Abstract
The title compound, [ZnCl2(C10H12N4O2)]·0.5H2O, was readily prepared by the reaction between ZnCl2 and 2-hydroxyimino-N′-[1-(2-pyridyl)ethylidene]propanohydrazide. The Zn atom has a distorted trigonal–bipyramidal geometry with two Cl atoms and one azomethine N atom in the equatorial plane and one pyridine N atom and one amide O atom in the axial positions. In the crystal structure, complex molecules are connected in pairs by N—H⋯Cl hydrogen bonds, formed between the amide NH of one molecule and the Cl atom of a neighboring one. Molecular pairs are connected by hydrogen bonds involving the uncoordinated water molecule, which lies on a twofold axis.
Related literature
For details of the structure and biological activity of zinc(II) complexes, see: Canary et al. (1998 ▶); Comba et al. (2002 ▶); Kasuga et al. (2003 ▶); Panosyan et al. (2003 ▶); Rodriuez-Argüelles et al. (1995 ▶); Sousa et al. (2003 ▶). For the preparation of 2-(oximato)propanehydrazide, see: Fritsky et al. (1998 ▶).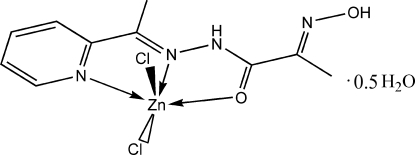
Experimental
Crystal data
[ZnCl2(C10H12N4O2)]·0.5H2O
M r = 365.51
Monoclinic,
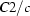
a = 14.5666 (8) Å
b = 12.9898 (8) Å
c = 15.8671 (8) Å
β = 109.873 (5)°
V = 2823.5 (3) Å3
Z = 8
Mo Kα radiation
μ = 2.13 mm−1
T = 100 (2) K
0.5 × 0.1 × 0.05 mm
Data collection
Kuma KM-4 CCD area-detector diffractometer
Absorption correction: multi-scan (PLATON; Spek, 2003 ▶) T min = 0.774, T max = 0.891
17382 measured reflections
3373 independent reflections
3069 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.070
S = 1.21
3373 reflections
191 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.45 e Å−3
Δρmin = −0.37 e Å−3
Data collection: KM-4 CCD Software (Kuma, 1999 ▶); cell refinement: SAINT (Bruker, 1999 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680706535X/hy2108sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680706535X/hy2108Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Zn1—N1 | 2.1409 (19) |
| Zn1—N2 | 2.1142 (18) |
| Zn1—O1 | 2.2348 (16) |
| Zn1—Cl2 | 2.2513 (6) |
| Zn1—Cl3 | 2.2195 (6) |
| N2—Zn1—N1 | 74.71 (7) |
| N2—Zn1—O1 | 72.57 (6) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H2⋯Cl3i | 0.80 (3) | 2.59 (3) | 3.310 (2) | 150 (3) |
| O1W—H1W⋯Cl2ii | 0.81 (3) | 2.44 (3) | 3.182 (2) | 153 (3) |
| O2—H1⋯O1Wiii | 0.88 (3) | 1.89 (4) | 2.7015 (19) | 153 (3) |
Symmetry codes: (i) 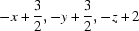 ; (ii)
; (ii) 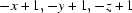 ; (iii)
; (iii) 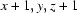 .
.
Acknowledgments
The authors are grateful to NATO for financial support (grant CBP. NUKR. CLG 982019).
supplementary crystallographic information
Comment
In the past few decades mononuclear zinc(II) complexes attract a lot of attention because of their biological and catalytical activity (Kasuga et al., 2003; Panosyan et al., 2003; Rodriuez-Argüelles et al., 1995; Sousa et al., 2003). On the other hand, many zinc mononuclear complexes containing additional vacant donor sets and chelate centers are taken as ligands for building of homo- and heteropolynuclear systems, which are widely used in bioinorganic modeling and catalysis. The properties of the obtained compounds are closely connected with their structure, so the best results one could obtain using well designed ligand systems. We report here the synthesis and structure of a zinc complex, [ZnCl2(C10H12N4O2)].0.5H2O, based on a novel polynucleative ligand 2-(oximato)-N'-(1-(pyridin-2-yl)ethylidene)propanehydrazide (L).
