Abstract
In the crystal structure of the title polymeric compound, [Cu(C12H12O4)(C10H8N2)]n, the asymmetric unit consists of one CuII ion, one 5-tert-butylisophthalate (tbip) and one 2,2′-bipyridine (bpy) ligand. The copper(II) ion is four-coordinated by two N atoms from bipy and two O atoms from two tbip ligands, leading to a distorted tetrahedral coordination. Each tbip ligand adopts a bis-monodentate coordination mode to connect two symmetry-related copper(II) ions, so forming a zigzag polymer chain parallel to [001]. The tert-butyl methyl groups are disordered over two positions with occupancies of 0.506 (6)/0.494 (6)
Related literature
For related literature on the synthesis of flexible organic ligands, see: Chang et al. (2005 ▶); Ma, Chen et al. (2008 ▶); Xu et al. (2006 ▶). For related literature on coordination polymers, see: Ma, Wang, Huo et al. (2008 ▶); Ma, Wang, Wang et al. (2008 ▶); Pan et al. (2006 ▶); Yang et al. (2002 ▶). For bond-length data, see: Allen et al. (1987 ▶).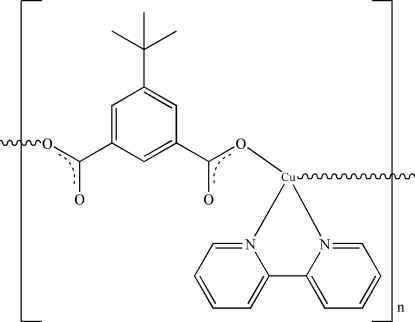
Experimental
Crystal data
[Cu(C12H12O4)(C10H8N2)]
M r = 439.94
Monoclinic,

a = 8.905 (2) Å
b = 20.875 (5) Å
c = 11.564 (3) Å
β = 98.188 (3)°
V = 2127.8 (9) Å3
Z = 4
Mo Kα radiation
μ = 1.06 mm−1
T = 296 (2) K
0.29 × 0.22 × 0.16 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 1997 ▶) T min = 0.716, T max = 0.845
15716 measured reflections
3949 independent reflections
3021 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.130
S = 1.05
3949 reflections
260 parameters
91 restraints
H-atom parameters constrained
Δρmax = 0.64 e Å−3
Δρmin = −0.55 e Å−3
Data collection: SMART (Bruker, 1997 ▶); cell refinement: SAINT (Bruker, 1997 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808035484/su2072sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808035484/su2072Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Cu1—O1 | 1.933 (2) |
| Cu1—O3i | 1.956 (2) |
| Cu1—N1 | 1.985 (3) |
| Cu1—N2 | 1.983 (3) |
| O1—Cu1—O3i | 88.17 (11) |
| O1—Cu1—N1 | 94.83 (11) |
| O3i—Cu1—N1 | 172.79 (11) |
| O1—Cu1—N2 | 173.34 (11) |
| O3i—Cu1—N2 | 96.69 (11) |
| N1—Cu1—N2 | 80.88 (11) |
Symmetry code: (i)  .
.
Acknowledgments
The authors thank Luo Yang Normal University for supporting this work.
supplementary crystallographic information
Comment
It is well known that organic ligands play a crucial role in the design and construction of desirable frameworks. The changes in flexibility, length and symmetry of organic ligands can result in a remarkable class of materials bearing diverse architectures and functions. Thus, the construction of target molecules is a challenge for synthetic chemists (Ma et al., 2008; Chang et al., 2005; Xu et al., 2006). Benzene-1,3-dicarboxylic acid (isophthalic acid, H2isop) and its derivatives, with special conformations such as, an angle of 120° between two carboxylic groups, present versatile coordination modes that can yield predetermined networks. Such ligands have been widely used to construct coordination polymers (Pan et al., 2006; Yang et al., 2002; Ma et al., 2008).
