Abstract
The title compound, C9H10BrNO4S, is an intermediate for the formation of benzothiazines. In the crystal structure, intermolecular N—H⋯O hydrogen bonds link the molecules, forming R 2 2(10) ring motifs, which are linked into a two-dimensional polymeric sheet through intermolecular C—H⋯O hydrogen bonds.
Related literature
For general background, see: Arshad et al. (2008 ▶); Tahir et al. (2008 ▶). For a related structure, see: Bornaghi et al. (2005 ▶). For ring motifs, see: Bernstein et al. (1995 ▶). For bond-length data, see: Allen et al. (1987 ▶).
Experimental
Crystal data
C9H10BrNO4S
M r = 308.15
Triclinic,

a = 6.0451 (2) Å
b = 7.0369 (2) Å
c = 13.8695 (5) Å
α = 83.866 (2)°
β = 81.190 (1)°
γ = 87.027 (2)°
V = 579.31 (3) Å3
Z = 2
Mo Kα radiation
μ = 3.73 mm−1
T = 296 (2) K
0.23 × 0.18 × 0.12 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.449, T max = 0.639
13148 measured reflections
2846 independent reflections
1893 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.116
S = 1.04
2846 reflections
145 parameters
H-atom parameters constrained
Δρmax = 0.53 e Å−3
Δρmin = −0.59 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2003 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808037550/hk2573sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808037550/hk2573Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.86 | 2.41 | 2.987 (4) | 125 |
| C3—H3A⋯O3ii | 0.97 | 2.50 | 3.410 (4) | 156 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
MNA gratefully acknowledges the Higher Education Commission, Islamabad, Pakistan, for providing a Scholarship under the Indigenous PhD Programme (PIN 042-120607-PS2-183).
supplementary crystallographic information
Comment
The glycine methyl ester is the simplest among the amino acids and the p-bromobenzenesulfonyl chloride is also comercially available. We have synthesized the title compound, (I), from their condensation. It has been prepared as an intermediate for the formation of benzothiazines, which are of our research interest (Arshad et al., 2008; Tahir et al., 2008).
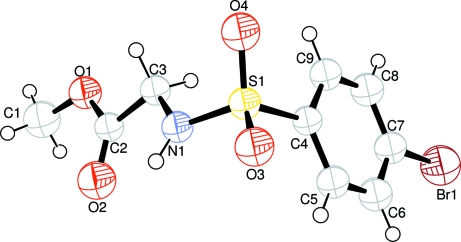
In the molecule of the title compound, (I), (Fig. 1) the bond lengths (Allen et al., 1987) and angles are within normal ranges, and are comparable with the corresponding values in N-(2-nitrophenylsulfonyl)glycine methyl ester, (II), (Bornaghi et al., 2005). Ring A (C4-C9) is, of course, planar, and the Br atom lies slightly out of the ring plane [0.069 (3) Å]. The S1-N1 bond is almost perpendicular to the ring plane, with N1-S1-C4-C5 [-83.4 (3)°] and/or N1/S1/C4/C9 [93.7 (3)°] torsion angles. The (O1/O2/C1-C3) moiety is planar, and it is oriented with respect to ring A at a dihedral angle of 87.74 (3)°.
In the crystal structure, intermolecular N-H···O hydrogen bonds (Table 1) link the molecules to form R22(10) ring motifs (Bernstein et al., 1995), in which they are linked to form a two dimensional polymeric sheet through intermolecular C-H···O hydrogen bonds (Table 1, Fig. 2).
Experimental
Glycine methyl ester hydrochloride (0.246 g, 1.95 mmol) was dissolved in water (10 ml) in round bottom flask. The pH of the solution was adjusted to 8–9 using sodium carbonate (1 N), and then 4-bromo benzene sulfonyl chloride (0.5 g, 1.95 mmol) was added. The mixture was stirred for 2 h at room temperature. During the reaction, pH was strictly maintained at 8–9 as HCl produced, which lowers the pH. Colorless solid product obtained was washed, dried and recrystalized from methanol for X-ray analysis (m.p. 393-394 K).