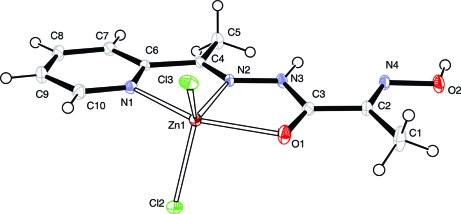
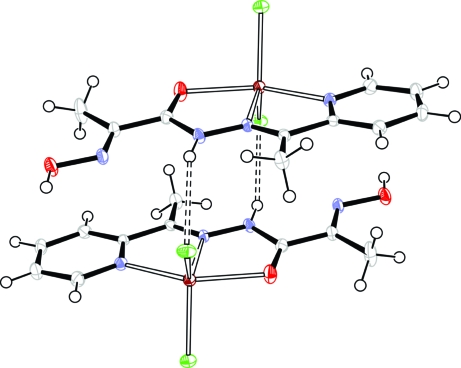
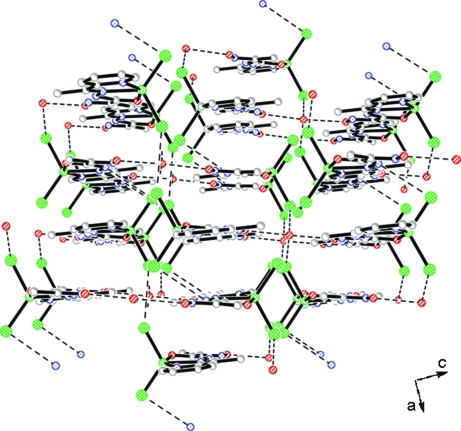
The title compound consists of neutral complex molecules and solvated water molecules (Fig. 1). The Zn atom has a distorted trigonal-bipyramidal geometry, defined by two Cl atoms and one azomethine N atom at the equatorial plane and a pyridine N atom and an amide O atom at the axial positions. The Zn—N, Zn—O and Zn—Cl bond lengths are comparable to previously reported zinc complexes with thiosemicarbasone and semicarbasone derivatives (Kasuga et al., 2003) and with pyridine complexed to the metal ion (Canary et al., 1998; Comba et al., 2002) (Table 1). The bite angles around the central atom deviate from an ideal square-planar configuration [e.g. N2—Zn1—O1 = 72.57 (6)°, N2—Zn1—N1 = 74.71 (7)°], that is a consequence of the formation of two almost flat five-membered chelate rings. In the crystal packing, the molecules are connected in pairs by N—H···Cl hydrogen bonds (Fig. 2), where the protonated amide group from one molecule acts as a donor and the Cl atom from the neighboring molecule as an acceptor (Table 2). The weak π-stacking interaction is observed between the pyridine ring and oximic group. The molecular pairs are connected to each other by hydrogen bonds involving the solvated water molecules, resulting in an extensive three-dimensional system (Fig. 3).
Experimental
2-(Oximato)-N'-(1-(pyridin-2-yl)ethylidene)propanehydrazide (L) was prepared according to early reported method (Fritsky et al., 1998). 2-Acetylpyridine (1.2 ml, 0.0107 mol) was added to a stirred warm ethanol/water solution (20 ml) of 2-(oximato)propanehydrazide (1.17 g, 0.01 mol). After stirring at 333 k for 6 h, the solution was cooled and a white precipitate was formed immediately. It was filtered off, washed with water and acetone and dried under vacuum (yield 72%, 1.59 g). 1H NMR, 400.13 MHz, (DMSO-d6): 12.118 (s, 1H, N—OH), 10.242 (s, 1H, NH), 8.608 (dt, 1H, py-6, J6,5 = 4.8 Hz, J6,4 = 1.8 Hz), 8.066 (d, 1H, py-3, J3,4 = 7.8 Hz), 7.861 (td, 1H, py-4, J4,5,3 = 7.8 Hz, J4,6 = 1.8 Hz), 7.415 (ddd, 1H, py-5, J5,6 = 4.8 Hz, J5,4 = 7.8 Hz, J = 1.2 Hz), 2.376 (s, 3H, CH3(py)), 1.985(s, 3H, CH3). IR (KBr, cm-1): 1660 (COamid), 1030 (NOoxim), 3340 (NHas). Analysis calculated for C10H12N4O2: C 54.54, H 5.49, N 25.44%; found: C 54.41, H 5.62, N 25.42%.
