Abstract
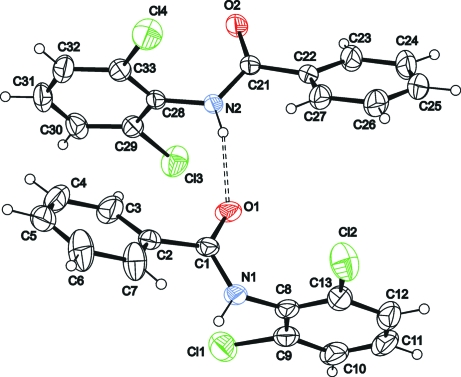
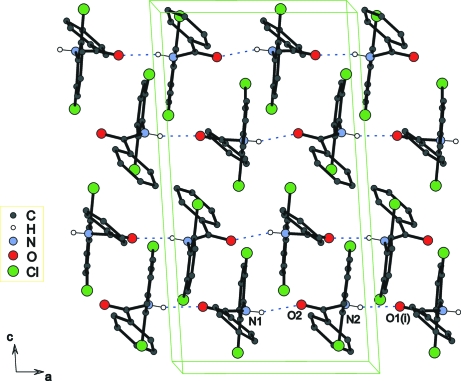
The conformation of the N—H and C=O bonds in the structure of the title compound (N26DCPBA), C13H9Cl2NO, are anti to each other, similar to that observed in N-phenylbenzamide (NPBA), N-(2-chlorophenyl)benzamide (N2CPBA), N-(2,3-dichlorophenyl)benzamide (N23DCPBA) and other benzanilides. The asymmetric unit of N26DCPBA contains two molecules. The bond parameters in N26DCPBA are similar to those in NPBA, N2CPBA, N23DCPBA and other benzanilides. The amide group, –NHCO–, makes a dihedral angle of 30.8 (1)° with the benzoyl ring in the first molecule and 35.1 (2)° in the second molecule of the asymmetric unit. The dihedral angle between the two benzene rings (benzoyl and aniline) is 56.8 (1)° in the first molecule and 59.1 (1)° in the second molecule. N—H⋯O hydrogen bonds give rise to infinite chains running along the a axis of the crystal structure.
Related literature
For related literature, see: Gowda et al. (2003 ▶, 2007a
▶,b
▶,c
▶, 2008 ▶).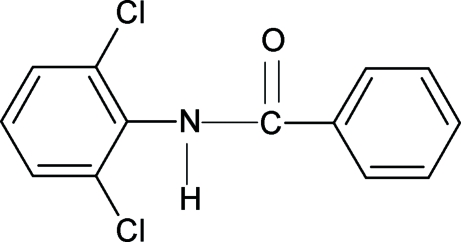
Experimental
Crystal data
C13H9Cl2NO
M r = 266.11
Monoclinic,
a = 10.0431 (2) Å
b = 13.7150 (3) Å
c = 18.4585 (4) Å
β = 93.623 (2)°
V = 2537.41 (9) Å3
Z = 8
Mo Kα radiation
μ = 0.49 mm−1
T = 295 (2) K
0.26 × 0.24 × 0.21 mm
Data collection
Oxford Diffraction Xcalibur System diffractometer
Absorption correction: analytical [(Oxford Diffraction, 2007 ▶); analytical numeric absorption correction using a multifaceted crystal model (Clark & Reid, 1995 ▶)] T min = 0.882, T max = 0.904
54956 measured reflections
4956 independent reflections
3810 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.043
wR(F 2) = 0.127
S = 1.10
4956 reflections
314 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.40 e Å−3
Δρmin = −0.35 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2007 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2007 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 2002 ▶); software used to prepare material for publication: SHELXL97], PLATON (Spek, 2003 ▶) and WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680800305X/om2209sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800305X/om2209Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O2 | 0.860 (19) | 2.09 (2) | 2.9035 (18) | 158 (2) |
| N2—H2N⋯O1i | 0.843 (18) | 2.060 (19) | 2.8831 (18) | 165.1 (19) |
Symmetry code: (i) 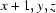 .
.