The title compound, (I), was prepared by hydrothermal synthesis using 5-tert-butyl isophthalic acid, 2,2'-bipyridine and copper(II) actate. The asymmetric unit of (I) consists of one copper(II) ion, one tbip and one bipy ligand molecules (Fig. 1). Each copper(II) ion is four-coordinated by two nitrogen atoms from one bipy molecule and two oxygen atoms from two tbip ligands (Table 1). The coordination geometry of the copper(II) ion is distorted tetrahedral. The Cu—O bond lengths [1.933 (2)–1.965 (2) Å] are within the range reported for tetrahedral environments, and the Cu—N bond lengths [1.983 (3)–1.985 (3) Å] are also similar to those found in other tetrahedral copper complexes of bipy (Allen et al., 1987). Each tbip ligand adopts the bis-monodentated coordination mode to connect two symmetry related copper(II) ions so forming a zigzag polymer chain (Fig. 2).
Experimental
A mixture of 5-tert-butyl isophthalic acid (0.1 mmol, 23.1 mg), 2,2'-bipyridine (0.1 mmol, 15.8 mg), Cu(OAc).2.4H2O (0.05 mmol, 11.5 mg), NaOH (0.1 mmol, 4.0 mg) and H2O (15 ml) was placed in a Teflon-lined stainless steel vessel, and heated to 160 °C for 4 days. It was then cooled to room temperature over a period of 24 h. Blue block-like crystals of compound (I) were obtained.
Refinement
The tertiary butyl methyl groups are disordered over two almost equally occupied positions: 0.506 (6)/0.494 (6). The H-atoms were included in calculated positions and treated as riding atoms: C—H = 0.93–0.96 Å with Uiso(H) = 1.2 or 1.5Ueq(parent C-atom).
Figures
Fig. 1.
A view of the asymmetric unit of compound (I), with thermal ellipsoids drawn at the 50% probability level. H atoms have been omitted for clarity. Atoms with label A are related to those without by symmetry operation (x, -y+1.5, z+0.5).
Fig. 2.
A partial view, along the a axis, of the crystal packing of compound (I) showing the zigzag polymer chain. H atoms have been omitted for clarity.
Crystal data
| [Cu(C12H12O4)(C10H8N2)] | F000 = 908 |
| Mr = 439.94 | Dx = 1.373 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3455 reflections |
| a = 8.905 (2) Å | θ = 2.5–22.5º |
| b = 20.875 (5) Å | µ = 1.06 mm−1 |
| c = 11.564 (3) Å | T = 296 (2) K |
| β = 98.188 (3)º | Block, blue |
| V = 2127.8 (9) Å3 | 0.29 × 0.22 × 0.16 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3949 independent reflections |
| Radiation source: fine-focus sealed tube | 3021 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.040 |
| T = 296(2) K | θmax = 25.5º |
| φ and ω scans | θmin = 2.5º |
