Abstract
The title compound, C16H16N2O5S, is a potent new fungicide. There are two molecules in the asymmetric unit which are linked by C—H⋯π interactions, forming a dimer. The two phenyl rings in each molecules are almost coplanar, with C—N—C—C torsion angles of 177.6 (2) and −172.5 (2)°. There are intermolecular and intramolecular N—H⋯O hydrogen bonds in the crystal structure.
Related literature
For the preparation and properties of substituted amides, see: Gong et al. (2008 ▶); Liu et al. (2007a
▶,b
▶); Wang et al. (2008 ▶).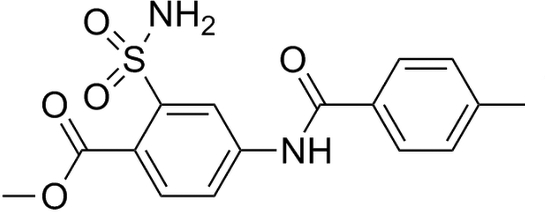
Experimental
Crystal data
C16H16N2O5S
M r = 348.37
Triclinic,

a = 9.1968 (16) Å
b = 11.078 (2) Å
c = 15.914 (3) Å
α = 75.894 (3)°
β = 87.124 (3)°
γ = 84.123 (3)°
V = 1563.7 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.24 mm−1
T = 294 (2) K
0.24 × 0.20 × 0.14 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.945, T max = 0.968
8144 measured reflections
5479 independent reflections
3677 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.120
S = 1.01
5479 reflections
461 parameters
8 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.21 e Å−3
Δρmin = −0.32 e Å−3
Data collection: SMART (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks 070704Bc, I. DOI: 10.1107/S160053680803599X/bq2101sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803599X/bq2101Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O7i | 0.890 (10) | 2.414 (15) | 3.256 (3) | 158 (2) |
| N2—H2A⋯O8ii | 0.888 (10) | 2.166 (12) | 2.993 (3) | 155 (2) |
| N3—H3A⋯O8iii | 0.889 (10) | 2.59 (2) | 3.254 (3) | 132 (2) |
| N4—H4A⋯O1iii | 0.886 (10) | 2.088 (11) | 2.958 (3) | 168 (3) |
| N2—H2B⋯O4 | 0.892 (10) | 2.149 (18) | 2.905 (3) | 142 (2) |
| N4—H4B⋯O9 | 0.888 (10) | 2.10 (2) | 2.789 (3) | 134 (2) |
| N4—H4B⋯O6iv | 0.888 (10) | 2.59 (2) | 3.298 (3) | 137 (2) |
| C7—H7A⋯Cg1ii | 0.96 | 2.87 | 3.7491 | 148 (2) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  . Cg1 is the centroid of the C17--C22 ring.
. Cg1 is the centroid of the C17--C22 ring.
supplementary crystallographic information
Comment
Amide derivative as a kind of highly bioactive compound has been studied broadly for many years. Whereas numerous references to the preparations and properties of a large variety of substituted amides exist in the literature (Liu et al., 2007a). These compounds had long been used in agriculture (Liu et al., 2007b) and medicine (Gong et al.,2008). In view of these facts and in continuation of our interest in the agriculture, we attempted to synthesize a series of amide derivatives, some of which have comparatively high fungicidal activity.
The molecular structure of title compound is showing in Fig.1. The X-ray analysis reveals that the benzene ring is planar. The carboxamide moiety is coplanar with the benzene ring [dihedral angle -1.5 (4)°]. The crystal structure is stabilized by the formation of inversion related dimers linked by C-H···π interactions (Table 1, Fig. 2).
Experimental
The title compound was prepared according to the similar reported procedure (Wang, et al., 2008). Dropwised 4-methylbenzoyl chloride (7.5 mmol) to methyl 4-amino-2-sulfamoylbenzoate (7.50 mmol) in THF (20 ml) solution, then refluxed for 4 h. Colorless single crystals suitable for X-ray diffraction were obtained by recrystallization from a mixture of ethyl acetate and petroleum ether.
Refinement
The structure was solved by direct methods and successive Fourier difference synthesis. The H atoms bonded to C and N atoms were positioned geometrically and refined using a riding model [aromatic C—H=0.93Å, aliphatic C—H = 0.97 (2)Å, N—H=0.86Å, Uiso(H) = 1.2Ueq(C)].
Figures
Fig. 1.
The structure of (I) with displacement ellipsoids drawn at the 30% probability level.
Fig. 2.
Partial packing diagram for (I). The dotted lines show the C-H···π interactions and H-bond. H atoms have been omitted for clarity.
Fig. 3.
Reaction scheme.
