Abstract
The title compound, [Co(C10H8N2)3]Cl3·4H2O, contains discrete [Co(bpy)3]3+ cations (bpy is 2,2′-bipyridine), Cl− anions and water molecules. The [Co(bpy)3]3+ complex cation exhibits C 2 symmetry with the twofold axis through the central Co atom and bisecting one bpy ligand and one of the Cl− anions. The four solvent water molecules and the remaining two Cl− anions lie on a mirror plane. Hydrogen-bond interactions define a two-dimensional layer structure parallel to (100), which consists of seven-membered [Cl2(H2O)5], eight-membered [Cl4(H2O)4] and ten-membered [Cl2(H2O)8] rings.
Related literature
For general background, see: Liu et al. (1996 ▶); Nauta & Miller (2000 ▶); Ludwig (2001 ▶); Saha & Bernal (2005 ▶); Reger et al. (2006 ▶); Li et al. (2007 ▶); Mir & Vittal (2007 ▶). For related structures, see: Hernández-Molina et al. (1998 ▶).
Experimental
Crystal data
[Co(C10H8N2)3]Cl3·4H2O
M r = 705.90
Orthorhombic,
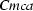
a = 20.171 (4) Å
b = 23.170 (5) Å
c = 13.316 (3) Å
V = 6223 (2) Å3
Z = 8
Mo Kα radiation
μ = 0.86 mm−1
T = 295 (2) K
0.12 × 0.10 × 0.08 mm
Data collection
Bruker P4 diffractometer
Absorption correction: ψ scan (XSCANS; Siemens, 1996 ▶) T min = 0.905, T max = 0.929
3430 measured reflections
2824 independent reflections
1980 reflections with I > 2σ(I)
R int = 0.049
3 standard reflections every 97 reflections intensity decay: none
Refinement
R[F 2 > 2σ(F 2)] = 0.056
wR(F 2) = 0.162
S = 1.03
2824 reflections
225 parameters
12 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 1.01 e Å−3
Δρmin = −0.66 e Å−3
Data collection: XSCANS (Siemens, 1996 ▶); cell refinement: XSCANS; data reduction: XSCANS; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808038154/bg2219sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808038154/bg2219Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1W2⋯O2 | 0.85 (4) | 2.04 (5) | 2.885 (9) | 176 (9) |
| O1—H1W1⋯Cl1 | 0.83 (4) | 2.30 (4) | 3.127 (7) | 180 (9) |
| O2—H2W1⋯Cl1i | 0.84 (4) | 2.48 (4) | 3.317 (7) | 175 (6) |
| O2—H2W2⋯Cl2 | 0.84 (3) | 2.33 (3) | 3.165 (5) | 173 (7) |
| O3—H3W1⋯Cl1ii | 0.85 (6) | 2.33 (6) | 3.161 (5) | 166 (6) |
| O3—H3W2⋯Cl2iii | 0.84 (4) | 2.27 (5) | 3.095 (5) | 170 (7) |
| O4—H4W1⋯O1 | 0.79 (5) | 2.06 (5) | 2.841 (8) | 169 (5) |
| O4—H4W2⋯O3 | 0.81 (3) | 2.00 (3) | 2.795 (8) | 168 (7) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
This project was sponsored by the K. C. Wong Magna Fund of Ningbo University, the Expert Project of Key Basic Research of the Ministry of Science and Technology of China (grant No. 2003CCA00800), the Ningbo Municipal Natural Science Foundation (grant No. 2006 A610061), and the Newer Training Program Foundation for Talent of the Science and Technology Department of Zhejiang Province (grant No. 2007R40G2070020).