Refinement
H atoms were positioned geometrically, with N-H = 0.86 Å (for NH) and C-H = 0.93, 0.97 and 0.96 Å for aromatic, methylene and methyl H, respectively, and constrained to ride on their parent atoms with Uiso(H) = xUeq(C,N), where x = 1.5 for methyl H and x = 1.2 for all other H atoms.
Figures
Fig. 1.
The molecular structure of the title molecule, with the atom-numbering scheme.
Fig. 2.
A partial packing diagram. Hydrogen bonds are shown as dashed lines.
Crystal data
| C9H10BrNO4S | Z = 2 |
| Mr = 308.15 | F000 = 308 |
| Triclinic, P1 | Dx = 1.767 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 6.0451 (2) Å | Cell parameters from 2846 reflections |
| b = 7.0369 (2) Å | θ = 1.5–28.3º |
| c = 13.8695 (5) Å | µ = 3.73 mm−1 |
| α = 83.866 (2)º | T = 296 (2) K |
| β = 81.190 (1)º | Prism, colorless |
| γ = 87.027 (2)º | 0.23 × 0.18 × 0.12 mm |
| V = 579.31 (3) Å3 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 2846 independent reflections |
| Radiation source: fine-focus sealed tube | 1893 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.038 |
| Detector resolution: 7.40 pixels mm-1 | θmax = 28.3º |
| T = 296(2) K | θmin = 1.5º |
| ω scans | h = −8→8 |
| Absorption correction: multi-scan(SADABS; Bruker, 2005) | k = −9→9 |
| Tmin = 0.449, Tmax = 0.639 | l = −18→18 |
| 13148 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.040 | H-atom parameters constrained |
| wR(F2) = 0.116 | w = 1/[σ2(Fo2) + (0.0448P)2 + 0.5395P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 2846 reflections | Δρmax = 0.53 e Å−3 |
| 145 parameters | Δρmin = −0.59 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.31108 (8) | 0.18201 (7) | 0.47759 (3) | 0.0859 (2) | |
| S1 | −0.15899 (12) | 0.80316 (11) | 0.19808 (6) | 0.0428 (2) | |
| O1 | 0.4939 (4) | 0.7999 (3) | −0.06466 (17) | 0.0535 (6) | |
| O2 | 0.2102 (5) | 0.6116 (4) | −0.06458 (19) | 0.0694 (8) | |
| O3 | −0.3871 (3) | 0.7563 (4) | 0.20253 (19) | 0.0589 (6) | |
| O4 | −0.1032 (4) | 0.9893 (3) | 0.21559 (19) | 0.0602 (6) | |
| N1 | −0.0358 (4) | 0.7668 (4) | 0.08966 (18) | 0.0426 (6) | |
| H1 | −0.1060 | 0.7168 | 0.0497 | 0.051* | |
| C1 | 0.6104 (7) | 0.7313 (6) | −0.1532 (3) | 0.0720 (12) | |
| H1A | 0.7505 | 0.7935 | −0.1714 | 0.108* | |
| H1B | 0.5207 | 0.7589 | −0.2048 | 0.108* | |
| H1C | 0.6379 | 0.5957 | −0.1425 | 0.108* | |
| C2 | 0.2962 (5) | 0.7298 (4) | −0.0293 (2) | 0.0411 (7) | |
| C3 | 0.1966 (5) | 0.8203 (5) | 0.0605 (2) | 0.0427 (7) | |
| H3A | 0.2818 | 0.7791 | 0.1133 | 0.051* | |
| H3B | 0.2032 | 0.9583 | 0.0477 | 0.051* | |
| C4 | −0.0381 (5) | 0.6363 (4) | 0.2804 (2) | 0.0385 (6) | |