Zinc(II) chloride (0.014 g, 0.1 mmol) in H2O (5 ml) was added to 10 ml of hot methanol solution of L (0.022 g, 0.1 mmol). The solution was left for slow evaporation at room temperature. After 5 days cubic crystals of the title compound suitable for X-ray analysis were obtained.
Refinement
H atoms on C atoms were positioned geometrically and refined as riding atoms, with C—H = 0.93Å (CH) and 0.96Å (CH3), and with Uiso(H) = 1.2Ueq(C) for aromatic and Uiso(H) = 1.5Ueq(C) for methyl group. H atoms on N and O atoms were located from a difference Fourier map and were refined isotropically.
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are shown at the 40% probability level. Water molecule is not shown.
Fig. 2.
A view of a pair of the complex molecules. Displacement ellipsoids are shown at the 40% probability level. Hydrogen bonds are indicated by dashed lines.
Fig. 3.
A packing diagram of the title compound along the b-axis direction. Hydrogen bonds are indicated by dashed lines. H atoms have been omitted for clarity.
Crystal data
| [ZnCl2(C10H12N4O2)]·0.5H2O | F000 = 1480 |
| Mr = 365.51 | Dx = 1.720 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 3373 reflections |
| a = 14.5666 (8) Å | θ = 3.1–28.5º |
| b = 12.9898 (8) Å | µ = 2.13 mm−1 |
| c = 15.8671 (8) Å | T = 100 (2) K |
| β = 109.873 (5)º | Cubic, yellow |
| V = 2823.5 (3) Å3 | 0.5 × 0.1 × 0.05 mm |
| Z = 8 |
Data collection
| Kum KM4 CCD area-detector diffractometer | 3373 independent reflections |
| Radiation source: fine-focus sealed tube | 3069 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.043 |
| T = 100(2) K | θmax = 28.5º |
| ω scans | θmin = 3.1º |
| Absorption correction: multi-scan(PLATON; Spek, 2003) | h = −19→19 |
| Tmin = 0.774, Tmax = 0.891 | k = −16→16 |
| 17382 measured reflections | l = −20→21 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.037 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.070 | w = 1/[σ2(Fo2) + (0.0282P)2 + 3.185P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.21 | (Δ/σ)max = 0.002 |
| 3373 reflections | Δρmax = 0.45 e Å−3 |
| 191 parameters | Δρmin = −0.37 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.8446 (2) | 0.45584 (19) | 0.90460 (16) | 0.0300 (6) | |
| H1A | 0.8849 | 0.3958 | 0.9225 | 0.045* | |