supplementary crystallographic information
Comment
Due to the central role that water plays in biological and chemical processes, research on its structure, properties and functions has attracted the scientist's attention (Liu, et al., 1996; Nauta & Miller, 2000; Ludwig, 2001; Mir & Vittal, 2007). However, there are only a few reports focused on the experimental identificaion and analysis of hydrogen-bond networks between water of crystallization and chloride counterions in crystalline materials. As a few examples we can mention [cis-α-(trine)CoCl2]Cl.3H2O (Saha & Bernal, 2005) where a two-dimensional layered structure containing [Cl2(H2O)4] six-membered rings build up. In the crystal structure of {p-C6H4[CH2OCH2C(pz)3]2[Ru(p-cymene)]2}Cl4.14H2O two-dimensional layers built from four-membered [Cl(H2O)3], five-membered [Cl(H2O)4], six-membered [Cl(H2O)5] and seven-membered [Cl3(H2O)4] rings formed through hydrogen bond self-assembly (Reger, et al., 2006). A hybrid water-chloride structure of 14 water molecules and 4 Cl- anions connected into layers is formed in the crystal structure of an europium complex [Eu3(BDC)3(phen)3Cl(H2O)6]Cl2.4H2O (Li, et al., 2007). The references suggest that a great variety of mixed water-chloride supramolecular self-assemblies is possible. Herein, we describe a structure of a cobalt(III) complex containing discrete tris(2,2'-bipyridine)cobalt(III) cations and infinite layers of water and chloride anions, and where seven-membered (Cl2(H2O)5), eight-membered (Cl4(H2O)4) and ten-membered (Cl2(H2O)8) rings can be found.
The crystal structure of the title compound is composed of [Co(bpy)3]3+ complex cations, Cl- anions and crystal H2O molecules (Fig. 1). Within the trivalent complex cations, the Co atoms are each surrounded by six N atoms of three chelating bpy ligands to complete a distorted octahedral coordination with d(Co—N) = 1,928 (3)–1.939 (3) Å, the cis and trans N—Co—N bond angles in the range 83.50 (19)–93.82 (14) and 175.23 (14)–176.38 (19)°, respectively. Such distances are similar to those found in other related structures (Hernández-Molina, et al., 1998). The complex cation exhibits C2 symmetry with the twofold axis through the central Co atom and bisecting the C5—C5i bond of one bpy ligand (i = -x + 1/2, y, -z + 3/2), as well as one of the Cl- anions (Cl3). Through a C—H···Cl weak hydrogen bond the [Co(phen)3]3+ moieties are interlink into one-dimensional chains along the [001] direction (Fig.2). There are no π-π stacking interactions in the chains.
The most starling feature of the solid-state structure of 1 is the hydrogen-bonding interactions of the four water molecules and the remaining two Cl- anions (Cl1 and Cl2), which lay on a mirror plane. The four crystal water molecules (O1, O2, O3,and O4) locate at the crystallographic 8f position and through intermolecular hydrogen bonds determine a linear water tetramer (H2O)4. The O···O distances span the range 2.795 (8)–2.885 (9) Å. Interestingly, such tetrameric water groups are further hydrogen-bond-interacting with the Cl- anions to complete a two-dimensional water-chloride framework parallel to (100). As Shown in Fig. 3, the two-dimensional layers are composed by seven- (five water molecules and two Cl- anions), eight- (four water molecules and four Cl- anions) and ten-membered (eight water molecules and two Cl- anions) rings through O—H···O and O—H···Cl interactions (Table 1). The O···Cl distances range from 3.095 (5) to 3.317 (7) Å and O—H···Cl angles span the range 166 (6) to 180 (9)°. The water-chloride layer can be viewed as building blocks, which are pillared by [Co(bpy)3]3+ spacers to produce an infinite three-dimensional supramolecular edifice. In this sense, the layer is different from already reported water clusters and other morphologies, which are situated within the host cavities or channels generated by organic and inorganic moieties (Fig. 4).
Experimental
Addition of 10 ml CH3OH containing 0.162 g (1.04 mmol) 2,2'-bipyridine (bpy) to an aqueous solution of 0.238 g (1.00 mmol) CoCl2.6H2O in 10 ml H2O gave a yellow solution, then dropwise added 3 ml 30% H2O2 solution to the mixture, stirred for ca half an hour. The resulting light-yellow solution (PH = 6.48) was allowed to stand at room temperature. After 8 days, a small amount of yellow block crystals had grown.
