Abstract
In the title compound, Na+·C6H4IO3S−·H2O, the Na atom is hexacoordinated by O atoms, forming a two-dimensional sheet-like structure in the bc plane, with the iodobenzene rings protruding above and below. Na⋯O contact distances are in the range 2.419 (2)–2.7218 (18) Å and O⋯Na⋯O angles are in the range 73.70 (5)–158.64 (7)°. The crystal structure is stabilized by O—H⋯O and C—H⋯O hydrogen bonds and C—H⋯π interactions. The I atom is disordered over two positions with occupancies of 0.78 (2) and 0.22 (2).
Related literature
For related literature on the synthesis of biologically active benzothiazine derivatives, see: Arshad et al. (2008 ▶); Chau & Kice (1977 ▶); Shafiq, Khan et al. (2008 ▶); Shafiq, Tahir et al. (2008 ▶); Tahir et al. (2008 ▶).
Experimental
Crystal data
Na+·C6H4IO3S−·H2O
M r = 324.06
Monoclinic,

a = 13.6141 (4) Å
b = 8.8233 (3) Å
c = 7.8493 (3) Å
β = 92.171 (1)°
V = 942.19 (6) Å3
Z = 4
Mo Kα radiation
μ = 3.64 mm−1
T = 296 (2) K
0.25 × 0.17 × 0.15 mm
Data collection
Bruker KAPPA APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.482, T max = 0.580
10141 measured reflections
2339 independent reflections
2135 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.019
wR(F 2) = 0.047
S = 1.05
2339 reflections
128 parameters
H-atom parameters constrained
Δρmax = 0.41 e Å−3
Δρmin = −0.42 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2003 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808039202/su2075sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808039202/su2075Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4A⋯O2i | 0.85 | 1.98 | 2.824 (2) | 174 |
| C6—H6⋯O1 | 0.93 | 2.42 | 2.834 (3) | 107 |
| C5—H5⋯Cgi | 0.93 | 2.79 | 3.661 (3) | 156 |
Symmetry code: (i)  . Cg is the centroid of the benzene ring.
. Cg is the centroid of the benzene ring.
Acknowledgments
MNA greatfully acknowledges the Higher Education Commission, Islamabad, Pakistan, for providing him a Scholarship under the Indigenous PhD Program (PIN 042–120607–PS2–183).
supplementary crystallographic information
Comment
In the continuation to our research work on the synthesis of biologically active benzothiazine derivatives (Arshad et al., 2008; Shafiq, Khan, Tahir & Siddiqui, 2008; Shafiq, Tahir, Khan, Ahmad & Siddiqui, 2008; Tahir et al., 2008), we synthesized the title compound, (I).
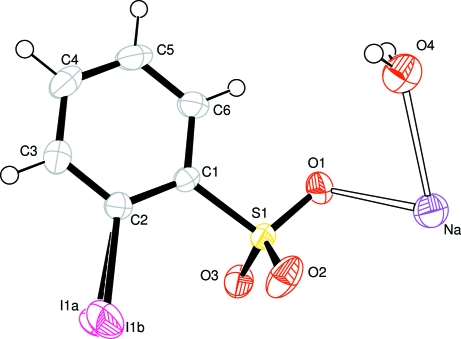
The molecular structure of compound (I) is illustrated in Fig. 1. The bond lengths and bond angles, as far as the 2-iodobenzenesulfonate is concerned, are similar to those reported for the potassium salt analogue, reported on recently (Arshad et al., 2008). The Na-atom is hexa-coordinated with O-atoms, involving four from the sulfonic groups and two water molecules. The range of the Na···O distances is 2.419 (2)–2.7218 (18) Å, whereas the range of O···Na···O angles is 73.70 (5)–158.64 (7)°. In this way a two-dimensional sheet-like structure in the bc plane is formed, with the iodobenzene rings protruding above and below.
It is interesting that one of the O-atoms of the sulfonic group is not involved in coordination with the Na-atom, but makes an intermolecular hydrogen bond with a water molecule, similar to the situation on the potassium salt analogue mentioned above. Also, one of the H-atoms of the H2O molecule is not involved in intra or intermolecular H-bonding.
In the crystal structure of compound (I) a two dimensional polymeric network extending along the b axis is formed (Fig. 2). There exist O-H···O and C-H···O hydrogen bonds, and C-H···π-interactions involving the centroid, Cg, of the benzene ring, see Table 1 for details.