| Absorption correction: multi-scan(SADABS; Bruker, 1997) | h = −10→10 |
| Tmin = 0.716, Tmax = 0.845 | k = −25→25 |
| 15716 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.045 | H-atom parameters constrained |
| wR(F2) = 0.130 | w = 1/[σ2(Fo2) + (0.065P)2 + 1.7017P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 3949 reflections | Δρmax = 0.64 e Å−3 |
| 260 parameters | Δρmin = −0.54 e Å−3 |
| 91 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| C10 | −0.2988 (8) | 0.7849 (4) | 0.0801 (8) | 0.0991 (17) | 0.50 |
| H10A | −0.3808 | 0.7876 | 0.1255 | 0.149* | 0.50 |
| H10B | −0.2580 | 0.7422 | 0.0851 | 0.149* | 0.50 |
| H10C | −0.3355 | 0.7948 | 0.0000 | 0.149* | 0.50 |
| C11 | −0.1382 (9) | 0.8222 (4) | 0.2581 (6) | 0.1033 (17) | 0.50 |
| H11A | −0.0577 | 0.8506 | 0.2893 | 0.155* | 0.50 |
| H11B | −0.1070 | 0.7786 | 0.2736 | 0.155* | 0.50 |
| H11C | −0.2266 | 0.8310 | 0.2943 | 0.155* | 0.50 |
| C12 | −0.2268 (8) | 0.9003 (3) | 0.1063 (7) | 0.1019 (19) | 0.50 |
| H12A | −0.3113 | 0.9085 | 0.1473 | 0.153* | 0.50 |
| H12B | −0.2570 | 0.9071 | 0.0242 | 0.153* | 0.50 |
| H12C | −0.1450 | 0.9288 | 0.1341 | 0.153* | 0.50 |
| C10' | −0.2432 (9) | 0.7777 (3) | 0.1868 (8) | 0.0991 (17) | 0.50 |
| H10D | −0.2073 | 0.7791 | 0.2691 | 0.149* | 0.50 |
| H10E | −0.2138 | 0.7378 | 0.1553 | 0.149* | 0.50 |
| H10F | −0.3518 | 0.7813 | 0.1743 | 0.149* | 0.50 |
| C11' | −0.1082 (9) | 0.8784 (4) | 0.2317 (7) | 0.1033 (17) | 0.50 |
| H11D | −0.0967 | 0.9208 | 0.2021 | 0.155* | 0.50 |
| H11E | −0.0112 | 0.8626 | 0.2671 | 0.155* | 0.50 |
| H11F | −0.1764 | 0.8796 | 0.2890 | 0.155* | 0.50 |
| C12' | −0.2878 (8) | 0.8749 (4) | 0.0548 (7) | 0.1019 (19) | 0.50 |
| H12D | −0.3658 | 0.8871 | 0.0998 | 0.153* | 0.50 |
| H12E | −0.3324 | 0.8523 | −0.0138 | 0.153* | 0.50 |
| H12F | −0.2371 | 0.9125 | 0.0323 | 0.153* | 0.50 |
| Cu1 | 0.30126 (5) | 0.570966 (18) | 0.17572 (3) | 0.03784 (16) | |
| O1 | 0.1606 (3) | 0.63474 (11) | 0.1038 (2) | 0.0497 (6) | |
| O2 | 0.3724 (3) | 0.67027 (13) | 0.0490 (3) | 0.0598 (7) | |
| O3 | 0.3259 (3) | 0.87708 (12) | −0.1821 (2) | 0.0507 (6) | |
| O4 | 0.1347 (3) | 0.94173 (12) | −0.1635 (2) | 0.0542 (7) | |
| N1 | 0.2646 (3) | 0.50997 (13) | 0.0430 (2) | 0.0368 (6) | |
| N2 | 0.4556 (3) | 0.50539 (13) | 0.2322 (2) | 0.0371 (6) | |
| C1 | 0.2394 (5) | 0.67826 (18) | 0.0641 (3) | 0.0510 (8) | |
| C2 | 0.1631 (4) | 0.74186 (15) | 0.0367 (3) | 0.0397 (8) | |
| C3 | 0.2214 (4) | 0.78533 (16) | −0.0351 (3) | 0.0394 (8) | |
| H3 | 0.3084 | 0.7756 | −0.0676 | 0.047* | |