Acknowledgments
MT and JK thank the Grant Agency of the Slovak Republic (Grant No. VEGA 1/0817/08) and the Structural Funds, Interreg IIIA, for financial support in purchasing the diffractometer.
supplementary crystallographic information
Comment
In the present work, the structure of N-(2,6-dichlorophenyl)-benzamide (N26DCPBA) has been determined to explore the effect of substituents on the structure of N-aromatic amides (Gowda et al., 2003,2007a,b,c,2008). The conformation of the N—H and C=O bonds in the structure of N26DCPBA (Fig.1) are anti to each other, similar to that observed in N-(phenyl)-benzamide (NPBA)(Gowda et al., 2003), N-(2-chlorophenyl)-benzamide (N2CPBA), N-(2,3-dichlorophenyl)-benzamide (N23DCPBA) and other benzanilides(Gowda et al., 2007a,b,c, 2008). The title compound crystallizes in the space group P21/c, with two molecules in the asymmetric unit. The bond parameters in N26DCPBA are similar to those in NPBA, N2CPBA, N23DCPBA and other benzanilides. The amide group –NHCO– has the dihedral angle of 30.8 (1)° with the benzoyl ring in the first molecule and 35.1 (2)° in the second molecule of the asymmetric unit. The dihedral angle between the two benzene rings (benzoyl and aniline) is 56.8 (1)° in the first molecule and 59.1 (1)° in the second molecule. One-dimensional chains of N26DCPBA along the base vector [1 0 0] formed by hydrogen bonds N1–H1N···O2(i) and N2–H2N···O1 (Table 1) as viewed down the b axis is shown in Fig.2. Symmetry code (i): x + 1,y,z.
Experimental
The title compound was prepared according to the literature method (Gowda et al., 2003). The purity of the compound was checked by determining its melting point. It was characterized by recording its infrared and NMR spectra. Single crystals of the title compound were obtained from an ethanolic solution and used for X-ray diffraction studies at room temperature.
Refinement
H atoms bonded to C atoms were placed in geometrically calculated positions and subsequently treated as riding with C—H bond distance 0.93 Å. H(N) atoms were visible in the difference map. In the refinement the N—H distance was restrained to 0.86 (4) Å. The Uiso(H) values were set at 1.2 Ueq(C,N).
Figures
Fig. 1.
Molecular structure of the title compound showing the atom labelling scheme. Numbers of the C atoms in the second molecule are increased by 20. Displacement ellipsoids are drawn at the 30% probability level. The hydrogen bond N2—H2N···O1 is shown as a dashed line.
Fig. 2.
Crystal structure of the title compound viewed down the b axis. One-dimensional chains along the base vector [1 0 0] are formed by hydrogen bonds N1–H1N···O2 and N2–H2N···O1(i). Symmetry code (i): x + 1,y,z. H atoms not involved in hydrogen bonding are omitted.
Crystal data