Crystal data
| C16H16N2O5S | Z = 4 |
| Mr = 348.37 | F000 = 728 |
| Triclinic, P1 | Dx = 1.480 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 9.1968 (16) Å | Cell parameters from 2452 reflections |
| b = 11.078 (2) Å | θ = 2.6–25.9º |
| c = 15.914 (3) Å | µ = 0.24 mm−1 |
| α = 75.894 (3)º | T = 294 (2) K |
| β = 87.124 (3)º | Clubbed, colorless |
| γ = 84.123 (3)º | 0.24 × 0.20 × 0.14 mm |
| V = 1563.7 (5) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 5479 independent reflections |
| Radiation source: fine-focus sealed tube | 3677 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.023 |
| T = 294(2) K | θmax = 25.0º |
| phi and ω scans | θmin = 1.9º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −10→8 |
| Tmin = 0.945, Tmax = 0.968 | k = −13→10 |
| 8144 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.120 | w = 1/[σ2(Fo2) + (0.0551P)2 + 0.5359P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.003 |
| 5479 reflections | Δρmax = 0.22 e Å−3 |
| 461 parameters | Δρmin = −0.32 e Å−3 |
| 8 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.48593 (7) | 0.28763 (6) | 0.70812 (4) | 0.0363 (2) | |
| S2 | 0.64701 (7) | 1.09651 (6) | 1.08377 (4) | 0.03159 (18) | |
| O1 | 0.1820 (2) | 0.58510 (18) | 1.01610 (12) | 0.0516 (6) | |
| O2 | 0.59114 (19) | 0.21940 (18) | 0.76916 (12) | 0.0465 (5) | |
| O3 | 0.5328 (2) | 0.36685 (19) | 0.62914 (12) | 0.0530 (6) | |
| O4 | 0.2127 (2) | 0.40235 (19) | 0.58979 (12) | 0.0556 (6) | |
| O5 | 0.1961 (2) | 0.60599 (18) | 0.58636 (12) | 0.0495 (5) | |
| O6 | 0.9616 (2) | 0.79515 (19) | 0.78099 (12) | 0.0543 (6) | |
| O7 | 0.55730 (19) | 1.17378 (17) | 1.01674 (11) | 0.0404 (5) | |
| O8 | 0.57554 (19) | 1.02353 (17) | 1.15791 (11) | 0.0431 (5) | |
| O9 | 0.8735 (2) | 0.96320 (19) | 1.21207 (12) | 0.0518 (6) | |
| O10 | 0.9933 (2) | 0.78427 (19) | 1.20227 (12) | 0.0571 (6) | |
| N1 | 0.3212 (3) | 0.4194 (2) | 0.98482 (14) | 0.0377 (6) | |
| N2 | 0.3947 (3) | 0.1860 (2) | 0.68291 (16) | 0.0423 (6) | |
| N3 | 0.7773 (2) | 0.9255 (2) | 0.81909 (14) | 0.0393 (6) | |
| N4 | 0.7427 (2) | 1.1888 (2) | 1.11465 (16) | 0.0376 (6) | |
| C1 | 0.3070 (3) | 0.4445 (2) | 1.13223 (16) | 0.0325 (6) | |
| C2 | 0.2253 (3) | 0.4934 (3) | 1.19400 (18) | 0.0480 (8) | |
| H2 | 0.1469 | 0.5530 | 1.1770 | 0.058* | |
| C3 | 0.2596 (3) | 0.4544 (3) | 1.28001 (18) | 0.0522 (8) | |
| H3 | 0.2023 | 0.4876 | 1.3203 | 0.063* | |
| C4 | 0.3756 (3) | 0.3677 (3) | 1.30870 (17) | 0.0410 (7) | |
| C5 | 0.4579 (3) | 0.3219 (3) | 1.24656 (19) | 0.0533 (8) | |
| H5 | 0.5386 | 0.2646 | 1.2633 | 0.064* | |
| C6 | 0.4239 (3) | 0.3587 (3) | 1.16056 (18) | 0.0485 (8) | |
| H6 | 0.4811 | 0.3249 | 1.1205 | 0.058* | |
| C7 | 0.4133 (4) | 0.3275 (3) | 1.40279 (18) | 0.0558 (9) | |
| H7A | 0.3940 | 0.2419 | 1.4253 | 0.084* | |