| H1B | 0.8500 | 0.4827 | 0.8501 | 0.045* | |
| H1C | 0.7778 | 0.4378 | 0.8949 | 0.045* | |
| C2 | 0.87720 (17) | 0.53560 (17) | 0.97658 (14) | 0.0155 (4) | |
| C3 | 0.86291 (17) | 0.64543 (17) | 0.95017 (14) | 0.0149 (4) | |
| C4 | 0.89866 (16) | 0.89516 (17) | 1.03894 (14) | 0.0127 (4) | |
| C5 | 0.96032 (18) | 0.89213 (18) | 1.13564 (14) | 0.0187 (5) | |
| H5A | 0.9937 | 0.8272 | 1.1490 | 0.028* | |
| H5B | 0.9197 | 0.9003 | 1.1718 | 0.028* | |
| H5C | 1.0073 | 0.9469 | 1.1483 | 0.028* | |
| C6 | 0.86661 (16) | 0.99579 (17) | 0.99301 (14) | 0.0124 (4) | |
| C7 | 0.89136 (17) | 1.08943 (18) | 1.03628 (15) | 0.0163 (5) | |
| H7 | 0.9293 | 1.0921 | 1.0967 | 0.020* | |
| C8 | 0.85855 (19) | 1.17985 (18) | 0.98807 (16) | 0.0194 (5) | |
| H8 | 0.8745 | 1.2437 | 1.0157 | 0.023* | |
| C9 | 0.80194 (19) | 1.17314 (18) | 0.89857 (16) | 0.0203 (5) | |
| H9 | 0.7788 | 1.2323 | 0.8650 | 0.024* | |
| C10 | 0.78027 (17) | 1.07659 (18) | 0.85973 (15) | 0.0181 (5) | |
| H10 | 0.7424 | 1.0722 | 0.7994 | 0.022* | |
| N1 | 0.81153 (14) | 0.98950 (15) | 0.90538 (12) | 0.0140 (4) | |
| N2 | 0.86837 (14) | 0.81687 (14) | 0.98712 (12) | 0.0127 (4) | |
| N3 | 0.88998 (14) | 0.71758 (14) | 1.01651 (13) | 0.0144 (4) | |
| N4 | 0.91703 (14) | 0.51964 (14) | 1.06044 (12) | 0.0161 (4) | |
| O1 | 0.82753 (14) | 0.67055 (12) | 0.87067 (10) | 0.0213 (4) | |
| O2 | 0.92836 (13) | 0.41557 (13) | 1.08008 (11) | 0.0211 (4) | |
| Cl2 | 0.88241 (4) | 0.85938 (4) | 0.75864 (3) | 0.01726 (13) | |
| Cl3 | 0.63307 (4) | 0.81753 (5) | 0.78506 (4) | 0.01874 (13) | |
| Zn1 | 0.793127 (19) | 0.838156 (19) | 0.848619 (16) | 0.01239 (8) | |
| H1 | 0.954 (3) | 0.413 (3) | 1.139 (2) | 0.048 (10)* | |
| O1W | 0.0000 | 0.34448 (19) | 0.2500 | 0.0206 (5) | |
| H1W | 0.045 (2) | 0.308 (3) | 0.250 (2) | 0.048 (11)* | |
| H2 | 0.905 (2) | 0.699 (2) | 1.068 (2) | 0.024 (8)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0471 (17) | 0.0123 (12) | 0.0180 (12) | −0.0014 (11) | −0.0052 (11) | −0.0005 (9) |
| C2 | 0.0178 (11) | 0.0114 (11) | 0.0162 (11) | −0.0004 (9) | 0.0045 (9) | 0.0004 (8) |
| C3 | 0.0163 (11) | 0.0135 (11) | 0.0150 (10) | −0.0008 (9) | 0.0054 (9) | −0.0011 (8) |
| C4 | 0.0134 (11) | 0.0124 (11) | 0.0129 (10) | −0.0022 (8) | 0.0054 (8) | −0.0001 (8) |