| C13H9Cl2NO | F000 = 1088 |
| Mr = 266.11 | Dx = 1.393 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 24495 reflections |
| a = 10.0431 (2) Å | θ = 3.1–29.5º |
| b = 13.7150 (3) Å | µ = 0.49 mm−1 |
| c = 18.4585 (4) Å | T = 295 (2) K |
| β = 93.623 (2)º | Block, colorless |
| V = 2537.41 (9) Å3 | 0.26 × 0.24 × 0.21 mm |
| Z = 8 |
Data collection
| Oxford Diffraction Xcalibur System diffractometer | 4956 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 3810 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.023 |
| Detector resolution: 10.434 pixels mm-1 | θmax = 26.0º |
| T = 295(2) K | θmin = 5.1º |
| φ scans, and ω scans with κ offsets | h = −12→12 |
| Absorption correction: analytical[(Oxford Diffraction, 2007); analytical numeric absorption correction using a multifaceted crystal model (Clark & Reid, 1995)] | k = −16→16 |
| Tmin = 0.883, Tmax = 0.904 | l = −22→22 |
| 54956 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.127 | w = 1/[σ2(Fo2) + (0.0646P)2 + 0.4979P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max = 0.001 |
| 4956 reflections | Δρmax = 0.40 e Å−3 |
| 314 parameters | Δρmin = −0.35 e Å−3 |
| 2 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0083 (11) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.29163 (15) | 0.88930 (13) | 0.13557 (10) | 0.0468 (4) | |
| C2 | 0.31340 (17) | 0.99515 (13) | 0.12131 (10) | 0.0496 (4) | |
| C3 | 0.2344 (2) | 1.06295 (18) | 0.1520 (2) | 0.0957 (10) | |
| H3 | 0.1718 | 1.0433 | 0.184 | 0.115* | |
| C4 | 0.2478 (3) | 1.1613 (2) | 0.1353 (2) | 0.1205 (13) | |
| H4 | 0.1942 | 1.2072 | 0.1564 | 0.145* | |
| C5 | 0.3392 (3) | 1.19053 (19) | 0.08819 (19) | 0.0957 (10) | |
| H5 | 0.3447 | 1.2559 | 0.0752 | 0.115* | |
| C6 | 0.4206 (4) | 1.1252 (2) | 0.06070 (14) | 0.0979 (9) | |
| H6 | 0.4856 | 1.1456 | 0.0304 | 0.118* | |
| C7 | 0.4090 (3) | 1.02747 (17) | 0.07699 (13) | 0.0798 (7) | |
| H7 | 0.4667 | 0.9828 | 0.0577 | 0.096* | |
| C8 | 0.39430 (16) | 0.73180 (13) | 0.15565 (12) | 0.0555 (5) | |
| C9 | 0.41165 (19) | 0.70599 (15) | 0.22837 (14) | 0.0661 (6) | |
| C10 | 0.4153 (2) | 0.60990 (18) | 0.25095 (17) | 0.0805 (7) | |
| H10 | 0.4284 | 0.5945 | 0.3 | 0.097* | |
| C11 | 0.3993 (2) | 0.53776 (18) | 0.19996 (19) | 0.0853 (8) | |
| H11 | 0.4011 | 0.4729 | 0.2146 | 0.102* | |
| C12 | 0.3805 (2) | 0.55993 (17) | 0.12743 (18) | 0.0822 (8) | |
| H12 | 0.3702 | 0.5102 | 0.0933 | 0.099* | |
| C13 | 0.3771 (2) | 0.65690 (16) | 0.10524 (14) | 0.0664 (6) | |
| N1 | 0.39953 (14) | 0.83121 (11) | 0.13436 (10) | 0.0535 (4) | |
| H1N | 0.4772 (19) | 0.8565 (16) | 0.1310 (11) | 0.064* | |
| O1 | 0.18181 (11) | 0.85646 (10) | 0.14687 (9) | 0.0635 (4) | |
| Cl1 | 0.42425 (7) | 0.79839 (5) | 0.29312 (4) | 0.0924 (2) | |