| H7B | 0.3551 | 0.3793 | 1.4345 | 0.084* | |
| H7C | 0.5150 | 0.3354 | 1.4087 | 0.084* | |
| C8 | 0.2647 (3) | 0.4900 (2) | 1.04024 (16) | 0.0340 (6) | |
| C9 | 0.2944 (3) | 0.4394 (2) | 0.89600 (16) | 0.0324 (6) | |
| C10 | 0.3841 (3) | 0.3679 (2) | 0.84935 (16) | 0.0334 (6) | |
| H10 | 0.4578 | 0.3106 | 0.8773 | 0.040* | |
| C11 | 0.3642 (3) | 0.3814 (2) | 0.76245 (16) | 0.0312 (6) | |
| C12 | 0.2557 (3) | 0.4688 (2) | 0.71804 (16) | 0.0345 (6) | |
| C13 | 0.1686 (3) | 0.5392 (3) | 0.76606 (17) | 0.0407 (7) | |
| H13 | 0.0970 | 0.5987 | 0.7379 | 0.049* | |
| C14 | 0.1844 (3) | 0.5242 (3) | 0.85342 (17) | 0.0400 (7) | |
| H14 | 0.1218 | 0.5706 | 0.8839 | 0.048* | |
| C15 | 0.2222 (3) | 0.4858 (3) | 0.62516 (17) | 0.0380 (7) | |
| C16 | 0.1575 (4) | 0.6338 (3) | 0.49659 (19) | 0.0649 (10) | |
| H16A | 0.2336 | 0.5978 | 0.4642 | 0.097* | |
| H16B | 0.1455 | 0.7227 | 0.4740 | 0.097* | |
| H16C | 0.0675 | 0.5993 | 0.4918 | 0.097* | |
| C17 | 0.8123 (3) | 0.9163 (3) | 0.66853 (17) | 0.0359 (6) | |
| C18 | 0.7012 (3) | 1.0074 (3) | 0.63916 (19) | 0.0544 (9) | |
| H18 | 0.6432 | 1.0407 | 0.6791 | 0.065* | |
| C19 | 0.6733 (3) | 1.0506 (3) | 0.55252 (19) | 0.0556 (9) | |
| H19 | 0.5980 | 1.1132 | 0.5353 | 0.067* | |
| C20 | 0.7543 (3) | 1.0035 (3) | 0.49037 (18) | 0.0427 (7) | |
| C21 | 0.8621 (4) | 0.9087 (3) | 0.51993 (19) | 0.0571 (9) | |
| H21 | 0.9166 | 0.8727 | 0.4800 | 0.069* | |
| C22 | 0.8920 (3) | 0.8655 (3) | 0.60655 (18) | 0.0509 (8) | |
| H22 | 0.9661 | 0.8017 | 0.6239 | 0.061* | |
| C23 | 0.7282 (4) | 1.0553 (3) | 0.39532 (18) | 0.0587 (9) | |
| H23A | 0.6304 | 1.0431 | 0.3829 | 0.088* | |
| H23B | 0.7967 | 1.0128 | 0.3623 | 0.088* | |
| H23C | 0.7408 | 1.1430 | 0.3801 | 0.088* | |
| C24 | 0.8570 (3) | 0.8714 (3) | 0.76026 (17) | 0.0356 (6) | |
| C25 | 0.8069 (3) | 0.9098 (2) | 0.90663 (16) | 0.0318 (6) | |
| C26 | 0.7350 (3) | 0.9965 (2) | 0.94941 (16) | 0.0318 (6) | |
| H26 | 0.6692 | 1.0599 | 0.9195 | 0.038* | |
| C27 | 0.7607 (3) | 0.9889 (2) | 1.03508 (15) | 0.0283 (6) | |
| C28 | 0.8636 (3) | 0.8951 (2) | 1.08109 (16) | 0.0309 (6) | |
| C29 | 0.9298 (3) | 0.8095 (2) | 1.03729 (17) | 0.0360 (6) | |
| H29 | 0.9963 | 0.7461 | 1.0665 | 0.043* | |
| C30 | 0.9014 (3) | 0.8142 (2) | 0.95255 (16) | 0.0355 (6) | |
| H30 | 0.9457 | 0.7531 | 0.9262 | 0.043* | |
| C31 | 0.9073 (3) | 0.8870 (2) | 1.17093 (16) | 0.0329 (6) | |
| C32 | 1.0496 (3) | 0.7699 (3) | 1.28726 (18) | 0.0553 (9) | |
| H32A | 1.0717 | 0.8498 | 1.2942 | 0.083* | |
| H32B | 1.1369 | 0.7134 | 1.2940 | 0.083* | |
| H32C | 0.9777 | 0.7371 | 1.3303 | 0.083* | |
| H1A | 0.382 (3) | 0.3537 (18) | 1.0090 (17) | 0.056 (9)* | |
| H2A | 0.374 (3) | 0.1235 (19) | 0.7272 (12) | 0.051 (9)* | |
| H2B | 0.319 (2) | 0.222 (2) | 0.6504 (14) | 0.059 (10)* | |