| C4 | 0.1487 (4) | 0.84380 (15) | −0.0584 (3) | 0.0378 (8) | |
| C5 | 0.0213 (4) | 0.85804 (16) | −0.0065 (3) | 0.0430 (8) | |
| H5 | −0.0261 | 0.8974 | −0.0220 | 0.052* | |
| C6 | −0.0377 (4) | 0.81567 (17) | 0.0678 (3) | 0.0460 (9) | |
| C7 | 0.0343 (4) | 0.75671 (16) | 0.0860 (3) | 0.0448 (9) | |
| H7 | −0.0050 | 0.7264 | 0.1325 | 0.054* | |
| C8 | 0.2057 (4) | 0.89150 (16) | −0.1391 (3) | 0.0422 (8) | |
| C9 | −0.1753 (6) | 0.8323 (2) | 0.1271 (5) | 0.0809 (12) | |
| C13 | 0.1605 (4) | 0.51736 (18) | −0.0526 (3) | 0.0469 (9) | |
| H13 | 0.1031 | 0.5547 | −0.0608 | 0.056* | |
| C14 | 0.1365 (4) | 0.4717 (2) | −0.1382 (3) | 0.0541 (10) | |
| H14 | 0.0625 | 0.4775 | −0.2026 | 0.065* | |
| C15 | 0.2233 (5) | 0.4176 (2) | −0.1271 (4) | 0.0575 (11) | |
| H15 | 0.2087 | 0.3860 | −0.1842 | 0.069* | |
| C16 | 0.3328 (4) | 0.40997 (18) | −0.0308 (3) | 0.0485 (9) | |
| H16 | 0.3938 | 0.3736 | −0.0230 | 0.058* | |
| C17 | 0.3503 (4) | 0.45690 (16) | 0.0533 (3) | 0.0352 (7) | |
| C18 | 0.4614 (4) | 0.45458 (16) | 0.1610 (3) | 0.0353 (7) | |
| C19 | 0.5640 (4) | 0.40508 (18) | 0.1901 (3) | 0.0478 (9) | |
| H19 | 0.5675 | 0.3703 | 0.1402 | 0.057* | |
| C20 | 0.6603 (5) | 0.4085 (2) | 0.2941 (4) | 0.0556 (10) | |
| H20 | 0.7289 | 0.3755 | 0.3156 | 0.067* | |
| C21 | 0.6549 (5) | 0.4606 (2) | 0.3664 (3) | 0.0548 (10) | |
| H21 | 0.7203 | 0.4638 | 0.4364 | 0.066* | |
| C22 | 0.5505 (4) | 0.50766 (18) | 0.3323 (3) | 0.0475 (9) | |
| H22 | 0.5457 | 0.5428 | 0.3811 | 0.057* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C10 | 0.081 (4) | 0.096 (3) | 0.134 (4) | 0.009 (3) | 0.061 (3) | 0.009 (3) |
| C11 | 0.101 (4) | 0.102 (4) | 0.118 (4) | 0.020 (3) | 0.058 (3) | −0.004 (3) |
| C12 | 0.087 (4) | 0.084 (3) | 0.144 (5) | 0.035 (3) | 0.048 (3) | 0.007 (3) |
| C10' | 0.081 (4) | 0.096 (3) | 0.134 (4) | 0.009 (3) | 0.061 (3) | 0.009 (3) |
| C11' | 0.101 (4) | 0.102 (4) | 0.118 (4) | 0.020 (3) | 0.058 (3) | −0.004 (3) |
| C12' | 0.087 (4) | 0.084 (3) | 0.144 (5) | 0.035 (3) | 0.048 (3) | 0.007 (3) |
| Cu1 | 0.0495 (3) | 0.0268 (2) | 0.0376 (3) | 0.00568 (18) | 0.00751 (18) | 0.00124 (16) |
| O1 | 0.0618 (16) | 0.0312 (13) | 0.0578 (16) | 0.0093 (12) | 0.0143 (13) | 0.0113 (11) |
| O2 | 0.0639 (14) | 0.0477 (13) | 0.0722 (16) | 0.0222 (12) | 0.0247 (13) | 0.0112 (12) |
| O3 | 0.0599 (17) | 0.0424 (14) | 0.0514 (16) | −0.0017 (12) | 0.0132 (13) | 0.0113 (12) |
| O4 | 0.0725 (18) | 0.0373 (14) | 0.0517 (16) | 0.0066 (13) | 0.0042 (13) | 0.0124 (12) |