| Cl2 | 0.35242 (9) | 0.68484 (5) | 0.01367 (4) | 0.1030 (3) | |
| C21 | 0.79317 (15) | 0.84885 (12) | 0.13788 (10) | 0.0457 (4) | |
| C22 | 0.81608 (17) | 0.75379 (13) | 0.10084 (11) | 0.0505 (4) | |
| C23 | 0.7373 (2) | 0.67477 (16) | 0.11555 (17) | 0.0803 (8) | |
| H23 | 0.6724 | 0.6804 | 0.1491 | 0.096* | |
| C24 | 0.7548 (3) | 0.58684 (18) | 0.0802 (2) | 0.1034 (11) | |
| H24 | 0.7023 | 0.5334 | 0.0906 | 0.124* | |
| C25 | 0.8487 (3) | 0.57822 (19) | 0.03020 (18) | 0.0922 (9) | |
| H25 | 0.8588 | 0.5193 | 0.0062 | 0.111* | |
| C26 | 0.9277 (3) | 0.65546 (18) | 0.01526 (13) | 0.0761 (7) | |
| H26 | 0.9919 | 0.6491 | −0.0186 | 0.091* | |
| C27 | 0.9124 (2) | 0.74372 (15) | 0.05076 (11) | 0.0588 (5) | |
| H27 | 0.9669 | 0.7963 | 0.0409 | 0.071* | |
| C28 | 0.89884 (14) | 0.99789 (11) | 0.18305 (9) | 0.0412 (4) | |
| C29 | 0.92285 (17) | 1.01514 (13) | 0.25670 (10) | 0.0501 (4) | |
| C30 | 0.9290 (2) | 1.10850 (16) | 0.28488 (12) | 0.0622 (5) | |
| H30 | 0.9468 | 1.1184 | 0.3344 | 0.075* | |
| C31 | 0.9085 (2) | 1.18598 (15) | 0.23909 (13) | 0.0670 (6) | |
| H31 | 0.9121 | 1.249 | 0.2578 | 0.08* | |
| C32 | 0.8826 (2) | 1.17230 (14) | 0.16579 (13) | 0.0653 (5) | |
| H32 | 0.8683 | 1.2255 | 0.1351 | 0.078* | |
| C33 | 0.87794 (18) | 1.07878 (13) | 0.13833 (10) | 0.0511 (4) | |
| N2 | 0.90279 (14) | 0.90255 (10) | 0.15368 (8) | 0.0450 (4) | |
| H2N | 0.9794 (18) | 0.8817 (14) | 0.1457 (10) | 0.054* | |
| O2 | 0.68208 (11) | 0.87638 (10) | 0.15285 (9) | 0.0677 (4) | |
| Cl3 | 0.94534 (8) | 0.91674 (5) | 0.31518 (3) | 0.0865 (2) | |
| Cl4 | 0.84784 (8) | 1.06116 (5) | 0.04585 (3) | 0.0869 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0336 (8) | 0.0493 (10) | 0.0569 (10) | −0.0039 (7) | −0.0009 (7) | 0.0105 (8) |
| C2 | 0.0427 (9) | 0.0468 (10) | 0.0576 (10) | −0.0041 (8) | −0.0106 (8) | 0.0080 (8) |
| C3 | 0.0452 (11) | 0.0592 (14) | 0.185 (3) | 0.0027 (10) | 0.0251 (15) | 0.0048 (16) |
| C4 | 0.0598 (15) | 0.0540 (15) | 0.247 (4) | 0.0129 (12) | 0.004 (2) | −0.001 (2) |
| C5 | 0.0892 (19) | 0.0524 (14) | 0.139 (3) | −0.0148 (13) | −0.0468 (18) | 0.0265 (16) |
| C6 | 0.163 (3) | 0.0603 (15) | 0.0716 (15) | −0.0325 (18) | 0.0185 (17) | 0.0136 (12) |
| C7 | 0.125 (2) | 0.0535 (12) | 0.0637 (13) | −0.0217 (13) | 0.0321 (13) | 0.0015 (10) |
| C8 | 0.0290 (8) | 0.0467 (10) | 0.0905 (15) | −0.0013 (7) | 0.0021 (8) | 0.0144 (10) |
| C9 | 0.0424 (10) | 0.0583 (12) | 0.0961 (16) | −0.0013 (9) | −0.0076 (10) | 0.0177 (11) |
| C10 | 0.0614 (13) | 0.0643 (15) | 0.114 (2) | 0.0050 (11) | −0.0110 (13) | 0.0311 (14) |
| C11 | 0.0612 (13) | 0.0495 (13) | 0.145 (3) | 0.0099 (10) | 0.0081 (14) | 0.0293 (15) |
| C12 | 0.0664 (14) | 0.0485 (12) | 0.133 (2) | −0.0020 (10) | 0.0192 (15) | −0.0005 (14) |