| H3A | 0.700 (2) | 0.979 (2) | 0.8004 (17) | 0.055 (9)* | |
| H4A | 0.775 (3) | 1.2487 (19) | 1.0724 (12) | 0.051 (9)* | |
| H4B | 0.806 (2) | 1.151 (2) | 1.1556 (13) | 0.062 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0364 (4) | 0.0364 (4) | 0.0364 (4) | 0.0022 (3) | 0.0045 (3) | −0.0123 (3) |
| S2 | 0.0301 (4) | 0.0323 (4) | 0.0305 (4) | 0.0073 (3) | −0.0017 (3) | −0.0079 (3) |
| O1 | 0.0721 (14) | 0.0415 (12) | 0.0356 (11) | 0.0238 (11) | −0.0059 (10) | −0.0095 (9) |
| O2 | 0.0377 (11) | 0.0493 (12) | 0.0521 (12) | 0.0131 (9) | −0.0062 (9) | −0.0173 (10) |
| O3 | 0.0604 (13) | 0.0513 (13) | 0.0464 (12) | −0.0103 (11) | 0.0193 (10) | −0.0120 (10) |
| O4 | 0.0787 (16) | 0.0478 (13) | 0.0415 (12) | 0.0046 (12) | −0.0141 (11) | −0.0149 (10) |
| O5 | 0.0683 (14) | 0.0400 (12) | 0.0349 (11) | 0.0076 (10) | −0.0095 (10) | −0.0024 (9) |
| O6 | 0.0574 (13) | 0.0596 (14) | 0.0436 (12) | 0.0261 (11) | −0.0084 (10) | −0.0197 (11) |
| O7 | 0.0395 (11) | 0.0411 (11) | 0.0379 (11) | 0.0156 (9) | −0.0102 (8) | −0.0101 (9) |
| O8 | 0.0397 (11) | 0.0453 (12) | 0.0382 (11) | 0.0025 (9) | 0.0088 (9) | −0.0030 (9) |
| O9 | 0.0670 (14) | 0.0497 (13) | 0.0380 (11) | 0.0193 (11) | −0.0147 (10) | −0.0166 (10) |
| O10 | 0.0750 (15) | 0.0503 (13) | 0.0406 (12) | 0.0300 (12) | −0.0232 (11) | −0.0106 (10) |
| N1 | 0.0463 (14) | 0.0334 (13) | 0.0311 (13) | 0.0108 (11) | −0.0048 (10) | −0.0087 (10) |
| N2 | 0.0500 (16) | 0.0340 (14) | 0.0429 (15) | 0.0016 (12) | −0.0002 (13) | −0.0115 (12) |
| N3 | 0.0349 (13) | 0.0495 (15) | 0.0336 (13) | 0.0132 (12) | −0.0062 (10) | −0.0162 (11) |
| N4 | 0.0408 (14) | 0.0331 (14) | 0.0387 (14) | 0.0032 (11) | −0.0059 (12) | −0.0099 (11) |
| C1 | 0.0370 (15) | 0.0282 (14) | 0.0319 (14) | −0.0013 (12) | −0.0018 (12) | −0.0070 (11) |
| C2 | 0.0487 (18) | 0.0529 (19) | 0.0390 (17) | 0.0212 (15) | −0.0050 (14) | −0.0144 (14) |
| C3 | 0.064 (2) | 0.057 (2) | 0.0338 (16) | 0.0158 (17) | 0.0003 (14) | −0.0156 (14) |
| C4 | 0.0524 (18) | 0.0353 (16) | 0.0341 (15) | −0.0038 (14) | −0.0037 (13) | −0.0057 (13) |
| C5 | 0.059 (2) | 0.0521 (19) | 0.0432 (18) | 0.0233 (16) | −0.0097 (15) | −0.0095 (15) |
| C6 | 0.0571 (19) | 0.0495 (18) | 0.0363 (16) | 0.0185 (15) | −0.0016 (14) | −0.0150 (14) |
| C7 | 0.074 (2) | 0.052 (2) | 0.0383 (17) | 0.0018 (17) | −0.0105 (16) | −0.0059 (15) |
| C8 | 0.0386 (15) | 0.0307 (15) | 0.0325 (14) | −0.0015 (13) | 0.0013 (12) | −0.0084 (12) |
| C9 | 0.0374 (15) | 0.0290 (14) | 0.0307 (14) | −0.0010 (12) | −0.0008 (12) | −0.0081 (11) |
| C10 | 0.0351 (15) | 0.0279 (14) | 0.0361 (15) | 0.0014 (12) | −0.0022 (12) | −0.0073 (12) |
| C11 | 0.0325 (14) | 0.0285 (14) | 0.0325 (14) | 0.0000 (12) | −0.0002 (11) | −0.0086 (11) |
| C12 | 0.0388 (15) | 0.0324 (15) | 0.0316 (14) | 0.0004 (12) | −0.0036 (12) | −0.0071 (12) |