| C5 | 0.0235 (13) | 0.0145 (12) | 0.0138 (10) | −0.0020 (9) | 0.0005 (9) | 0.0002 (9) |
| C6 | 0.0122 (10) | 0.0130 (11) | 0.0135 (10) | 0.0006 (8) | 0.0063 (8) | 0.0016 (8) |
| C7 | 0.0181 (12) | 0.0163 (11) | 0.0141 (10) | −0.0019 (9) | 0.0050 (9) | −0.0016 (8) |
| C8 | 0.0256 (13) | 0.0108 (11) | 0.0220 (12) | −0.0017 (9) | 0.0086 (10) | −0.0027 (9) |
| C9 | 0.0243 (13) | 0.0140 (12) | 0.0211 (12) | 0.0019 (9) | 0.0058 (10) | 0.0033 (9) |
| C10 | 0.0196 (12) | 0.0179 (12) | 0.0152 (11) | 0.0031 (9) | 0.0039 (9) | 0.0019 (9) |
| N1 | 0.0153 (9) | 0.0132 (9) | 0.0130 (9) | −0.0003 (7) | 0.0042 (7) | −0.0008 (7) |
| N2 | 0.0150 (9) | 0.0103 (9) | 0.0130 (9) | 0.0011 (7) | 0.0051 (7) | 0.0016 (7) |
| N3 | 0.0202 (10) | 0.0115 (9) | 0.0097 (9) | −0.0001 (8) | 0.0028 (8) | 0.0021 (7) |
| N4 | 0.0208 (10) | 0.0103 (9) | 0.0169 (9) | 0.0011 (8) | 0.0062 (8) | 0.0019 (7) |
| O1 | 0.0333 (10) | 0.0136 (8) | 0.0122 (7) | 0.0026 (7) | 0.0015 (7) | 0.0002 (6) |
| O2 | 0.0322 (10) | 0.0126 (8) | 0.0164 (8) | 0.0014 (7) | 0.0054 (7) | 0.0043 (6) |
| Cl2 | 0.0152 (3) | 0.0210 (3) | 0.0172 (3) | −0.0004 (2) | 0.0076 (2) | −0.0007 (2) |
| Cl3 | 0.0154 (3) | 0.0255 (3) | 0.0158 (3) | −0.0057 (2) | 0.0059 (2) | −0.0026 (2) |
| Zn1 | 0.01436 (14) | 0.01195 (13) | 0.01015 (12) | −0.00066 (10) | 0.00323 (9) | −0.00064 (9) |
| O1W | 0.0272 (14) | 0.0137 (12) | 0.0202 (12) | 0.000 | 0.0070 (11) | 0.000 |
Geometric parameters (Å, °)
| C1—C2 | 1.495 (3) | C7—H7 | 0.9300 |
| C1—H1A | 0.9600 | C8—C9 | 1.381 (3) |
| C1—H1B | 0.9600 | C8—H8 | 0.9300 |
| C1—H1C | 0.9600 | C9—C10 | 1.386 (3) |
| C2—N4 | 1.275 (3) | C9—H9 | 0.9300 |
| C2—C3 | 1.482 (3) | C10—N1 | 1.337 (3) |
| C3—O1 | 1.233 (3) | C10—H10 | 0.9300 |
| C3—N3 | 1.364 (3) | Zn1—N1 | 2.1409 (19) |
| C4—N2 | 1.288 (3) | N2—N3 | 1.371 (3) |
| C4—C5 | 1.492 (3) | Zn1—N2 | 2.1142 (18) |
| C4—C6 | 1.492 (3) | N3—H2 | 0.80 (3) |
| C5—H5A | 0.9600 | N4—O2 | 1.384 (2) |
| C5—H5B | 0.9600 | Zn1—O1 | 2.2348 (16) |
| C5—H5C | 0.9600 | O2—H1 | 0.88 (3) |
| C6—N1 | 1.351 (3) | Zn1—Cl2 | 2.2513 (6) |
| C6—C7 | 1.383 (3) | Zn1—Cl3 | 2.2195 (6) |
| C7—C8 | 1.394 (3) | O1W—H1W | 0.81 (3) |
| C2—C1—H1A | 109.5 | C7—C8—H8 | 120.5 |
| C2—C1—H1B | 109.5 | C8—C9—C10 | 118.7 (2) |
| H1A—C1—H1B | 109.5 | C8—C9—H9 | 120.6 |