| C13 | 0.0478 (10) | 0.0548 (12) | 0.0976 (16) | −0.0053 (9) | 0.0116 (10) | 0.0053 (11) |
| N1 | 0.0312 (7) | 0.0452 (8) | 0.0843 (11) | −0.0067 (6) | 0.0043 (7) | 0.0129 (8) |
| O1 | 0.0320 (6) | 0.0573 (8) | 0.1014 (11) | −0.0022 (5) | 0.0056 (6) | 0.0213 (7) |
| Cl1 | 0.1026 (5) | 0.0785 (4) | 0.0927 (5) | −0.0146 (3) | −0.0221 (4) | 0.0098 (3) |
| Cl2 | 0.1321 (6) | 0.0836 (5) | 0.0940 (5) | −0.0293 (4) | 0.0131 (4) | −0.0028 (4) |
| C21 | 0.0333 (8) | 0.0438 (9) | 0.0596 (10) | 0.0012 (7) | 0.0003 (7) | −0.0075 (8) |
| C22 | 0.0382 (9) | 0.0435 (9) | 0.0679 (12) | 0.0031 (7) | −0.0111 (8) | −0.0117 (8) |
| C23 | 0.0445 (11) | 0.0524 (12) | 0.145 (2) | −0.0056 (9) | 0.0107 (12) | −0.0206 (13) |
| C24 | 0.0651 (15) | 0.0515 (14) | 0.192 (3) | −0.0098 (11) | −0.0021 (18) | −0.0332 (17) |
| C25 | 0.0757 (16) | 0.0631 (15) | 0.134 (2) | 0.0173 (13) | −0.0220 (16) | −0.0463 (15) |
| C26 | 0.0871 (16) | 0.0718 (15) | 0.0681 (14) | 0.0202 (13) | −0.0060 (12) | −0.0257 (11) |
| C27 | 0.0664 (12) | 0.0531 (11) | 0.0563 (11) | 0.0077 (9) | −0.0004 (9) | −0.0102 (9) |
| C28 | 0.0288 (7) | 0.0404 (9) | 0.0547 (10) | −0.0010 (6) | 0.0049 (7) | −0.0089 (7) |
| C29 | 0.0464 (9) | 0.0506 (10) | 0.0536 (10) | 0.0008 (8) | 0.0050 (8) | −0.0060 (8) |
| C30 | 0.0624 (12) | 0.0631 (13) | 0.0613 (12) | −0.0083 (10) | 0.0060 (10) | −0.0219 (10) |
| C31 | 0.0693 (13) | 0.0448 (11) | 0.0887 (16) | −0.0085 (9) | 0.0194 (11) | −0.0236 (11) |
| C32 | 0.0701 (13) | 0.0409 (10) | 0.0862 (16) | −0.0001 (9) | 0.0148 (11) | 0.0002 (10) |
| C33 | 0.0468 (9) | 0.0513 (10) | 0.0554 (10) | 0.0013 (8) | 0.0054 (8) | −0.0051 (8) |
| N2 | 0.0290 (6) | 0.0427 (8) | 0.0634 (9) | 0.0026 (6) | 0.0029 (6) | −0.0147 (6) |
| O2 | 0.0316 (6) | 0.0583 (8) | 0.1137 (12) | −0.0017 (6) | 0.0088 (7) | −0.0256 (8) |
| Cl3 | 0.1228 (5) | 0.0719 (4) | 0.0638 (4) | 0.0113 (3) | −0.0019 (3) | 0.0093 (3) |
| Cl4 | 0.1163 (5) | 0.0878 (4) | 0.0553 (3) | 0.0093 (4) | −0.0057 (3) | 0.0019 (3) |
Geometric parameters (Å, °)
| C1—O1 | 1.2214 (19) | C21—O2 | 1.2258 (19) |
| C1—N1 | 1.346 (2) | C21—N2 | 1.341 (2) |
| C1—C2 | 1.494 (2) | C21—C22 | 1.497 (2) |
| C2—C3 | 1.368 (3) | C22—C23 | 1.379 (3) |
| C2—C7 | 1.373 (3) | C22—C27 | 1.386 (3) |
| C3—C4 | 1.391 (4) | C23—C24 | 1.388 (3) |
| C3—H3 | 0.93 | C23—H23 | 0.93 |
| C4—C5 | 1.364 (5) | C24—C25 | 1.366 (4) |
| C4—H4 | 0.93 | C24—H24 | 0.93 |
| C5—C6 | 1.335 (4) | C25—C26 | 1.362 (4) |
| C5—H5 | 0.93 | C25—H25 | 0.93 |
| C6—C7 | 1.380 (3) | C26—C27 | 1.390 (3) |
| C6—H6 | 0.93 | C26—H26 | 0.93 |
| C7—H7 | 0.93 | C27—H27 | 0.93 |
| C8—C9 | 1.389 (3) | C28—C29 | 1.386 (2) |
| C8—C13 | 1.389 (3) | C28—C33 | 1.391 (3) |
| C8—N1 | 1.421 (2) | C28—N2 | 1.417 (2) |
| C9—C10 | 1.382 (3) | C29—C30 | 1.382 (3) |
| C9—Cl1 | 1.741 (3) | C29—Cl3 | 1.734 (2) |