Experimental
The title compound was prepared following the method used by Chau & Kice (1977). Suitable crystals for X-ray analysis were obtained from the reaction mixture.
Refinement
The Iodine atom was disordered over two positions (I1A/I1B) with occupancies of 0.78 (2)/0.22 (2). The water H-atoms were located in a difference Fourier map and were refined with distance restraints: O-H = 0.82 (2) Å with Uiso(H) = 1.2Ueq(O). The C-bound H-atoms were positioned geometrically and treated as riding atoms: C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
Molecular structure of compound (I), with the atom numbering scheme. The thermal ellipsoids are drawn at the 50% probability level.
Fig. 2.
A view along the c axis of the crystal packing in compound (I), showing the coordination of the O-atoms with the sodium atom and the hydrogen bonding involving the water molecules (H-atoms not involved in hydrogen bonding have been removed for clarity).
Crystal data
| Na+·C6H4IO3S–·H2O | F000 = 616 |
| Mr = 324.06 | Dx = 2.285 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2339 reflections |
| a = 13.6141 (4) Å | θ = 1.5–28.3º |
| b = 8.8233 (3) Å | µ = 3.64 mm−1 |
| c = 7.8493 (3) Å | T = 296 (2) K |
| β = 92.171 (1)º | Prismatic, colorless |
| V = 942.19 (6) Å3 | 0.25 × 0.17 × 0.15 mm |
| Z = 4 |
Data collection
| Bruker KAPPA APEXII CCD diffractometer | 2339 independent reflections |
| Radiation source: fine-focus sealed tube | 2135 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.024 |
| Detector resolution: 7.40 pixels mm-1 | θmax = 28.3º |
| T = 296(2) K | θmin = 1.5º |
| ω scans | h = −18→18 |
| Absorption correction: multi-scan(SADABS; Bruker, 2005) | k = −11→11 |
| Tmin = 0.482, Tmax = 0.580 | l = −10→10 |
| 10141 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.019 | H-atom parameters constrained |
| wR(F2) = 0.047 | w = 1/[σ2(Fo2) + (0.0176P)2 + 0.7234P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.009 |
| 2339 reflections | Δρmax = 0.42 e Å−3 |
| 128 parameters | Δρmin = −0.42 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| I1A | 0.19343 (9) | −0.1923 (2) | 0.55095 (18) | 0.0385 (2) | 0.78 (2) |
| I1B | 0.1984 (5) | −0.1716 (17) | 0.5644 (12) | 0.0628 (13) | 0.22 (2) |
| S1 | 0.37585 (3) | 0.02639 (5) | 0.34970 (6) | 0.0236 (1) | |
| Na | 0.51091 (7) | 0.29771 (10) | 0.49492 (12) | 0.0402 (3) | |
| O1 | 0.42883 (11) | 0.14214 (18) | 0.2606 (2) | 0.0369 (5) | |
| O2 | 0.38308 (12) | 0.0514 (2) | 0.53146 (19) | 0.0461 (6) | |
| O3 | 0.40017 (11) | −0.12539 (18) | 0.2976 (2) | 0.0390 (5) | |
| O4 | 0.39172 (13) | 0.4952 (2) | 0.38765 (19) | 0.0413 (5) | |
| C1 | 0.24956 (13) | 0.0530 (2) | 0.2877 (2) | 0.0227 (5) | |