| N1 | 0.0414 (16) | 0.0347 (15) | 0.0340 (15) | 0.0035 (12) | 0.0042 (12) | 0.0052 (12) |
| N2 | 0.0448 (16) | 0.0336 (15) | 0.0322 (14) | −0.0010 (12) | 0.0029 (12) | 0.0018 (12) |
| C1 | 0.0611 (16) | 0.0381 (16) | 0.0573 (18) | 0.0190 (15) | 0.0207 (15) | 0.0079 (14) |
| C2 | 0.051 (2) | 0.0288 (17) | 0.0407 (19) | 0.0085 (15) | 0.0102 (16) | 0.0041 (14) |
| C3 | 0.046 (2) | 0.0351 (18) | 0.0373 (18) | 0.0047 (15) | 0.0085 (15) | 0.0008 (15) |
| C4 | 0.046 (2) | 0.0293 (16) | 0.0363 (18) | 0.0006 (14) | 0.0008 (15) | 0.0015 (14) |
| C5 | 0.046 (2) | 0.0299 (18) | 0.051 (2) | 0.0079 (15) | −0.0001 (16) | 0.0030 (15) |
| C6 | 0.047 (2) | 0.0335 (18) | 0.059 (2) | 0.0083 (16) | 0.0106 (17) | 0.0025 (17) |
| C7 | 0.051 (2) | 0.0324 (18) | 0.055 (2) | 0.0062 (16) | 0.0187 (17) | 0.0105 (16) |
| C8 | 0.058 (2) | 0.0337 (18) | 0.0317 (17) | −0.0047 (17) | −0.0048 (16) | 0.0022 (14) |
| C9 | 0.075 (3) | 0.062 (2) | 0.118 (3) | 0.021 (2) | 0.054 (3) | 0.011 (2) |
| C13 | 0.047 (2) | 0.049 (2) | 0.043 (2) | 0.0062 (17) | 0.0025 (16) | 0.0034 (17) |
| C14 | 0.047 (2) | 0.070 (3) | 0.043 (2) | −0.009 (2) | −0.0010 (17) | −0.0015 (19) |
| C15 | 0.062 (3) | 0.060 (3) | 0.049 (2) | −0.009 (2) | 0.0051 (19) | −0.020 (2) |
| C16 | 0.052 (2) | 0.042 (2) | 0.052 (2) | 0.0024 (17) | 0.0075 (18) | −0.0099 (17) |
| C17 | 0.0374 (18) | 0.0351 (17) | 0.0343 (17) | −0.0003 (14) | 0.0090 (14) | 0.0010 (14) |
| C18 | 0.0388 (18) | 0.0322 (17) | 0.0367 (18) | 0.0022 (14) | 0.0112 (14) | 0.0039 (14) |
| C19 | 0.053 (2) | 0.043 (2) | 0.048 (2) | 0.0117 (18) | 0.0132 (17) | 0.0019 (17) |
| C20 | 0.055 (2) | 0.058 (2) | 0.053 (2) | 0.020 (2) | 0.0061 (19) | 0.012 (2) |
| C21 | 0.057 (2) | 0.061 (3) | 0.044 (2) | 0.004 (2) | −0.0044 (18) | 0.0116 (19) |
| C22 | 0.057 (2) | 0.043 (2) | 0.041 (2) | −0.0016 (18) | 0.0012 (17) | 0.0005 (16) |
Geometric parameters (Å, °)
| C10—C9 | 1.521 (9) | N1—C17 | 1.341 (4) |
| C10—H10A | 0.9600 | N1—C13 | 1.347 (4) |
| C10—H10B | 0.9600 | N2—C22 | 1.333 (4) |
| C10—H10C | 0.9600 | N2—C18 | 1.348 (4) |
| C11—C9 | 1.519 (9) | C1—C2 | 1.505 (5) |
| C11—H11A | 0.9600 | C2—C3 | 1.381 (5) |
| C11—H11B | 0.9600 | C2—C7 | 1.386 (5) |
| C11—H11C | 0.9600 | C3—C4 | 1.390 (5) |
| C12—C9 | 1.501 (7) | C3—H3 | 0.9300 |
| C12—H12A | 0.9600 | C4—C5 | 1.389 (5) |
| C12—H12B | 0.9600 | C4—C8 | 1.502 (5) |
| C12—H12C | 0.9600 | C5—C6 | 1.387 (5) |
| C10'—C9 | 1.504 (8) | C5—H5 | 0.9300 |
| C10'—H10D | 0.9600 | C6—C7 | 1.390 (5) |
| C10'—H10E | 0.9600 | C6—C9 | 1.527 (6) |
| C10'—H10F | 0.9600 | C7—H7 | 0.9300 |