| C5 | −0.1158 (6) | 0.4529 (5) | 0.2982 (2) | 0.0482 (8) | |
| H5 | −0.2363 | 0.4206 | 0.2696 | 0.058* | |
| C6 | −0.0151 (6) | 0.3180 (5) | 0.3582 (2) | 0.0534 (8) | |
| H6 | −0.0672 | 0.1944 | 0.3709 | 0.064* | |
| C7 | 0.1645 (6) | 0.3690 (6) | 0.3992 (2) | 0.0528 (9) | |
| C8 | 0.2420 (6) | 0.5515 (6) | 0.3828 (3) | 0.0549 (9) | |
| H8 | 0.3625 | 0.5834 | 0.4115 | 0.066* | |
| C9 | 0.1398 (5) | 0.6868 (5) | 0.3233 (2) | 0.0484 (8) | |
| H9 | 0.1898 | 0.8111 | 0.3121 | 0.058* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0822 (3) | 0.1005 (4) | 0.0689 (3) | 0.0267 (3) | −0.0172 (2) | 0.0133 (2) |
| S1 | 0.0313 (4) | 0.0443 (5) | 0.0512 (5) | −0.0016 (3) | −0.0027 (3) | −0.0019 (3) |
| O1 | 0.0403 (12) | 0.0621 (15) | 0.0588 (14) | −0.0191 (10) | 0.0057 (10) | −0.0201 (12) |
| O2 | 0.0744 (17) | 0.0703 (18) | 0.0649 (16) | −0.0429 (14) | 0.0103 (13) | −0.0240 (14) |
| O3 | 0.0288 (11) | 0.0740 (17) | 0.0704 (16) | −0.0032 (10) | −0.0048 (10) | 0.0059 (13) |
| O4 | 0.0603 (15) | 0.0420 (14) | 0.0770 (18) | 0.0016 (11) | −0.0034 (13) | −0.0112 (12) |
| N1 | 0.0335 (13) | 0.0531 (16) | 0.0423 (14) | −0.0158 (11) | −0.0078 (10) | 0.0004 (12) |
| C1 | 0.060 (2) | 0.078 (3) | 0.076 (3) | −0.016 (2) | 0.020 (2) | −0.031 (2) |
| C2 | 0.0409 (16) | 0.0361 (16) | 0.0465 (17) | −0.0116 (12) | −0.0062 (13) | −0.0003 (13) |
| C3 | 0.0363 (16) | 0.0476 (18) | 0.0453 (17) | −0.0140 (13) | −0.0052 (12) | −0.0053 (14) |
| C4 | 0.0320 (14) | 0.0470 (17) | 0.0359 (15) | −0.0035 (12) | −0.0008 (11) | −0.0067 (13) |
| C5 | 0.0478 (18) | 0.052 (2) | 0.0467 (18) | −0.0114 (15) | −0.0110 (14) | −0.0048 (15) |
| C6 | 0.061 (2) | 0.050 (2) | 0.0488 (19) | −0.0070 (16) | −0.0077 (16) | −0.0017 (15) |
| C7 | 0.0484 (19) | 0.071 (2) | 0.0348 (16) | 0.0117 (17) | 0.0007 (14) | −0.0016 (15) |
| C8 | 0.0398 (17) | 0.076 (3) | 0.050 (2) | −0.0057 (16) | −0.0110 (14) | −0.0062 (18) |
| C9 | 0.0386 (17) | 0.055 (2) | 0.0524 (19) | −0.0110 (14) | −0.0041 (14) | −0.0102 (16) |
Geometric parameters (Å, °)
| Br1—C7 | 1.889 (3) | C3—N1 | 1.458 (4) |
| S1—O4 | 1.424 (2) | C3—H3A | 0.9700 |
| S1—O3 | 1.425 (2) | C3—H3B | 0.9700 |
| S1—N1 | 1.613 (3) | C4—C5 | 1.381 (4) |
| S1—C4 | 1.759 (3) | C4—C9 | 1.385 (4) |
| O1—C2 | 1.320 (3) | C5—C6 | 1.374 (5) |
| O1—C1 | 1.436 (4) | C5—H5 | 0.9300 |
| O2—C2 | 1.188 (4) | C6—C7 | 1.381 (5) |
| N1—H1 | 0.8600 | C6—H6 | 0.9300 |
| C1—H1A | 0.9600 | C7—C8 | 1.374 (5) |
| C1—H1B | 0.9600 | C8—C9 | 1.377 (5) |
| C1—H1C | 0.9600 | C8—H8 | 0.9300 |
| C2—C3 | 1.489 (4) | C9—H9 | 0.9300 |