| C2 | 0.17328 (14) | −0.0292 (2) | 0.3573 (2) | 0.0251 (5) | |
| C3 | 0.07722 (15) | −0.0064 (3) | 0.2991 (3) | 0.0328 (6) | |
| C4 | 0.05587 (16) | 0.0976 (3) | 0.1724 (3) | 0.0386 (7) | |
| C5 | 0.13033 (18) | 0.1803 (3) | 0.1044 (3) | 0.0412 (7) | |
| C6 | 0.22673 (16) | 0.1593 (3) | 0.1618 (3) | 0.0332 (6) | |
| H3 | 0.02673 | −0.06165 | 0.34590 | 0.0393* | |
| H4 | −0.00875 | 0.11181 | 0.13299 | 0.0463* | |
| H4A | 0.38645 | 0.48643 | 0.28018 | 0.0496* | |
| H4B | 0.33495 | 0.51683 | 0.41658 | 0.0496* | |
| H5 | 0.11586 | 0.25088 | 0.01914 | 0.0495* | |
| H6 | 0.27653 | 0.21667 | 0.11578 | 0.0398* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| I1A | 0.0425 (3) | 0.0384 (5) | 0.0345 (3) | −0.0046 (4) | 0.0010 (2) | 0.0146 (3) |
| I1B | 0.0625 (16) | 0.071 (3) | 0.0543 (18) | −0.0264 (19) | −0.0055 (11) | 0.0310 (15) |
| S1 | 0.0214 (2) | 0.0248 (2) | 0.0243 (2) | 0.0002 (2) | −0.0019 (2) | −0.0017 (2) |
| Na | 0.0466 (5) | 0.0325 (5) | 0.0407 (5) | 0.0011 (4) | −0.0087 (4) | −0.0053 (4) |
| O1 | 0.0285 (7) | 0.0346 (8) | 0.0478 (9) | −0.0059 (6) | 0.0027 (6) | 0.0055 (7) |
| O2 | 0.0349 (8) | 0.0784 (13) | 0.0245 (7) | −0.0004 (8) | −0.0055 (6) | −0.0061 (8) |
| O3 | 0.0301 (8) | 0.0252 (8) | 0.0612 (10) | 0.0051 (6) | −0.0043 (7) | −0.0076 (7) |
| O4 | 0.0447 (9) | 0.0519 (10) | 0.0274 (7) | 0.0000 (8) | 0.0018 (7) | −0.0009 (7) |
| C1 | 0.0224 (8) | 0.0227 (9) | 0.0227 (8) | 0.0025 (7) | −0.0012 (7) | −0.0016 (7) |
| C2 | 0.0277 (9) | 0.0240 (9) | 0.0234 (9) | 0.0001 (7) | −0.0006 (7) | −0.0011 (7) |
| C3 | 0.0261 (9) | 0.0349 (12) | 0.0373 (11) | −0.0030 (8) | 0.0008 (8) | −0.0037 (9) |
| C4 | 0.0283 (10) | 0.0430 (13) | 0.0438 (12) | 0.0083 (9) | −0.0091 (9) | −0.0036 (10) |
| C5 | 0.0405 (12) | 0.0413 (14) | 0.0413 (12) | 0.0096 (10) | −0.0066 (10) | 0.0141 (10) |
| C6 | 0.0333 (10) | 0.0316 (11) | 0.0346 (11) | 0.0019 (8) | 0.0017 (8) | 0.0091 (9) |
Geometric parameters (Å, °)
| I1A—C2 | 2.104 (2) | O4—H4B | 0.8400 |
| I1B—C2 | 2.073 (12) | O4—H4A | 0.8500 |
| S1—O1 | 1.4459 (16) | C1—C6 | 1.389 (3) |
| S1—O2 | 1.4433 (16) | C1—C2 | 1.395 (3) |
| S1—O3 | 1.4424 (16) | C2—C3 | 1.384 (3) |
| S1—C1 | 1.7846 (18) | C3—C4 | 1.376 (4) |
| Na—O1 | 2.5219 (18) | C4—C5 | 1.373 (3) |
| Na—O4 | 2.505 (2) | C5—C6 | 1.384 (3) |
| Na—O3i | 2.7218 (18) | C3—H3 | 0.9300 |
| Na—O3ii | 2.5060 (18) | C4—H4 | 0.9300 |
| Na—O4iii | 2.419 (2) | C5—H5 | 0.9300 |
| Na—O1iv | 2.4609 (18) | C6—H6 | 0.9300 |
| I1A···O2 | 3.368 (2) | O4···O1iv | 3.191 (2) |
| I1A···O3 | 3.557 (2) | O4···O1i | 3.036 (2) |
| I1A···C1v | 3.750 (2) | O1···H4Aviii | 2.8900 |
| I1A···I1Avi | 4.055 (2) | O1···H6 | 2.4200 |
| I1A···I1Av | 4.055 (2) | O2···H6iv | 2.6100 |
| I1A···C2v | 3.456 (2) | O2···H4Aiv | 1.9800 |