Refinement
H atoms bonded to C atoms were palced in geometrically calculated position and were refined using a riding model, with Uiso(H) = 1.2 Ueq(C). H atoms attached to O atoms were found in a difference Fourier synthesis and were refined using a riding model, with the O—H distances fixed as initially found and with Uiso(H) values set at 1.5 Ueq(O).
Figures
Fig. 1.
ORTEP view of the title compound. The dispalcement ellipsoids are drawn at 40% probability level. [Symmetry code: (i) -x + 1/2, y, -z + 3/2]
Fig. 2.
The one-dimensional chain of the [Co(phen)3]3+ cations along with [001] direction.
Fig. 3.
The two-dimensional water-chloride framework parallel to (100). [Symmetry codes: (ii) -x, -y + 3/2, z - 1/2; (iii) -x, -y + 3/2, z + 1/2; (iv): x, -y + 1, -z + 1]
Fig. 4.
Packing diagram of the supramolecular edifice viewed along the crystallographic c axis.
Crystal data
| [Co(C10H8N2)3]Cl3·4H2O | F000 = 2912 |
| Mr = 705.90 | Dx = 1.507 Mg m−3 |
| Orthorhombic, Cmca | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -C 2bc 2 | Cell parameters from 25 reflections |
| a = 20.171 (4) Å | θ = 5.0–12.5º |
| b = 23.170 (5) Å | µ = 0.86 mm−1 |
| c = 13.316 (3) Å | T = 295 (2) K |
| V = 6223 (2) Å3 | Block, yellow |
| Z = 8 | 0.12 × 0.10 × 0.08 mm |
Data collection
| Bruker P4 diffractometer | Rint = 0.049 |
| Radiation source: fine-focus sealed tube | θmax = 25.0º |
| Monochromator: graphite | θmin = 1.8º |
| T = 295(2) K | h = −23→1 |
| θ/2θ scans | k = −1→27 |
| Absorption correction: ψ scan(XSCANS; Siemens, 1996) | l = −1→15 |
| Tmin = 0.905, Tmax = 0.929 | 3 standard reflections |
| 3430 measured reflections | every 97 reflections |
| 2824 independent reflections | intensity decay: none |
| 1980 reflections with I > 2σ(I) |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.056 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.162 | w = 1/[σ2(Fo2) + (0.1023P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 2824 reflections | Δρmax = 1.01 e Å−3 |
| 225 parameters | Δρmin = −0.66 e Å−3 |
| 12 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Co1 | 0.2500 | 0.40805 (3) | 0.7500 | 0.0230 (2) | |
| Cl1 | 0.0000 | 0.80958 (8) | 0.57842 (16) | 0.0580 (5) | |
| Cl2 | 0.0000 | 0.52160 (8) | 0.83302 (13) | 0.0487 (5) | |
| Cl3 | 0.2500 | 0.71245 (7) | 0.7500 | 0.0508 (5) | |
| N1 | 0.19627 (15) | 0.47048 (14) | 0.6973 (2) | 0.0254 (7) | |