| C13 | 0.0416 (16) | 0.0408 (17) | 0.0379 (16) | 0.0123 (13) | −0.0122 (13) | −0.0106 (13) |
| C14 | 0.0426 (17) | 0.0390 (16) | 0.0394 (16) | 0.0094 (13) | −0.0034 (13) | −0.0161 (13) |
| C15 | 0.0367 (16) | 0.0418 (17) | 0.0330 (15) | 0.0038 (13) | −0.0025 (12) | −0.0070 (13) |
| C16 | 0.086 (3) | 0.065 (2) | 0.0357 (18) | 0.008 (2) | −0.0110 (17) | −0.0007 (16) |
| C17 | 0.0353 (15) | 0.0380 (16) | 0.0372 (15) | −0.0008 (13) | −0.0023 (12) | −0.0156 (12) |
| C18 | 0.059 (2) | 0.066 (2) | 0.0393 (17) | 0.0270 (17) | −0.0078 (15) | −0.0258 (16) |
| C19 | 0.058 (2) | 0.061 (2) | 0.0457 (19) | 0.0245 (17) | −0.0130 (15) | −0.0193 (16) |
| C20 | 0.0486 (18) | 0.0449 (17) | 0.0388 (16) | −0.0052 (15) | −0.0019 (13) | −0.0175 (14) |
| C21 | 0.069 (2) | 0.063 (2) | 0.0392 (18) | 0.0207 (18) | 0.0022 (15) | −0.0224 (16) |
| C22 | 0.0561 (19) | 0.0534 (19) | 0.0408 (17) | 0.0197 (16) | −0.0012 (14) | −0.0171 (15) |
| C23 | 0.074 (2) | 0.064 (2) | 0.0374 (17) | 0.0037 (19) | −0.0082 (16) | −0.0148 (16) |
| C24 | 0.0330 (15) | 0.0386 (16) | 0.0371 (15) | 0.0042 (13) | −0.0043 (12) | −0.0154 (13) |
| C25 | 0.0278 (14) | 0.0353 (15) | 0.0339 (14) | −0.0016 (12) | −0.0014 (11) | −0.0121 (12) |
| C26 | 0.0270 (14) | 0.0320 (15) | 0.0344 (15) | 0.0048 (11) | −0.0037 (11) | −0.0066 (12) |
| C27 | 0.0279 (13) | 0.0247 (13) | 0.0309 (14) | −0.0005 (11) | 0.0008 (11) | −0.0049 (11) |
| C28 | 0.0295 (14) | 0.0308 (14) | 0.0308 (14) | −0.0002 (12) | −0.0002 (11) | −0.0050 (11) |
| C29 | 0.0356 (15) | 0.0324 (15) | 0.0365 (15) | 0.0080 (13) | −0.0046 (12) | −0.0051 (12) |
| C30 | 0.0361 (15) | 0.0345 (15) | 0.0353 (15) | 0.0058 (12) | 0.0001 (12) | −0.0110 (12) |
| C31 | 0.0287 (14) | 0.0359 (16) | 0.0310 (14) | −0.0011 (12) | 0.0008 (11) | −0.0030 (12) |
| C32 | 0.062 (2) | 0.060 (2) | 0.0378 (17) | 0.0126 (17) | −0.0176 (15) | −0.0043 (15) |
Geometric parameters (Å, °)
| S1—O3 | 1.422 (2) | C9—C14 | 1.388 (3) |
| S1—O2 | 1.4275 (19) | C9—C10 | 1.397 (3) |
| S1—N2 | 1.606 (2) | C10—C11 | 1.374 (3) |
| S1—C11 | 1.784 (2) | C10—H10 | 0.9300 |
| S2—O8 | 1.4296 (19) | C11—C12 | 1.405 (3) |
| S2—O7 | 1.4318 (17) | C12—C13 | 1.391 (4) |
| S2—N4 | 1.592 (2) | C12—C15 | 1.488 (4) |
| S2—C27 | 1.797 (2) | C13—C14 | 1.373 (3) |
| O1—C8 | 1.226 (3) | C13—H13 | 0.9300 |
| O4—C15 | 1.206 (3) | C14—H14 | 0.9300 |
| O5—C15 | 1.327 (3) | C16—H16A | 0.9600 |
| O5—C16 | 1.441 (3) | C16—H16B | 0.9600 |
| O6—C24 | 1.216 (3) | C16—H16C | 0.9600 |
| O9—C31 | 1.197 (3) | C17—C18 | 1.374 (4) |
| O10—C31 | 1.322 (3) | C17—C22 | 1.393 (4) |
| O10—C32 | 1.439 (3) | C17—C24 | 1.487 (4) |
| N1—C8 | 1.366 (3) | C18—C19 | 1.373 (4) |
| N1—C9 | 1.407 (3) | C18—H18 | 0.9300 |
| N1—H1A | 0.890 (10) | C19—C20 | 1.379 (4) |
| N2—H2A | 0.888 (10) | C19—H19 | 0.9300 |