| I1A···C3v | 3.688 (3) | C1···I1Avi | 3.750 (2) |
| I1B···O3 | 3.540 (8) | C1···I1Bvi | 3.846 (14) |
| I1B···O2 | 3.211 (11) | C2···I1Avi | 3.456 (2) |
| I1B···C3v | 3.797 (13) | C2···I1Bvi | 3.526 (13) |
| I1B···C1v | 3.846 (14) | C3···I1Avi | 3.688 (3) |
| I1B···C2v | 3.526 (13) | C3···I1Bvi | 3.797 (13) |
| I1A···H4vii | 3.3300 | C4···C4x | 3.506 (3) |
| S1···O2ii | 3.4468 (17) | C2···H5iv | 2.8900 |
| O1···O3i | 3.149 (2) | C3···H5iv | 2.8800 |
| O1···O4viii | 3.036 (2) | C4···H4x | 3.0700 |
| O1···O4ix | 3.191 (2) | C6···H4Bix | 2.9200 |
| O2···S1ii | 3.4468 (17) | H4···I1Axi | 3.3300 |
| O2···I1B | 3.211 (11) | H4···C4x | 3.0700 |
| O2···I1A | 3.368 (2) | H4A···O2ix | 1.9800 |
| O2···O4iv | 2.824 (2) | H4B···C6iv | 2.9200 |
| O3···I1B | 3.540 (8) | H5···C2ix | 2.8900 |
| O3···O1viii | 3.149 (2) | H5···C3ix | 2.8800 |
| O3···I1A | 3.557 (2) | H6···O1 | 2.4200 |
| O4···O2ix | 2.824 (2) | H6···O2ix | 2.6100 |
| O1—S1—O2 | 110.70 (10) | Na—O4—Naiii | 93.37 (7) |
| O1—S1—O3 | 113.24 (9) | Naiii—O4—H4A | 118.00 |
| O1—S1—C1 | 105.56 (9) | Naiii—O4—H4B | 103.00 |
| O2—S1—O3 | 114.48 (10) | Na—O4—H4A | 107.00 |
| O2—S1—C1 | 106.16 (9) | Na—O4—H4B | 131.00 |
| O3—S1—C1 | 105.90 (9) | H4A—O4—H4B | 104.00 |
| O1—Na—O4 | 82.53 (6) | C2—C1—C6 | 118.68 (17) |
| O1—Na—O3i | 73.70 (5) | S1—C1—C6 | 118.04 (14) |
| O1—Na—O3ii | 109.48 (6) | S1—C1—C2 | 123.27 (13) |
| O1—Na—O4iii | 155.53 (7) | I1A—C2—C3 | 115.71 (15) |
| O1—Na—O1iv | 122.18 (6) | I1A—C2—C1 | 124.06 (14) |
| O3i—Na—O4 | 81.15 (6) | I1B—C2—C3 | 118.2 (2) |
| O3ii—Na—O4 | 158.64 (7) | C1—C2—C3 | 120.23 (17) |
| O4—Na—O4iii | 86.63 (6) | I1B—C2—C1 | 121.3 (2) |
| O1iv—Na—O4 | 79.96 (6) | C2—C3—C4 | 120.5 (2) |
| O3i—Na—O3ii | 118.69 (6) | C3—C4—C5 | 119.8 (2) |
| O3i—Na—O4iii | 83.01 (6) | C4—C5—C6 | 120.5 (2) |
| O1iv—Na—O3i | 153.11 (6) | C1—C6—C5 | 120.4 (2) |
| O3ii—Na—O4iii | 88.08 (6) | C2—C3—H3 | 120.00 |
| O1iv—Na—O3ii | 78.69 (6) | C4—C3—H3 | 120.00 |
| O1iv—Na—O4iii | 76.94 (6) | C3—C4—H4 | 120.00 |
| S1—O1—Na | 104.30 (8) | C5—C4—H4 | 120.00 |
| S1—O1—Naix | 144.40 (10) | C4—C5—H5 | 120.00 |
| Na—O1—Naix | 107.32 (6) | C6—C5—H5 | 120.00 |
| S1—O3—Naviii | 125.79 (9) | C1—C6—H6 | 120.00 |
| S1—O3—Naii | 119.31 (9) | C5—C6—H6 | 120.00 |
| Naviii—O3—Naii | 100.23 (6) | ||
| O2—S1—O1—Na | 4.83 (11) | O3i—Na—O4—Naiii | −83.44 (6) |
| O2—S1—O1—Naix | 156.95 (15) | O3ii—Na—O4—Naiii | 75.95 (19) |
| O3—S1—O1—Na | −125.30 (9) | O4iii—Na—O4—Naiii | 0.00 (7) |
| O3—S1—O1—Naix | 26.83 (19) | O1iv—Na—O4—Naiii | 77.33 (6) |
| C1—S1—O1—Na | 119.29 (8) | O1—Na—O3i—S1i | 136.29 (11) |
| C1—S1—O1—Naix | −88.58 (16) | O1—Na—O3i—Naix | −1.72 (6) |
| O1—S1—O3—Naviii | −18.05 (13) | O4—Na—O3i—S1i | 51.55 (11) |
| O1—S1—O3—Naii | 112.93 (10) | O4—Na—O3i—Naix | −86.47 (6) |