| C2—C1—H1C | 109.5 | C10—C9—H9 | 120.6 |
| H1A—C1—H1C | 109.5 | N1—C10—C9 | 122.7 (2) |
| H1B—C1—H1C | 109.5 | N1—C10—H10 | 118.6 |
| N4—C2—C3 | 115.0 (2) | C9—C10—H10 | 118.6 |
| N4—C2—C1 | 126.8 (2) | C10—N1—C6 | 118.69 (19) |
| C3—C2—C1 | 118.27 (19) | C10—N1—Zn1 | 125.33 (15) |
| O1—C3—N3 | 121.2 (2) | C6—N1—Zn1 | 115.73 (15) |
| O1—C3—C2 | 120.9 (2) | C4—N2—N3 | 122.44 (18) |
| N3—C3—C2 | 117.88 (19) | C4—N2—Zn1 | 120.30 (15) |
| N2—C4—C5 | 126.3 (2) | N3—N2—Zn1 | 117.03 (13) |
| N2—C4—C6 | 113.42 (18) | C3—N3—N2 | 114.28 (18) |
| C5—C4—C6 | 120.29 (19) | C3—N3—H2 | 119 (2) |
| C4—C5—H5A | 109.5 | N2—N3—H2 | 125 (2) |
| C4—C5—H5B | 109.5 | C2—N4—O2 | 111.77 (18) |
| H5A—C5—H5B | 109.5 | C3—O1—Zn1 | 114.29 (14) |
| C4—C5—H5C | 109.5 | N4—O2—H1 | 104 (2) |
| H5A—C5—H5C | 109.5 | N2—Zn1—N1 | 74.71 (7) |
| H5B—C5—H5C | 109.5 | N2—Zn1—Cl3 | 123.36 (5) |
| N1—C6—C7 | 121.8 (2) | N1—Zn1—Cl3 | 105.30 (5) |
| N1—C6—C4 | 115.30 (19) | N2—Zn1—O1 | 72.57 (6) |
| C7—C6—C4 | 122.86 (19) | N1—Zn1—O1 | 147.07 (6) |
| C6—C7—C8 | 119.1 (2) | Cl3—Zn1—O1 | 95.66 (5) |
| C6—C7—H7 | 120.5 | N2—Zn1—Cl2 | 117.91 (5) |
| C8—C7—H7 | 120.5 | N1—Zn1—Cl2 | 97.91 (5) |
| C9—C8—C7 | 119.0 (2) | Cl3—Zn1—Cl2 | 118.08 (2) |
| C9—C8—H8 | 120.5 | O1—Zn1—Cl2 | 94.10 (5) |
| N4—C2—C3—O1 | 178.0 (2) | Zn1—N2—N3—C3 | 2.7 (2) |
| C1—C2—C3—O1 | −2.0 (4) | C3—C2—N4—O2 | −179.79 (19) |
| N4—C2—C3—N3 | −2.1 (3) | C1—C2—N4—O2 | 0.2 (4) |
| C1—C2—C3—N3 | 177.9 (2) | N3—C3—O1—Zn1 | −8.3 (3) |
| N2—C4—C6—N1 | 0.0 (3) | C2—C3—O1—Zn1 | 171.56 (16) |
| C5—C4—C6—N1 | 179.6 (2) | C4—N2—Zn1—N1 | −6.55 (17) |
| N2—C4—C6—C7 | −179.8 (2) | N3—N2—Zn1—N1 | 178.85 (17) |
| C5—C4—C6—C7 | −0.1 (3) | C4—N2—Zn1—Cl3 | −105.03 (17) |
| N1—C6—C7—C8 | 0.0 (3) | N3—N2—Zn1—Cl3 | 80.36 (16) |
| C4—C6—C7—C8 | 179.7 (2) | C4—N2—Zn1—O1 | 169.73 (19) |
| C6—C7—C8—C9 | 0.3 (4) | N3—N2—Zn1—O1 | −4.88 (15) |
| C7—C8—C9—C10 | −0.4 (4) | C4—N2—Zn1—Cl2 | 84.41 (17) |
| C8—C9—C10—N1 | 0.3 (4) | N3—N2—Zn1—Cl2 | −90.20 (15) |
| C9—C10—N1—C6 | −0.1 (3) | C10—N1—Zn1—N2 | −179.8 (2) |
| C9—C10—N1—Zn1 | −173.99 (18) | C6—N1—Zn1—N2 | 6.09 (15) |
| C7—C6—N1—C10 | −0.1 (3) | C10—N1—Zn1—Cl3 | −58.74 (19) |
| C4—C6—N1—C10 | −179.9 (2) | C6—N1—Zn1—Cl3 | 127.17 (14) |