| N2—H2B | 0.892 (10) | C20—C21 | 1.379 (4) |
| N3—C24 | 1.374 (3) | C20—C23 | 1.503 (4) |
| N3—C25 | 1.398 (3) | C21—C22 | 1.376 (4) |
| N3—H3A | 0.889 (10) | C21—H21 | 0.9300 |
| N4—H4A | 0.886 (10) | C22—H22 | 0.9300 |
| N4—H4B | 0.888 (10) | C23—H23A | 0.9600 |
| C1—C6 | 1.376 (4) | C23—H23B | 0.9600 |
| C1—C2 | 1.387 (3) | C23—H23C | 0.9600 |
| C1—C8 | 1.485 (3) | C25—C30 | 1.382 (3) |
| C2—C3 | 1.373 (4) | C25—C26 | 1.402 (3) |
| C2—H2 | 0.9300 | C26—C27 | 1.376 (3) |
| C3—C4 | 1.376 (4) | C26—H26 | 0.9300 |
| C3—H3 | 0.9300 | C27—C28 | 1.417 (3) |
| C4—C5 | 1.379 (4) | C28—C29 | 1.384 (3) |
| C4—C7 | 1.502 (4) | C28—C31 | 1.484 (3) |
| C5—C6 | 1.372 (4) | C29—C30 | 1.374 (3) |
| C5—H5 | 0.9300 | C29—H29 | 0.9300 |
| C6—H6 | 0.9300 | C30—H30 | 0.9300 |
| C7—H7A | 0.9600 | C32—H32A | 0.9600 |
| C7—H7B | 0.9600 | C32—H32B | 0.9600 |
| C7—H7C | 0.9600 | C32—H32C | 0.9600 |
| O3—S1—O2 | 119.98 (13) | C12—C13—H13 | 118.8 |
| O3—S1—N2 | 106.99 (13) | C13—C14—C9 | 119.7 (2) |
| O2—S1—N2 | 106.49 (13) | C13—C14—H14 | 120.2 |
| O3—S1—C11 | 107.35 (12) | C9—C14—H14 | 120.2 |
| O2—S1—C11 | 107.47 (11) | O4—C15—O5 | 123.6 (2) |
| N2—S1—C11 | 108.08 (12) | O4—C15—C12 | 125.2 (3) |
| O8—S2—O7 | 117.81 (11) | O5—C15—C12 | 111.1 (2) |
| O8—S2—N4 | 108.82 (12) | O5—C16—H16A | 109.5 |
| O7—S2—N4 | 105.66 (12) | O5—C16—H16B | 109.5 |
| O8—S2—C27 | 106.89 (11) | H16A—C16—H16B | 109.5 |
| O7—S2—C27 | 106.71 (11) | O5—C16—H16C | 109.5 |
| N4—S2—C27 | 110.93 (12) | H16A—C16—H16C | 109.5 |
| C15—O5—C16 | 116.1 (2) | H16B—C16—H16C | 109.5 |
| C31—O10—C32 | 117.0 (2) | C18—C17—C22 | 116.9 (3) |
| C8—N1—C9 | 127.6 (2) | C18—C17—C24 | 125.2 (2) |
| C8—N1—H1A | 114.6 (19) | C22—C17—C24 | 117.8 (2) |
| C9—N1—H1A | 117.9 (19) | C19—C18—C17 | 121.9 (3) |
| S1—N2—H2A | 114.3 (18) | C19—C18—H18 | 119.1 |
| S1—N2—H2B | 111.7 (18) | C17—C18—H18 | 119.1 |
| H2A—N2—H2B | 114.7 (16) | C18—C19—C20 | 121.7 (3) |
| C24—N3—C25 | 127.0 (2) | C18—C19—H19 | 119.2 |
| C24—N3—H3A | 118.2 (19) | C20—C19—H19 | 119.2 |
| C25—N3—H3A | 114.8 (19) | C21—C20—C19 | 116.5 (3) |
| S2—N4—H4A | 114.9 (17) | C21—C20—C23 | 121.9 (3) |
| S2—N4—H4B | 114.4 (18) | C19—C20—C23 | 121.5 (3) |
| H4A—N4—H4B | 116.4 (16) | C22—C21—C20 | 122.3 (3) |
| C6—C1—C2 | 117.4 (2) | C22—C21—H21 | 118.9 |
| C6—C1—C8 | 124.5 (2) | C20—C21—H21 | 118.9 |
| C2—C1—C8 | 118.1 (2) | C21—C22—C17 | 120.6 (3) |
| C3—C2—C1 | 120.5 (3) | C21—C22—H22 | 119.7 |
| C3—C2—H2 | 119.7 | C17—C22—H22 | 119.7 |
| C1—C2—H2 | 119.7 | C20—C23—H23A | 109.5 |
| C2—C3—C4 | 122.3 (3) | C20—C23—H23B | 109.5 |
| C2—C3—H3 | 118.9 | H23A—C23—H23B | 109.5 |
| C4—C3—H3 | 118.9 | C20—C23—H23C | 109.5 |
| C3—C4—C5 | 116.7 (3) | H23A—C23—H23C | 109.5 |
| C3—C4—C7 | 121.8 (3) | H23B—C23—H23C | 109.5 |