| O2—S1—O3—Naviii | −146.24 (10) | O1—Na—O3ii—S1ii | 19.44 (12) |
| O2—S1—O3—Naii | −15.26 (13) | O1—Na—O3ii—Naiv | −122.08 (6) |
| C1—S1—O3—Naviii | 97.16 (10) | O4—Na—O3ii—S1ii | 141.17 (16) |
| C1—S1—O3—Naii | −131.86 (9) | O4—Na—O3ii—Naiv | −0.4 (2) |
| O1—S1—C1—C2 | −174.65 (15) | O1—Na—O4iii—Naiii | 63.66 (18) |
| O1—S1—C1—C6 | 6.44 (18) | O4—Na—O4iii—Naiii | 0.00 (6) |
| O2—S1—C1—C2 | −57.08 (17) | O1—Na—O1iv—S1iv | −100.39 (17) |
| O2—S1—C1—C6 | 124.01 (17) | O1—Na—O1iv—Naiv | 107.94 (8) |
| O3—S1—C1—C2 | 65.01 (16) | O4—Na—O1iv—S1iv | −25.90 (16) |
| O3—S1—C1—C6 | −113.91 (17) | O4—Na—O1iv—Naiv | −177.57 (7) |
| O4—Na—O1—S1 | −111.84 (9) | S1—C1—C2—I1A | 2.0 (2) |
| O4—Na—O1—Naix | 84.73 (7) | S1—C1—C2—C3 | −177.78 (16) |
| O3i—Na—O1—S1 | 165.24 (9) | C6—C1—C2—I1A | −179.11 (16) |
| O3i—Na—O1—Naix | 1.81 (6) | C6—C1—C2—C3 | 1.1 (3) |
| O3ii—Na—O1—S1 | 49.96 (10) | S1—C1—C6—C5 | 177.54 (18) |
| O3ii—Na—O1—Naix | −113.48 (7) | C2—C1—C6—C5 | −1.4 (3) |
| O4iii—Na—O1—S1 | −176.30 (14) | I1A—C2—C3—C4 | −179.9 (2) |
| O4iii—Na—O1—Naix | 20.27 (19) | C1—C2—C3—C4 | −0.1 (3) |
| O1iv—Na—O1—S1 | −38.71 (11) | C2—C3—C4—C5 | −0.6 (4) |
| O1iv—Na—O1—Naix | 157.85 (7) | C3—C4—C5—C6 | 0.3 (4) |
| O1—Na—O4—Naiii | −158.02 (6) | C4—C5—C6—C1 | 0.7 (4) |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x+1, −y, −z+1; (iii) −x+1, −y+1, −z+1; (iv) x, −y+1/2, z+1/2; (v) x, −y−1/2, z+1/2; (vi) x, −y−1/2, z−1/2; (vii) −x, y−1/2, −z+1/2; (viii) −x+1, y−1/2, −z+1/2; (ix) x, −y+1/2, z−1/2; (x) −x, −y, −z; (xi) −x, y+1/2, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4A···O2ix | 0.85 | 1.98 | 2.824 (2) | 174 |
| C6—H6···O1 | 0.93 | 2.42 | 2.834 (3) | 107 |
| C5—H5···Cgix | 0.93 | 2.79 | 3.661 (3) | 156 |
Symmetry codes: (ix) x, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2075).
References
- Arshad, M. N., Khan, I. U., Ahmad, S., Shafiq, M. & Stoeckli-Evans, H. (2008). Acta Cryst. E64, m994. [DOI] [PMC free article] [PubMed]
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chau, M. M. & Kice, J. L. (1977). J. Org. Chem.42, 3265–3270.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Shafiq, M., Khan, I. U., Tahir, M. N. & Siddiqui, W. A. (2008). Acta Cryst. E64, o558. [DOI] [PMC free article] [PubMed]
- Shafiq, M., Tahir, M. N., Khan, I. U., Ahmad, S. & Siddiqui, W. A. (2008). Acta Cryst. E64, o1270. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Tahir, M. N., Shafiq, M., Khan, I. U., Siddiqui, W. A. & Arshad, M. N. (2008). Acta Cryst. E64, o557. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808039202/su2075sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808039202/su2075Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report