| O4—S1—O3 | 120.49 (15) | N1—C3—H3B | 109.6 |
| O4—S1—N1 | 106.96 (14) | C2—C3—H3B | 109.6 |
| O3—S1—N1 | 106.53 (14) | H3A—C3—H3B | 108.1 |
| O4—S1—C4 | 107.89 (15) | C5—C4—C9 | 120.5 (3) |
| O3—S1—C4 | 107.66 (14) | C5—C4—S1 | 119.4 (2) |
| N1—S1—C4 | 106.55 (14) | C9—C4—S1 | 120.1 (2) |
| C2—O1—C1 | 117.5 (3) | C6—C5—C4 | 120.1 (3) |
| C3—N1—S1 | 118.9 (2) | C6—C5—H5 | 120.0 |
| C3—N1—H1 | 120.6 | C4—C5—H5 | 120.0 |
| S1—N1—H1 | 120.6 | C5—C6—C7 | 118.9 (3) |
| O1—C1—H1A | 109.5 | C5—C6—H6 | 120.6 |
| O1—C1—H1B | 109.5 | C7—C6—H6 | 120.6 |
| H1A—C1—H1B | 109.5 | C8—C7—C6 | 121.6 (3) |
| O1—C1—H1C | 109.5 | C8—C7—Br1 | 119.2 (3) |
| H1A—C1—H1C | 109.5 | C6—C7—Br1 | 119.2 (3) |
| H1B—C1—H1C | 109.5 | C7—C8—C9 | 119.4 (3) |
| O2—C2—O1 | 124.5 (3) | C7—C8—H8 | 120.3 |
| O2—C2—C3 | 125.0 (3) | C9—C8—H8 | 120.3 |
| O1—C2—C3 | 110.4 (2) | C8—C9—C4 | 119.5 (3) |
| N1—C3—C2 | 110.1 (2) | C8—C9—H9 | 120.2 |
| N1—C3—H3A | 109.6 | C4—C9—H9 | 120.2 |
| C2—C3—H3A | 109.6 | ||
| C1—O1—C2—O2 | 1.4 (5) | C4—C5—C6—C7 | −0.4 (5) |
| C1—O1—C2—C3 | −178.3 (3) | C5—C6—C7—C8 | 1.1 (5) |
| O2—C2—C3—N1 | −9.3 (5) | C5—C6—C7—Br1 | −177.6 (3) |
| O1—C2—C3—N1 | 170.5 (3) | C6—C7—C8—C9 | −0.5 (5) |
| O4—S1—C4—C5 | 162.0 (2) | Br1—C7—C8—C9 | 178.1 (2) |
| O3—S1—C4—C5 | 30.5 (3) | C7—C8—C9—C4 | −0.6 (5) |
| N1—S1—C4—C5 | −83.4 (3) | C5—C4—C9—C8 | 1.2 (5) |
| O4—S1—C4—C9 | −20.8 (3) | S1—C4—C9—C8 | −175.9 (2) |
| O3—S1—C4—C9 | −152.3 (3) | C2—C3—N1—S1 | 165.0 (2) |
| N1—S1—C4—C9 | 93.7 (3) | O4—S1—N1—C3 | 44.2 (3) |
| C9—C4—C5—C6 | −0.7 (5) | O3—S1—N1—C3 | 174.3 (2) |
| S1—C4—C5—C6 | 176.5 (3) | C4—S1—N1—C3 | −71.0 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.86 | 2.41 | 2.987 (4) | 125 |
| C3—H3A···O3ii | 0.97 | 2.50 | 3.410 (4) | 156 |
Symmetry codes: (i) −x, −y+1, −z; (ii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HK2573).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Arshad, M. N., Tahir, M. N., Khan, I. U., Shafiq, M. & Siddiqui, W. A. (2008). Acta Cryst. E64, o2045. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bornaghi, L. F., Poulsen, S.-A., Healy, P. C. & White, A. R. (2005). Acta Cryst. E61, o323–o325.
- Bruker (2005). SADABS Bruker AXS Inc. Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc. Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Tahir, M. N., Shafiq, M., Khan, I. U., Siddiqui, W. A. & Arshad, M. N. (2008). Acta Cryst. E64, o557. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808037550/hk2573sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808037550/hk2573Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report