| C11'—C9 | 1.595 (9) | C8—Cu1ii | 2.538 (4) |
| C11'—H11D | 0.9600 | C13—C14 | 1.368 (5) |
| C11'—H11E | 0.9600 | C13—H13 | 0.9300 |
| C11'—H11F | 0.9600 | C14—C15 | 1.365 (6) |
| C12'—C9 | 1.502 (8) | C14—H14 | 0.9300 |
| C12'—H12D | 0.9600 | C15—C16 | 1.382 (6) |
| C12'—H12E | 0.9600 | C15—H15 | 0.9300 |
| C12'—H12F | 0.9600 | C16—C17 | 1.373 (5) |
| Cu1—O1 | 1.933 (2) | C16—H16 | 0.9300 |
| Cu1—O3i | 1.956 (2) | C17—C18 | 1.477 (5) |
| Cu1—N1 | 1.985 (3) | C18—C19 | 1.388 (5) |
| Cu1—N2 | 1.983 (3) | C19—C20 | 1.376 (5) |
| Cu1—C8i | 2.538 (4) | C19—H19 | 0.9300 |
| O1—C1 | 1.273 (4) | C20—C21 | 1.377 (6) |
| O2—C1 | 1.233 (5) | C20—H20 | 0.9300 |
| O3—C8 | 1.279 (5) | C21—C22 | 1.372 (5) |
| O3—Cu1ii | 1.956 (2) | C21—H21 | 0.9300 |
| O4—C8 | 1.236 (4) | C22—H22 | 0.9300 |
| C9—C10—H10A | 109.5 | C6—C5—C4 | 122.4 (3) |
| C9—C10—H10B | 109.5 | C6—C5—H5 | 118.8 |
| H10A—C10—H10B | 109.5 | C4—C5—H5 | 118.8 |
| C9—C10—H10C | 109.5 | C5—C6—C7 | 116.8 (3) |
| H10A—C10—H10C | 109.5 | C5—C6—C9 | 122.1 (3) |
| H10B—C10—H10C | 109.5 | C7—C6—C9 | 121.0 (4) |
| C9—C11—H11A | 109.5 | C2—C7—C6 | 121.8 (3) |
| C9—C11—H11B | 109.5 | C2—C7—H7 | 119.1 |
| H11A—C11—H11B | 109.5 | C6—C7—H7 | 119.1 |
| C9—C11—H11C | 109.5 | O4—C8—O3 | 122.7 (3) |
| H11A—C11—H11C | 109.5 | O4—C8—C4 | 119.8 (4) |
| H11B—C11—H11C | 109.5 | O3—C8—C4 | 117.5 (3) |
| C9—C12—H12A | 109.5 | O4—C8—Cu1ii | 76.6 (2) |
| C9—C12—H12B | 109.5 | O3—C8—Cu1ii | 49.07 (17) |
| H12A—C12—H12B | 109.5 | C4—C8—Cu1ii | 155.9 (2) |
| C9—C12—H12C | 109.5 | C12—C9—C10' | 131.1 (4) |
| H12A—C12—H12C | 109.5 | C12'—C9—C10' | 115.1 (5) |
| H12B—C12—H12C | 109.5 | C12—C9—C11 | 108.0 (5) |
| C9—C10'—H10D | 109.5 | C12'—C9—C11 | 132.1 (5) |
| C9—C10'—H10E | 109.5 | C10'—C9—C11 | 58.7 (3) |
| H10D—C10'—H10E | 109.5 | C12—C9—C10 | 111.7 (5) |
| C9—C10'—H10F | 109.5 | C12'—C9—C10 | 78.2 (4) |
| H10D—C10'—H10F | 109.5 | C10'—C9—C10 | 49.7 (2) |
| H10E—C10'—H10F | 109.5 | C11—C9—C10 | 108.0 (4) |
| C9—C11'—H11D | 109.5 | C12—C9—C6 | 112.9 (4) |
| C9—C11'—H11E | 109.5 | C12'—C9—C6 | 113.5 (5) |
| H11D—C11'—H11E | 109.5 | C10'—C9—C6 | 115.8 (4) |
| C9—C11'—H11F | 109.5 | C11—C9—C6 | 110.0 (5) |
| H11D—C11'—H11F | 109.5 | C10—C9—C6 | 106.1 (5) |
| H11E—C11'—H11F | 109.5 | C12—C9—C11' | 67.9 (3) |
| C9—C12'—H12D | 109.5 | C12'—C9—C11' | 102.3 (4) |
| C9—C12'—H12E | 109.5 | C10'—C9—C11' | 103.9 (5) |
| H12D—C12'—H12E | 109.5 | C11—C9—C11' | 47.3 (2) |
| C9—C12'—H12F | 109.5 | C10—C9—C11' | 146.9 (5) |
| H12D—C12'—H12F | 109.5 | C6—C9—C11' | 103.9 (5) |