| N2 | 0.19583 (15) | 0.34934 (14) | 0.6871 (3) | 0.0278 (8) | |
| N3 | 0.29820 (15) | 0.40542 (14) | 0.6251 (2) | 0.0267 (8) | |
| C1 | 0.13885 (18) | 0.46643 (19) | 0.6478 (3) | 0.0328 (10) | |
| H1A | 0.1227 | 0.4300 | 0.6316 | 0.039* | |
| C2 | 0.1025 (2) | 0.51433 (19) | 0.6198 (3) | 0.0367 (11) | |
| H2A | 0.0628 | 0.5100 | 0.5852 | 0.044* | |
| C3 | 0.1256 (2) | 0.5687 (2) | 0.6436 (3) | 0.0382 (11) | |
| H3A | 0.1016 | 0.6014 | 0.6257 | 0.046* | |
| C4 | 0.1849 (2) | 0.57363 (18) | 0.6944 (3) | 0.0370 (11) | |
| H4A | 0.2019 | 0.6098 | 0.7102 | 0.044* | |
| C5 | 0.21896 (19) | 0.52388 (17) | 0.7216 (3) | 0.0267 (9) | |
| C6 | 0.1430 (2) | 0.32196 (18) | 0.7267 (4) | 0.0368 (11) | |
| H6A | 0.1316 | 0.3286 | 0.7934 | 0.044* | |
| C7 | 0.1052 (2) | 0.28387 (19) | 0.6695 (4) | 0.0453 (12) | |
| H7A | 0.0695 | 0.2646 | 0.6983 | 0.054* | |
| C8 | 0.1204 (2) | 0.27501 (19) | 0.5722 (4) | 0.0486 (13) | |
| H8A | 0.0939 | 0.2512 | 0.5330 | 0.058* | |
| C9 | 0.1750 (2) | 0.30103 (18) | 0.5309 (4) | 0.0407 (11) | |
| H9A | 0.1867 | 0.2940 | 0.4645 | 0.049* | |
| C10 | 0.2128 (2) | 0.33844 (17) | 0.5904 (3) | 0.0300 (9) | |
| C11 | 0.27255 (19) | 0.36858 (17) | 0.5557 (3) | 0.0284 (9) | |
| C12 | 0.3015 (2) | 0.3604 (2) | 0.4646 (3) | 0.0410 (11) | |
| H12A | 0.2824 | 0.3358 | 0.4176 | 0.049* | |
| C13 | 0.3601 (2) | 0.3892 (2) | 0.4427 (4) | 0.0452 (12) | |
| H13A | 0.3813 | 0.3835 | 0.3815 | 0.054* | |
| C14 | 0.3861 (2) | 0.4261 (2) | 0.5121 (3) | 0.0397 (11) | |
| H14A | 0.4251 | 0.4459 | 0.4984 | 0.048* | |
| C15 | 0.3545 (2) | 0.43360 (18) | 0.6017 (3) | 0.0329 (10) | |
| H15A | 0.3725 | 0.4591 | 0.6482 | 0.039* | |
| O1 | 0.0000 | 0.6769 (3) | 0.6215 (5) | 0.092 (2) | |
| H1W1 | 0.0000 | 0.7122 (17) | 0.610 (8) | 0.138* | |
| H1W2 | 0.0000 | 0.673 (5) | 0.685 (3) | 0.138* | |
| O2 | 0.0000 | 0.6582 (2) | 0.8357 (5) | 0.0643 (15) | |
| H2W1 | 0.0000 | 0.664 (3) | 0.898 (3) | 0.096* | |
| H2W2 | 0.0000 | 0.6220 (14) | 0.829 (6) | 0.096* | |
| O3 | 0.0000 | 0.6013 (2) | 0.2581 (4) | 0.0701 (17) | |
| H3W1 | 0.0000 | 0.620 (3) | 0.203 (4) | 0.105* | |
| H3W2 | 0.0000 | 0.5664 (14) | 0.241 (6) | 0.105* | |
| O4 | 0.0000 | 0.5921 (2) | 0.4674 (4) | 0.0566 (13) | |
| H4W1 | 0.0000 | 0.619 (2) | 0.504 (4) | 0.085* | |
| H4W2 | 0.0000 | 0.600 (3) | 0.408 (2) | 0.085* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Co1 | 0.0243 (4) | 0.0150 (4) | 0.0297 (4) | 0.000 | −0.0016 (3) | 0.000 |