| C10—C11 | 1.368 (4) | C30—C31 | 1.365 (3) |
| C10—H10 | 0.93 | C30—H30 | 0.93 |
| C11—C12 | 1.374 (4) | C31—C32 | 1.374 (3) |
| C11—H11 | 0.93 | C31—H31 | 0.93 |
| C12—C13 | 1.391 (3) | C32—C33 | 1.379 (3) |
| C12—H12 | 0.93 | C32—H32 | 0.93 |
| C13—Cl2 | 1.736 (3) | C33—Cl4 | 1.732 (2) |
| N1—H1N | 0.860 (19) | N2—H2N | 0.843 (18) |
| O1—C1—N1 | 121.41 (16) | O2—C21—N2 | 121.88 (16) |
| O1—C1—C2 | 122.15 (16) | O2—C21—C22 | 122.66 (15) |
| N1—C1—C2 | 116.43 (14) | N2—C21—C22 | 115.46 (14) |
| C3—C2—C7 | 118.2 (2) | C23—C22—C27 | 119.09 (18) |
| C3—C2—C1 | 119.51 (18) | C23—C22—C21 | 119.21 (18) |
| C7—C2—C1 | 122.29 (18) | C27—C22—C21 | 121.68 (16) |
| C2—C3—C4 | 120.1 (3) | C22—C23—C24 | 119.9 (2) |
| C2—C3—H3 | 120 | C22—C23—H23 | 120 |
| C4—C3—H3 | 120 | C24—C23—H23 | 120 |
| C5—C4—C3 | 120.3 (3) | C25—C24—C23 | 120.4 (2) |
| C5—C4—H4 | 119.9 | C25—C24—H24 | 119.8 |
| C3—C4—H4 | 119.9 | C23—C24—H24 | 119.8 |
| C6—C5—C4 | 119.9 (2) | C26—C25—C24 | 120.4 (2) |
| C6—C5—H5 | 120.1 | C26—C25—H25 | 119.8 |
| C4—C5—H5 | 120.1 | C24—C25—H25 | 119.8 |
| C5—C6—C7 | 120.5 (3) | C25—C26—C27 | 119.9 (2) |
| C5—C6—H6 | 119.8 | C25—C26—H26 | 120.1 |
| C7—C6—H6 | 119.8 | C27—C26—H26 | 120.1 |
| C2—C7—C6 | 121.0 (3) | C22—C27—C26 | 120.3 (2) |
| C2—C7—H7 | 119.5 | C22—C27—H27 | 119.9 |
| C6—C7—H7 | 119.5 | C26—C27—H27 | 119.9 |
| C9—C8—C13 | 117.49 (18) | C29—C28—C33 | 117.15 (15) |
| C9—C8—N1 | 120.5 (2) | C29—C28—N2 | 121.70 (16) |
| C13—C8—N1 | 122.0 (2) | C33—C28—N2 | 121.06 (16) |
| C10—C9—C8 | 122.3 (2) | C30—C29—C28 | 121.88 (18) |
| C10—C9—Cl1 | 119.2 (2) | C30—C29—Cl3 | 119.07 (15) |
| C8—C9—Cl1 | 118.51 (16) | C28—C29—Cl3 | 119.05 (13) |
| C11—C10—C9 | 118.8 (3) | C31—C30—C29 | 119.13 (19) |
| C11—C10—H10 | 120.6 | C31—C30—H30 | 120.4 |
| C9—C10—H10 | 120.6 | C29—C30—H30 | 120.4 |
| C10—C11—C12 | 120.9 (2) | C30—C31—C32 | 120.99 (18) |
| C10—C11—H11 | 119.6 | C30—C31—H31 | 119.5 |
| C12—C11—H11 | 119.6 | C32—C31—H31 | 119.5 |
| C11—C12—C13 | 119.8 (3) | C31—C32—C33 | 119.3 (2) |
| C11—C12—H12 | 120.1 | C31—C32—H32 | 120.4 |
| C13—C12—H12 | 120.1 | C33—C32—H32 | 120.4 |
| C8—C13—C12 | 120.7 (2) | C32—C33—C28 | 121.57 (18) |
| C8—C13—Cl2 | 119.53 (16) | C32—C33—Cl4 | 119.47 (16) |
| C12—C13—Cl2 | 119.8 (2) | C28—C33—Cl4 | 118.95 (14) |
| C1—N1—C8 | 121.29 (14) | C21—N2—C28 | 123.19 (14) |
| C1—N1—H1N | 119.9 (14) | C21—N2—H2N | 121.5 (13) |
| C8—N1—H1N | 117.2 (15) | C28—N2—H2N | 115.3 (13) |
| O1—C1—C2—C3 | 30.4 (3) | O2—C21—C22—C23 | −34.2 (3) |
| N1—C1—C2—C3 | −150.5 (2) | N2—C21—C22—C23 | 145.9 (2) |
| O1—C1—C2—C7 | −148.3 (2) | O2—C21—C22—C27 | 144.2 (2) |
| N1—C1—C2—C7 | 30.8 (3) | N2—C21—C22—C27 | −35.7 (3) |
| C7—C2—C3—C4 | 3.0 (4) | C27—C22—C23—C24 | −0.2 (4) |
| C1—C2—C3—C4 | −175.7 (3) | C21—C22—C23—C24 | 178.3 (2) |