| H12E—C12'—H12F | 109.5 | N1—C13—C14 | 122.2 (4) |
| O1—Cu1—O3i | 88.17 (11) | N1—C13—H13 | 118.9 |
| O1—Cu1—N1 | 94.83 (11) | C14—C13—H13 | 118.9 |
| O3i—Cu1—N1 | 172.79 (11) | C15—C14—C13 | 118.7 (4) |
| O1—Cu1—N2 | 173.34 (11) | C15—C14—H14 | 120.6 |
| O3i—Cu1—N2 | 96.69 (11) | C13—C14—H14 | 120.6 |
| N1—Cu1—N2 | 80.88 (11) | C14—C15—C16 | 119.8 (4) |
| O1—Cu1—C8i | 82.87 (11) | C14—C15—H15 | 120.1 |
| O3i—Cu1—C8i | 29.60 (11) | C16—C15—H15 | 120.1 |
| N2—Cu1—C8i | 103.60 (11) | C17—C16—C15 | 119.0 (4) |
| N1—Cu1—C8i | 144.33 (12) | C17—C16—H16 | 120.5 |
| C1—O1—Cu1 | 106.8 (2) | C15—C16—H16 | 120.5 |
| C8—O3—Cu1ii | 101.3 (2) | N1—C17—C16 | 121.4 (3) |
| C17—N1—C13 | 118.9 (3) | N1—C17—C18 | 114.0 (3) |
| C17—N1—Cu1 | 115.6 (2) | C16—C17—C18 | 124.6 (3) |
| C13—N1—Cu1 | 125.4 (2) | N2—C18—C19 | 121.3 (3) |
| C22—N2—C18 | 118.9 (3) | N2—C18—C17 | 114.2 (3) |
| C22—N2—Cu1 | 125.8 (2) | C19—C18—C17 | 124.5 (3) |
| C18—N2—Cu1 | 115.3 (2) | C20—C19—C18 | 118.7 (4) |
| O2—C1—O1 | 123.0 (3) | C20—C19—H19 | 120.7 |
| O2—C1—C2 | 120.3 (3) | C18—C19—H19 | 120.7 |
| O1—C1—C2 | 116.7 (3) | C19—C20—C21 | 120.0 (4) |
| C3—C2—C7 | 120.3 (3) | C19—C20—H20 | 120.0 |
| C3—C2—C1 | 120.6 (3) | C21—C20—H20 | 120.0 |
| C7—C2—C1 | 119.1 (3) | C22—C21—C20 | 118.2 (4) |
| C2—C3—C4 | 119.3 (3) | C22—C21—H21 | 120.9 |
| C2—C3—H3 | 120.4 | C20—C21—H21 | 120.9 |
| C4—C3—H3 | 120.4 | N2—C22—C21 | 123.0 (4) |
| C5—C4—C3 | 119.4 (3) | N2—C22—H22 | 118.5 |
| C5—C4—C8 | 119.7 (3) | C21—C22—H22 | 118.5 |
| C3—C4—C8 | 120.9 (3) | ||
| O3i—Cu1—O1—C1 | 84.9 (3) | C3—C4—C8—Cu1ii | 47.6 (8) |
| N1—Cu1—O1—C1 | −101.7 (3) | C5—C6—C9—C12 | −6.0 (7) |
| C8i—Cu1—O1—C1 | 114.1 (3) | C7—C6—C9—C12 | 174.1 (5) |
| O1—Cu1—N1—C17 | 176.4 (2) | C5—C6—C9—C12' | 32.7 (7) |
| N2—Cu1—N1—C17 | 1.5 (2) | C7—C6—C9—C12' | −147.2 (5) |
| C8i—Cu1—N1—C17 | −99.1 (3) | C5—C6—C9—C10' | 169.2 (6) |
| N2—Cu1—N1—C13 | −180.0 (3) | C7—C6—C9—C10' | −10.7 (8) |
| C8i—Cu1—N1—C13 | 79.4 (3) | C5—C6—C9—C11 | −126.8 (5) |
| O3i—Cu1—N2—C22 | −7.2 (3) | C7—C6—C9—C11 | 53.3 (7) |
| N1—Cu1—N2—C22 | 179.6 (3) | C5—C6—C9—C10 | 116.6 (5) |
| C8i—Cu1—N2—C22 | −36.5 (3) | C7—C6—C9—C10 | −63.3 (6) |
| O3i—Cu1—N2—C18 | 172.5 (2) | C5—C6—C9—C11' | −77.6 (6) |
| N1—Cu1—N2—C18 | −0.6 (2) | C7—C6—C9—C11' | 102.6 (5) |
| C8i—Cu1—N2—C18 | 143.3 (2) | C17—N1—C13—C14 | 1.8 (5) |
| Cu1—O1—C1—O2 | 17.2 (5) | Cu1—N1—C13—C14 | −176.7 (3) |
| Cu1—O1—C1—C2 | −162.2 (3) | N1—C13—C14—C15 | −1.4 (6) |
| O2—C1—C2—C3 | 18.7 (6) | C13—C14—C15—C16 | −0.1 (6) |