| Cl1 | 0.0727 (12) | 0.0362 (10) | 0.0651 (12) | 0.000 | 0.000 | −0.0075 (9) |
| Cl2 | 0.0512 (9) | 0.0463 (10) | 0.0486 (10) | 0.000 | 0.000 | −0.0087 (8) |
| Cl3 | 0.0791 (12) | 0.0197 (8) | 0.0537 (10) | 0.000 | 0.0244 (9) | 0.000 |
| N1 | 0.0284 (17) | 0.0208 (17) | 0.0269 (17) | −0.0004 (14) | 0.0009 (14) | 0.0003 (15) |
| N2 | 0.0262 (17) | 0.0184 (17) | 0.039 (2) | −0.0013 (14) | −0.0021 (14) | 0.0005 (15) |
| N3 | 0.0277 (17) | 0.0194 (16) | 0.0331 (18) | 0.0035 (14) | 0.0010 (14) | 0.0009 (15) |
| C1 | 0.026 (2) | 0.030 (2) | 0.042 (2) | 0.0006 (18) | −0.0036 (19) | −0.002 (2) |
| C2 | 0.031 (2) | 0.043 (3) | 0.037 (2) | 0.0062 (19) | −0.0053 (19) | 0.007 (2) |
| C3 | 0.037 (2) | 0.032 (2) | 0.046 (3) | 0.0104 (19) | −0.002 (2) | 0.007 (2) |
| C4 | 0.043 (3) | 0.016 (2) | 0.052 (3) | 0.0019 (18) | 0.000 (2) | 0.003 (2) |
| C5 | 0.0297 (19) | 0.022 (2) | 0.028 (2) | −0.0002 (18) | 0.0029 (17) | 0.0022 (17) |
| C6 | 0.034 (2) | 0.022 (2) | 0.054 (3) | −0.0020 (19) | −0.001 (2) | 0.001 (2) |
| C7 | 0.034 (2) | 0.026 (2) | 0.076 (4) | −0.0095 (19) | −0.005 (2) | −0.002 (2) |
| C8 | 0.045 (3) | 0.029 (3) | 0.072 (4) | −0.005 (2) | −0.018 (3) | −0.015 (3) |
| C9 | 0.044 (3) | 0.028 (2) | 0.051 (3) | 0.000 (2) | −0.008 (2) | −0.012 (2) |
| C10 | 0.031 (2) | 0.022 (2) | 0.038 (2) | 0.0031 (17) | −0.0054 (19) | −0.0041 (19) |
| C11 | 0.034 (2) | 0.021 (2) | 0.030 (2) | 0.0027 (17) | −0.0067 (18) | −0.0040 (18) |
| C12 | 0.049 (3) | 0.037 (2) | 0.037 (3) | 0.007 (2) | −0.005 (2) | −0.006 (2) |
| C13 | 0.044 (3) | 0.056 (3) | 0.036 (3) | 0.014 (2) | 0.008 (2) | 0.003 (2) |
| C14 | 0.038 (2) | 0.037 (2) | 0.044 (3) | 0.000 (2) | 0.006 (2) | 0.007 (2) |
| C15 | 0.033 (2) | 0.027 (2) | 0.040 (3) | −0.0023 (18) | 0.0043 (19) | 0.000 (2) |
| O1 | 0.168 (7) | 0.043 (3) | 0.064 (4) | 0.000 | 0.000 | 0.000 (3) |
| O2 | 0.069 (3) | 0.053 (3) | 0.071 (4) | 0.000 | 0.000 | 0.016 (3) |
| O3 | 0.112 (5) | 0.038 (3) | 0.060 (4) | 0.000 | 0.000 | 0.004 (3) |
| O4 | 0.078 (3) | 0.043 (3) | 0.048 (3) | 0.000 | 0.000 | −0.006 (3) |
Geometric parameters (Å, °)
| Co1—N3i | 1.928 (3) | C7—C8 | 1.347 (7) |
| Co1—N3 | 1.928 (3) | C7—H7A | 0.9300 |
| Co1—N2i | 1.935 (3) | C8—C9 | 1.370 (7) |
| Co1—N2 | 1.935 (3) | C8—H8A | 0.9300 |
| Co1—N1 | 1.939 (3) | C9—C10 | 1.401 (6) |