| C2—C3—C4—C5 | 0.2 (5) | C22—C23—C24—C25 | −0.8 (4) |
| C3—C4—C5—C6 | −3.3 (5) | C23—C24—C25—C26 | 1.0 (5) |
| C4—C5—C6—C7 | 3.0 (5) | C24—C25—C26—C27 | −0.4 (4) |
| C3—C2—C7—C6 | −3.3 (4) | C23—C22—C27—C26 | 0.9 (3) |
| C1—C2—C7—C6 | 175.4 (2) | C21—C22—C27—C26 | −177.54 (18) |
| C5—C6—C7—C2 | 0.4 (4) | C25—C26—C27—C22 | −0.6 (3) |
| C13—C8—C9—C10 | 1.5 (3) | C33—C28—C29—C30 | −1.4 (3) |
| N1—C8—C9—C10 | −176.03 (18) | N2—C28—C29—C30 | 175.06 (16) |
| C13—C8—C9—Cl1 | −176.52 (14) | C33—C28—C29—Cl3 | 178.37 (13) |
| N1—C8—C9—Cl1 | 5.9 (2) | N2—C28—C29—Cl3 | −5.1 (2) |
| C8—C9—C10—C11 | −1.0 (3) | C28—C29—C30—C31 | 1.2 (3) |
| Cl1—C9—C10—C11 | 177.08 (18) | Cl3—C29—C30—C31 | −178.63 (16) |
| C9—C10—C11—C12 | 0.3 (4) | C29—C30—C31—C32 | −0.3 (3) |
| C10—C11—C12—C13 | −0.3 (4) | C30—C31—C32—C33 | −0.3 (3) |
| C9—C8—C13—C12 | −1.5 (3) | C31—C32—C33—C28 | 0.0 (3) |
| N1—C8—C13—C12 | 176.04 (18) | C31—C32—C33—Cl4 | −179.03 (16) |
| C9—C8—C13—Cl2 | 179.02 (14) | C29—C28—C33—C32 | 0.8 (3) |
| N1—C8—C13—Cl2 | −3.4 (2) | N2—C28—C33—C32 | −175.69 (17) |
| C11—C12—C13—C8 | 0.9 (3) | C29—C28—C33—Cl4 | 179.88 (12) |
| C11—C12—C13—Cl2 | −179.60 (17) | N2—C28—C33—Cl4 | 3.4 (2) |
| O1—C1—N1—C8 | −8.5 (3) | O2—C21—N2—C28 | −5.4 (3) |
| C2—C1—N1—C8 | 172.35 (18) | C22—C21—N2—C28 | 174.49 (16) |
| C9—C8—N1—C1 | −84.0 (2) | C29—C28—N2—C21 | 98.8 (2) |
| C13—C8—N1—C1 | 98.5 (2) | C33—C28—N2—C21 | −84.8 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O2 | 0.860 (19) | 2.09 (2) | 2.9035 (18) | 158 (2) |
| N2—H2N···O1i | 0.843 (18) | 2.060 (19) | 2.8831 (18) | 165.1 (19) |
Symmetry codes: (i) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: OM2209).
References
- Brandenburg, K. (2002). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887–897.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Gowda, B. T., Jyothi, K., Paulus, H. & Fuess, H. (2003). Z. Naturforsch. Teil A, 58, 225–230.
- Gowda, B. T., Sowmya, B. P., Kožíšek, J., Tokarčík, M. & Fuess, H. (2007a). Acta Cryst. E63, o2906. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Sowmya, B. P., Tokarčík, M., Kožíšek, J. & Fuess, H. (2007b). Acta Cryst. E63, o3326. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Sowmya, B. P., Tokarčík, M., Kožíšek, J. & Fuess, H. (2007c). Acta Cryst. E63, o3365. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Tokarčík, M., Kožíšek, J., Sowmya, B. P. & Fuess, H. (2008). Acta Cryst. E64, o462. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2007). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Abingdon, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680800305X/om2209sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680800305X/om2209Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report