| C7—C6—N1—Zn1 | 174.42 (17) | C10—N1—Zn1—O1 | 173.63 (16) |
| C4—C6—N1—Zn1 | −5.4 (2) | C6—N1—Zn1—O1 | −0.5 (2) |
| C5—C4—N2—N3 | 0.4 (3) | C10—N1—Zn1—Cl2 | 63.31 (19) |
| C6—C4—N2—N3 | 179.97 (19) | C6—N1—Zn1—Cl2 | −110.78 (15) |
| C5—C4—N2—Zn1 | −173.95 (18) | C3—O1—Zn1—N2 | 6.96 (17) |
| C6—C4—N2—Zn1 | 5.7 (3) | C3—O1—Zn1—N1 | 13.6 (2) |
| O1—C3—N3—N2 | 4.1 (3) | C3—O1—Zn1—Cl3 | −116.27 (17) |
| C2—C3—N3—N2 | −175.80 (19) | C3—O1—Zn1—Cl2 | 124.96 (17) |
| C4—N2—N3—C3 | −171.8 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H2···Cl3i | 0.80 (3) | 2.59 (3) | 3.310 (2) | 150 (3) |
| O1W—H1W···Cl2ii | 0.81 (3) | 2.44 (3) | 3.182 (2) | 153 (3) |
| O2—H1···O1Wiii | 0.88 (3) | 1.89 (4) | 2.7015 (19) | 153 (3) |
Symmetry codes: (i) −x+3/2, −y+3/2, −z+2; (ii) −x+1, −y+1, −z+1; (iii) x+1, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HY2108).
References
- Bruker (1999). SAINT Version 6.02. Bruker AXS Inc., Madison, Wisconsin, USA.
- Canary, J. W., Allen, C. S., Castagnetto, J. M., Chiu, Y. H., Toscano, P. J. & Wang, Y. H. (1998). Inorg. Chem.37, 6255–6262.
- Comba, P., Kerscher, M., Merz, M., Muller, V., Pritzkow, H., Remenyi, R., Schiek, W. & Xiong, Y. (2002). Chem. Eur. J.8, 5750–5760. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Fritsky, I. O., Kozłowski, H., Sadler, P. J., Yefetova, O. P., Świątek-Kozłowska, J., Kalibabchuk, V. A. & Glowiak, T. (1998). J. Chem. Soc. Dalton Trans. pp. 3269–3274.
- Kasuga, N. C., Sekino, K., Ishikawa, M., Honda, A., Yokoyama, M., Nakano, S., Shimada, N., Koumo, S. & Nomiya, K. (2003). J. Inorg. Biochem.96, 298–310. [DOI] [PubMed]
- Kuma (1999). KM-4 CCD Software Version 1.61. Kuma Diffraction, Wrocław, Poland.
- Panosyan, F. B., Lough, A. J. & Chin, J. (2003). Acta Cryst. E59, m827–m829.
- Rodriuez-Argüelles, M. C., Ferrari, M. B., Fava, G. G., Pelizzi, C., Tarasconi, P., Albertini, R., Dall’Aglio, P. P., Lunghi, P. & Pinelli, S. (1995). J. Inorg. Biochem.58, 157–175. [DOI] [PubMed]
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
- Sousa, G. F. de & Deflon, V. M. (2003). Transition Met. Chem.28, 74–78.
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680706535X/hy2108sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680706535X/hy2108Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report