| Co1—N1i | 1.939 (3) | C9—H9A | 0.9300 |
| N1—C1 | 1.336 (5) | C10—C11 | 1.467 (6) |
| N1—C5 | 1.358 (5) | C11—C12 | 1.360 (6) |
| N2—C6 | 1.348 (5) | C12—C13 | 1.388 (7) |
| N2—C10 | 1.355 (5) | C12—H12A | 0.9300 |
| N3—C15 | 1.347 (5) | C13—C14 | 1.363 (7) |
| N3—C11 | 1.360 (5) | C13—H13A | 0.9300 |
| C1—C2 | 1.381 (6) | C14—C15 | 1.363 (6) |
| C1—H1A | 0.9300 | C14—H14A | 0.9300 |
| C2—C3 | 1.379 (6) | C15—H15A | 0.9300 |
| C2—H2A | 0.9300 | O1—H1W1 | 0.83 (3) |
| C3—C4 | 1.379 (6) | O1—H1W2 | 0.85 (3) |
| C3—H3A | 0.9300 | O2—H2W1 | 0.85 (3) |
| C4—C5 | 1.390 (6) | O2—H2W2 | 0.84 (3) |
| C4—H4A | 0.9300 | O3—H3W1 | 0.85 (3) |
| C5—C5i | 1.462 (8) | O3—H3W2 | 0.84 (3) |
| C6—C7 | 1.394 (6) | O4—H4W1 | 0.80 (3) |
| C6—H6A | 0.9300 | O4—H4W2 | 0.81 (3) |
| N3i—Co1—N3 | 176.38 (19) | N1—C5—C5i | 114.3 (2) |
| N3i—Co1—N2i | 83.63 (14) | C4—C5—C5i | 123.9 (3) |
| N3—Co1—N2i | 93.82 (14) | N2—C6—C7 | 121.1 (4) |
| N3i—Co1—N2 | 93.82 (14) | N2—C6—H6A | 119.4 |
| N3—Co1—N2 | 83.63 (14) | C7—C6—H6A | 119.4 |
| N2i—Co1—N2 | 90.69 (19) | C8—C7—C6 | 119.8 (4) |
| N3i—Co1—N1 | 93.09 (13) | C8—C7—H7A | 120.1 |
| N3—Co1—N1 | 89.61 (13) | C6—C7—H7A | 120.1 |
| N2i—Co1—N1 | 175.23 (14) | C7—C8—C9 | 120.2 (4) |
| N2—Co1—N1 | 92.99 (13) | C7—C8—H8A | 119.9 |
| N3i—Co1—N1i | 89.61 (13) | C9—C8—H8A | 119.9 |
| N3—Co1—N1i | 93.09 (13) | C8—C9—C10 | 118.9 (5) |
| N2i—Co1—N1i | 92.99 (13) | C8—C9—H9A | 120.6 |
| N2—Co1—N1i | 175.23 (14) | C10—C9—H9A | 120.6 |
| N1—Co1—N1i | 83.50 (19) | N2—C10—C9 | 121.0 (4) |
| C1—N1—C5 | 118.3 (3) | N2—C10—C11 | 114.7 (3) |
| C1—N1—Co1 | 127.6 (3) | C9—C10—C11 | 124.3 (4) |
| C5—N1—Co1 | 113.9 (2) | N3—C11—C12 | 122.0 (4) |
| C6—N2—C10 | 119.0 (4) | N3—C11—C10 | 113.4 (3) |
| C6—N2—Co1 | 127.4 (3) | C12—C11—C10 | 124.6 (4) |
| C10—N2—Co1 | 113.5 (3) | C11—C12—C13 | 119.1 (4) |
| C15—N3—C11 | 117.9 (4) | C11—C12—H12A | 120.5 |
| C15—N3—Co1 | 127.6 (3) | C13—C12—H12A | 120.5 |
| C11—N3—Co1 | 114.5 (3) | C14—C13—C12 | 119.1 (4) |
| N1—C1—C2 | 122.5 (4) | C14—C13—H13A | 120.5 |
| N1—C1—H1A | 118.8 | C12—C13—H13A | 120.5 |
| C2—C1—H1A | 118.8 | C13—C14—C15 | 119.6 (4) |
| C3—C2—C1 | 119.5 (4) | C13—C14—H14A | 120.2 |
| C3—C2—H2A | 120.3 | C15—C14—H14A | 120.2 |
| C1—C2—H2A | 120.3 | N3—C15—C14 | 122.3 (4) |
| C2—C3—C4 | 118.8 (4) | N3—C15—H15A | 118.9 |
| C2—C3—H3A | 120.6 | C14—C15—H15A | 118.9 |