| C5—C4—C7 | 121.5 (3) | O6—C24—N3 | 122.3 (2) |
| C6—C5—C4 | 121.6 (3) | O6—C24—C17 | 121.7 (2) |
| C6—C5—H5 | 119.2 | N3—C24—C17 | 116.0 (2) |
| C4—C5—H5 | 119.2 | C30—C25—N3 | 123.7 (2) |
| C5—C6—C1 | 121.5 (3) | C30—C25—C26 | 118.9 (2) |
| C5—C6—H6 | 119.3 | N3—C25—C26 | 117.3 (2) |
| C1—C6—H6 | 119.3 | C27—C26—C25 | 120.8 (2) |
| C4—C7—H7A | 109.5 | C27—C26—H26 | 119.6 |
| C4—C7—H7B | 109.5 | C25—C26—H26 | 119.6 |
| H7A—C7—H7B | 109.5 | C26—C27—C28 | 120.5 (2) |
| C4—C7—H7C | 109.5 | C26—C27—S2 | 115.51 (18) |
| H7A—C7—H7C | 109.5 | C28—C27—S2 | 123.87 (18) |
| H7B—C7—H7C | 109.5 | C29—C28—C27 | 117.0 (2) |
| O1—C8—N1 | 122.2 (2) | C29—C28—C31 | 119.0 (2) |
| O1—C8—C1 | 121.4 (2) | C27—C28—C31 | 123.9 (2) |
| N1—C8—C1 | 116.4 (2) | C30—C29—C28 | 122.8 (2) |
| C14—C9—C10 | 119.1 (2) | C30—C29—H29 | 118.6 |
| C14—C9—N1 | 123.9 (2) | C28—C29—H29 | 118.6 |
| C10—C9—N1 | 117.0 (2) | C29—C30—C25 | 119.8 (2) |
| C11—C10—C9 | 120.5 (2) | C29—C30—H30 | 120.1 |
| C11—C10—H10 | 119.7 | C25—C30—H30 | 120.1 |
| C9—C10—H10 | 119.7 | O9—C31—O10 | 121.9 (2) |
| C10—C11—C12 | 121.0 (2) | O9—C31—C28 | 126.0 (2) |
| C10—C11—S1 | 117.22 (19) | O10—C31—C28 | 112.1 (2) |
| C12—C11—S1 | 121.70 (19) | O10—C32—H32A | 109.5 |
| C13—C12—C11 | 117.1 (2) | O10—C32—H32B | 109.5 |
| C13—C12—C15 | 118.1 (2) | H32A—C32—H32B | 109.5 |
| C11—C12—C15 | 124.7 (2) | O10—C32—H32C | 109.5 |
| C14—C13—C12 | 122.5 (2) | H32A—C32—H32C | 109.5 |
| C14—C13—H13 | 118.8 | H32B—C32—H32C | 109.5 |
| C6—C1—C2—C3 | −1.4 (5) | C22—C17—C18—C19 | −2.7 (5) |
| C8—C1—C2—C3 | 179.8 (3) | C24—C17—C18—C19 | 175.1 (3) |
| C1—C2—C3—C4 | 0.9 (5) | C17—C18—C19—C20 | 0.9 (5) |
| C2—C3—C4—C5 | 0.5 (5) | C18—C19—C20—C21 | 1.6 (5) |
| C2—C3—C4—C7 | 179.0 (3) | C18—C19—C20—C23 | −177.0 (3) |
| C3—C4—C5—C6 | −1.4 (5) | C19—C20—C21—C22 | −2.2 (5) |
| C7—C4—C5—C6 | −179.8 (3) | C23—C20—C21—C22 | 176.4 (3) |
| C4—C5—C6—C1 | 0.9 (5) | C20—C21—C22—C17 | 0.4 (5) |
| C2—C1—C6—C5 | 0.5 (5) | C18—C17—C22—C21 | 2.1 (5) |
| C8—C1—C6—C5 | 179.2 (3) | C24—C17—C22—C21 | −175.9 (3) |
| C9—N1—C8—O1 | −1.5 (4) | C25—N3—C24—O6 | 4.5 (5) |
| C9—N1—C8—C1 | 177.6 (2) | C25—N3—C24—C17 | −172.5 (2) |
| C6—C1—C8—O1 | −163.4 (3) | C18—C17—C24—O6 | −175.9 (3) |
| C2—C1—C8—O1 | 15.2 (4) | C22—C17—C24—O6 | 1.8 (4) |
| C6—C1—C8—N1 | 17.4 (4) | C18—C17—C24—N3 | 1.1 (4) |
| C2—C1—C8—N1 | −164.0 (3) | C22—C17—C24—N3 | 178.8 (3) |
| C8—N1—C9—C14 | −11.7 (4) | C24—N3—C25—C30 | −16.3 (4) |
| C8—N1—C9—C10 | 169.0 (3) | C24—N3—C25—C26 | 164.2 (3) |
| C14—C9—C10—C11 | 0.2 (4) | C30—C25—C26—C27 | 1.8 (4) |
| N1—C9—C10—C11 | 179.6 (2) | N3—C25—C26—C27 | −178.7 (2) |
| C9—C10—C11—C12 | 1.3 (4) | C25—C26—C27—C28 | 1.7 (4) |
| C9—C10—C11—S1 | 179.0 (2) | C25—C26—C27—S2 | −173.9 (2) |
| O3—S1—C11—C10 | −134.1 (2) | O8—S2—C27—C26 | 123.5 (2) |
