.26 (11) |
| C2—C1—C9—C10 | 7.07 (19) | C14—C15—C16—C17 | 0.2 (2) |
| C11—C1—C9—C10 | −171.92 (13) | Cl1—C15—C16—C17 | −177.50 (11) |
| C3—C4—C10—C5 | 175.05 (14) | C15—C16—C17—C12 | 0.8 (2) |
| C3—C4—C10—C9 | −2.3 (2) | C13—C12—C17—C16 | −0.5 (2) |
| C6—C5—C10—C4 | −177.05 (14) | C11—C12—C17—C16 | 172.96 (13) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1 | 0.94 (2) | 1.71 (2) | 2.5573 (16) | 148 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2069).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst.38, 381–388.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Higashi, T. (1999). NUMABS Rigaku Corporation, Tokyo, Japan.
- Mitsui, R., Nakaema, K., Noguchi, K., Okamoto, A. & Yonezawa, N. (2008). Acta Cryst. E64, o1278. [DOI] [PMC free article] [PubMed]
- Nakaema, K., Okamoto, A., Noguchi, K. & Yonezawa, N. (2007). Acta Cryst. E63, o4120.
- Nakaema, K., Watanabe, S., Okamoto, A., Noguchi, K. & Yonezawa, N. (2008). Acta Cryst. E64, o807. [DOI] [PMC free article] [PubMed]
- Rigaku (1998). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2004). CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808039603/su2069sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808039603/su2069Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report