| C4—C3—H3A | 120.6 | H1W1—O1—H1W2 | 107 (7) |
| C3—C4—C5 | 119.2 (4) | H2W1—O2—H2W2 | 106 (5) |
| C3—C4—H4A | 120.4 | H3W1—O3—H3W2 | 106 (5) |
| C5—C4—H4A | 120.4 | H4W1—O4—H4W2 | 114 (5) |
| N1—C5—C4 | 121.8 (4) |
Symmetry codes: (i) −x+1/2, y, −z+3/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1W2···O2 | 0.85 (4) | 2.04 (5) | 2.885 (9) | 176 (9) |
| O1—H1W1···Cl1 | 0.83 (4) | 2.30 (4) | 3.127 (7) | 180 (9) |
| O2—H2W1···Cl1ii | 0.84 (4) | 2.48 (4) | 3.317 (7) | 175 (6) |
| O2—H2W2···Cl2 | 0.84 (3) | 2.33 (3) | 3.165 (5) | 173 (7) |
| O3—H3W1···Cl1iii | 0.85 (6) | 2.33 (6) | 3.161 (5) | 166 (6) |
| O3—H3W2···Cl2iv | 0.84 (4) | 2.27 (5) | 3.095 (5) | 170 (7) |
| O4—H4W1···O1 | 0.79 (5) | 2.06 (5) | 2.841 (8) | 169 (5) |
| O4—H4W2···O3 | 0.81 (3) | 2.00 (3) | 2.795 (8) | 168 (7) |
| C1—H1A···N2 | 0.93 | 2.49 | 2.993 (5) | 114 |
| C4—H4A···Cl3 | 0.93 | 2.62 | 3.552 (4) | 179 |
| C6—H6A···N3i | 0.93 | 2.52 | 3.007 (6) | 113 |
| C8—H8A···Cl1iv | 0.93 | 2.79 | 3.710 (5) | 172 |
| C12—H12A···Cl3v | 0.93 | 2.58 | 3.477 (4) | 162 |
| C14—H14A···Cl2v | 0.93 | 2.77 | 3.526 (4) | 139 |
| C15—H15A···N1i | 0.93 | 2.49 | 2.991 (5) | 114 |
Symmetry codes: (ii) −x, −y+3/2, z+1/2; (iii) −x, −y+3/2, z−1/2; (iv) x, −y+1, −z+1; (i) −x+1/2, y, −z+3/2; (v) −x+1/2, −y+1, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BG2219).
References
- Hernández-Molina, M., Lloret, F., Ruiz-Pérez, C. & Julve, M. (1998). Inorg. Chem.37, 4131–4135. [DOI] [PubMed]
- Li, P., Qiu, Y. C., Liu, J. Q., Ling, Y., Cai, Y. P. & Yue, S. T. (2007). Inorg. Chem. Commun.10, 705–708.
- Liu, K., Brown, M. G., Carter, C., Saykally, R. J., Gregory, J. K. & Clary, D. C. (1996). Nature (London), 381, 501–503.
- Ludwig, R. (2001). Angew. Chem. Int. Ed.40, 1808–1827.
- Mir, M. H. & Vittal, J. J. (2007). Angew. Chem. Int. Ed.46, 5925–5928. [DOI] [PubMed]
- Nauta, K. & Miller, R. E. (2000). Science, 287, 293–295. [DOI] [PubMed]
- Reger, D. L., Semeniuc, R. F., Pettinari, C., Luna-Giles, F. & Smith, M. D. (2006). Cryst. Growth Des.6, 1068–1070.
- Saha, M. K. & Bernal, I. (2005). Inorg. Chem. Commun.8, 871–873.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siemens (1996). XSCANS Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808038154/bg2219sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808038154/bg2219Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report