| O2—S1—C11—C10 | −3.8 (3) | O7—S2—C27—C26 | −3.4 (2) |
| N2—S1—C11—C10 | 110.8 (2) | N4—S2—C27—C26 | −118.0 (2) |
| O3—S1—C11—C12 | 43.6 (3) | O8—S2—C27—C28 | −52.0 (2) |
| O2—S1—C11—C12 | 174.0 (2) | O7—S2—C27—C28 | −178.9 (2) |
| N2—S1—C11—C12 | −71.5 (2) | N4—S2—C27—C28 | 66.5 (2) |
| C10—C11—C12—C13 | −0.9 (4) | C26—C27—C28—C29 | −3.2 (4) |
| S1—C11—C12—C13 | −178.6 (2) | S2—C27—C28—C29 | 172.1 (2) |
| C10—C11—C12—C15 | −177.1 (3) | C26—C27—C28—C31 | 174.4 (2) |
| S1—C11—C12—C15 | 5.2 (4) | S2—C27—C28—C31 | −10.2 (4) |
| C11—C12—C13—C14 | −1.0 (4) | C27—C28—C29—C30 | 1.2 (4) |
| C15—C12—C13—C14 | 175.5 (3) | C31—C28—C29—C30 | −176.5 (2) |
| C12—C13—C14—C9 | 2.5 (4) | C28—C29—C30—C25 | 2.3 (4) |
| C10—C9—C14—C13 | −2.1 (4) | N3—C25—C30—C29 | 176.7 (3) |
| N1—C9—C14—C13 | 178.6 (3) | C26—C25—C30—C29 | −3.7 (4) |
| C16—O5—C15—O4 | −1.3 (4) | C32—O10—C31—O9 | −2.3 (4) |
| C16—O5—C15—C12 | −178.0 (2) | C32—O10—C31—C28 | 176.5 (2) |
| C13—C12—C15—O4 | −132.5 (3) | C29—C28—C31—O9 | 169.9 (3) |
| C11—C12—C15—O4 | 43.7 (4) | C27—C28—C31—O9 | −7.7 (4) |
| C13—C12—C15—O5 | 44.2 (4) | C29—C28—C31—O10 | −8.9 (4) |
| C11—C12—C15—O5 | −139.6 (3) | C27—C28—C31—O10 | 173.6 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O7i | 0.890 (10) | 2.414 (15) | 3.256 (3) | 158 (2) |
| N2—H2A···O8ii | 0.888 (10) | 2.166 (12) | 2.993 (3) | 155 (2) |
| N3—H3A···O8iii | 0.889 (10) | 2.59 (2) | 3.254 (3) | 132 (2) |
| N4—H4A···O1iii | 0.886 (10) | 2.088 (11) | 2.958 (3) | 168 (3) |
| N2—H2B···O4 | 0.892 (10) | 2.149 (18) | 2.905 (3) | 142 (2) |
| N4—H4B···O9 | 0.888 (10) | 2.10 (2) | 2.789 (3) | 134 (2) |
| N4—H4B···O6iv | 0.888 (10) | 2.59 (2) | 3.298 (3) | 137 (2) |
| C7—H7A···Cg1ii | 0.96 | 2.87 | 3.7491 | 148 (2) |
Symmetry codes: (i) x, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+2, −z+2; (iv) −x+2, −y+2, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BQ2101).
References
- Bruker. (2004). SAINT and SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Gong, Y., Barbay, J. K., Buntinx, M., Li, J., Van Wauweb, J., Claes, C., Van Lommen, G., Hornby, P. J. & He, W. (2008). Bioorg. Med. Chem. Lett.18, 3852–3855. [DOI] [PubMed]
- Liu, X. H., Chen, P. Q., He, F. Q., Wang, S. H., Song, H. B. & Li, Z. M. (2007a). Struct. Chem.18, 563–568.
- Liu, X. H., Chen, P. Q., Wang, B. L., Li, Y. H., Wang, S. H. & Li, Z. M. (2007b). Bioorg. Med. Chem. Lett.17, 3784–3788. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, M. Y., Guo, W. C., Lan, F., Li, Y. H. & Li, Z. M. (2008). Chin. J. Org. Chem.287, 649–656.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks 070704Bc, I. DOI: 10.1107/S160053680803599X/bq2101sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680803599X/bq2101Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