| O1—C1—C2—C3 | −162.0 (3) | C14—C15—C16—C17 | 1.1 (6) |
| O2—C1—C2—C7 | −160.4 (4) | C13—N1—C17—C16 | −0.7 (5) |
| O1—C1—C2—C7 | 19.0 (5) | Cu1—N1—C17—C16 | 178.0 (3) |
| C7—C2—C3—C4 | −0.7 (5) | C13—N1—C17—C18 | 179.3 (3) |
| C1—C2—C3—C4 | −179.7 (3) | Cu1—N1—C17—C18 | −2.1 (4) |
| C2—C3—C4—C5 | 1.7 (5) | C15—C16—C17—N1 | −0.7 (6) |
| C2—C3—C4—C8 | −178.1 (3) | C15—C16—C17—C18 | 179.3 (3) |
| C3—C4—C5—C6 | −0.5 (5) | C22—N2—C18—C19 | −0.1 (5) |
| C8—C4—C5—C6 | 179.3 (3) | Cu1—N2—C18—C19 | −179.8 (3) |
| C4—C5—C6—C7 | −1.7 (6) | C22—N2—C18—C17 | 179.5 (3) |
| C4—C5—C6—C9 | 178.4 (4) | Cu1—N2—C18—C17 | −0.3 (4) |
| C3—C2—C7—C6 | −1.7 (6) | N1—C17—C18—N2 | 1.5 (4) |
| C1—C2—C7—C6 | 177.4 (4) | C16—C17—C18—N2 | −178.5 (3) |
| C5—C6—C7—C2 | 2.8 (6) | N1—C17—C18—C19 | −178.9 (3) |
| C9—C6—C7—C2 | −177.3 (4) | C16—C17—C18—C19 | 1.1 (5) |
| Cu1ii—O3—C8—O4 | −22.7 (4) | N2—C18—C19—C20 | 0.4 (5) |
| Cu1ii—O3—C8—C4 | 155.6 (2) | C17—C18—C19—C20 | −179.2 (3) |
| C5—C4—C8—O4 | −3.7 (5) | C18—C19—C20—C21 | −0.8 (6) |
| C3—C4—C8—O4 | 176.1 (3) | C19—C20—C21—C22 | 0.9 (6) |
| C5—C4—C8—O3 | 178.0 (3) | C18—N2—C22—C21 | 0.2 (5) |
| C3—C4—C8—O3 | −2.2 (5) | Cu1—N2—C22—C21 | 180.0 (3) |
| C5—C4—C8—Cu1ii | −132.2 (6) | C20—C21—C22—N2 | −0.6 (6) |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) x, −y+3/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2072).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (1997). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Chang, F., Wang, Z.-M., Sun, H.-L., Wen, G.-H. & Zhang, X.-X. (2005). Dalton Trans. pp. 2976–2978. [DOI] [PubMed]
- Ma, C.-B., Chen, C.-N., Liu, Q.-T., Liao, D.-Z. & Li, L.-C. (2008). Eur. J. Inorg. Chem. pp. 1865–1870.
- Ma, L.-F., Wang, L.-Y., Huo, X.-K., Wang, Y.-Y., Fan, Y.-T., Wang, J.-G. & Chen, S. H. (2008). Cryst. Growth Des.8, 620–628.
- Ma, L.-F., Wang, Y.-Y., Wang, L.-Y., Liu, J.-Q., Wu, Y.-P., Wang, J.-G., Shi, Q.-Z. & Peng, S. M. (2008). Eur. J. Inorg. Chem. pp. 693–703.
- Pan, L., Parker, B., Huang, X. Y., Oison, D. H., Lee, J. Y. & Li, J. (2006). J. Am. Chem. Soc.128, 4180–4181. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xu, Y.-Q., Yuan, D.-Q., Wu, B.-L., Han, L., Wu, M.-Y., Jiang, F.-L. & Hong, M.-C. (2006). Cryst. Growth Des.6, 1168–1174.
- Yang, S.-Y., Long, L.-S., Huang, R.-B. & Zheng, L.-S. (2002). Chem. Commun. pp. 472–473. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536808035484/su2072sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808035484/su2072Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




