Abstract
A 14 nt DNA sequence 5′-AGAATGTGGCAAAG-3′ from the zinc finger repeat of the human KRAB zinc finger protein gene ZNF91 bearing the intercalator 2-methoxy,6-chloro,9-amino acridine (Acr) attached to the sugar–phosphate backbone in various positions has been shown to form a specific triple helix (triplex) with a 16 bp hairpin (intramolecular) or a two-stranded (intermolecular) duplex having the identical sequence in the same (parallel) orientation. Intramolecular targets with the identical sequence in the antiparallel orientation and a non-specific target sequence were tested as controls. Apparent binding constants for formation of the triplex were determined by quantitating electrophoretic band shifts. Binding of the single-stranded oligonucleotide probe sequence to the target led to an increase in the fluorescence anisotropy of acridine. The parallel orientation of the two identical sequence segments was confirmed by measurement of fluorescence resonance energy transfer between the acridine on the 5′-end of the probe strand as donor and BODIPY-Texas Red on the 3′-amino group of either strand of the target duplex as acceptor. There was full protection from OsO4-bipyridine modification of thymines in the probe strand of the triplex, in accordance with the presumed triplex formation, which excluded displacement of the homologous duplex strand by the probe–intercalator conjugate. The implications of these results for the existence of protein-independent parallel triplexes are discussed.
INTRODUCTION
Sequence-specific recognition of double-stranded (ds) DNA by oligonucleotide-directed triple helix (triplex) formation has been under intense investigation (1–5). For homopurine/homopyrimidine triplex structures recognition of the third strand occurs through the major groove by formation of Hoogsteen-type or reversed Hoogsteen-type hydrogen bonds between the bases of the oligopyrimidine or oligopurine third strand and the purine-rich strand of the duplex target (for reviews see 6,7). In these triplexes, Py·PuPy or Pu·PuPy, the third strand is oriented antiparallel to the corresponding homologous strand of the target. For this reason we shall refer to these conventional triplex structures as antiparallel.
The current interest in triple-helical DNA arises from its potential in diagnostic and therapeutic clinical applications, such as sequence-specific drug targeting, DNA cleavage, mutagenesis, genome mapping and artificial gene regulation (8–12). The objective of these applications is to form a DNA triplex structure accommodating an arbitrary sequence of all four nucleotides under physiological conditions. Conventional antiparallel triplexes, however, are limited primarily to runs of pyrimidines or purines.
Another type of DNA triplex that accommodates an arbitrary DNA sequence has been proposed as the putative intermediate in homologous recombination (13–15). This DNA triplex is presumed to occur in vivo, albeit transiently, in all organisms undergoing homologous recombination mediated by meiosis-specific proteins in eukaryotes or by RecA in bacteria. A molecular model of such a DNA structure has been calculated and denoted the parallel triplex or recombinant, R-form DNA (14). It accommodates two identical DNA sequences in the third strand and the duplex target, designated by Zhurkin the R (recombinant) and W (Watson) strands, respectively, in a parallel orientation. The complementary C (Crick) strand forms conventional Watson–Crick hydrogen bonds with the W strand. In the proposed model the R strand lies in the major groove of the Watson–Crick duplex, allowing isomorphous bonding schemes for all four possible triplets. Another model for the parallel triplex recombination helix in which the R strand lies in the minor groove has also been proposed (16–18).
It has been demonstrated experimentally that the RecA-induced association of an oligonucleotide probe with a duplex DNA target persists after removal of the protein, provided that the homologous region is sufficiently long (19,20). The first experimental evidence for parallel triplex formation from an arbitrary mixed sequence in a protein-free system was obtained for a model oligonucleotide able to fold back twice into an intrastrand parallel triplex (21). The entropy of mixing was significantly decreased by covalent binding of the strands with non-nucleotide linkers, leading to increased stability of the parallel triplex. Experiments with intrastrand parallel triplexes demonstrated that high ionic strength, divalent cations such as Mg2+ and Mn2+ and spermidine are stabilizing factors (21). In this study we have investigated ligands promoting the formation of a protein-free intermolecular parallel triplex.
Propidium iodide as an external ligand stabilizes antiparallel triplexes (22). Propidium iodide (23), ethidium bromide (EtBr) or one to three molecules of acridine orange per 10 base triplet (24) also stabilize intrastrand parallel helices. In the past decade an approach for effective stabilization of antiparallel triplexes has been developed, based on attachment of the intercalator to the oligonucleotide probe as a covalent adduct (25,26). In fact, a third strand oligonucleotide containing an internally incorporated acridine intercalator was found to stabilize an imperfect target sequence (27).
In this paper we present studies on triplex formation of the 14 base oligonucleotide (ODN) 5′-AGAATGTGGCAAAG-3′ sequence from the human KRAB zinc finger protein cDNA, ZNF91, with a 16 bp double-helical target DNA (either as a hairpin or ds duplex). The sequence comprises all four bases and is able to form a triplex with the 16 bp double-helical target DNA having the identical sequence in parallel orientation, but not with the same sequence in the reverse (antiparallel) configuration. We designed seven ODN probes of the same DNA sequence but with various intercalators attached at different sites and studied their interactions with three different target duplex hairpins (homologous parallel and antiparallel and non-homologous targets) and with the homologous duplex. The studies provide insight into the stability of mixed sequence triplexes as well as information about the influence of intercalating moieties on the equilibrium between triplex formation and third strand duplex invasion.
MATERIALS AND METHODS
Oligonucleotides
The 5′-AGAATGTGGCAAAG-3′ sequence is from the zinc finger repeat of the human cDNA ZNF91 (28). The ODNs were purchased from Eppendorf Biotronik (Berlin, Germany) and purified by HPLC. 2-Methoxy,6-chloro,9-amino acridine (Acr) was incorporated in the synthesis as acridine CPG and/or as acridine phosphoramidite (Glen Research) at the 3′-end or within the sequence, respectively. The commercial acridine CPG and acridine phosphoramidites were attached to the phosphate groups by a flexible pentamethylene linker. Psoralen was incorporated at the 5′-end of the ODN during synthesis as the psoralen phosphoramidite (Glen Research). An additional triethyleneglycol spacer (l) was introduced at the 3′-end for acridine or at the 5′-end for psoralen to lengthen the linker. Amino groups were attached to the 5′-ends or 3′-ends via hexaethyleneglycol spacers (L). The sequences of the ODNs are shown in Figure 1. The methidium intercalation unit (Met) was incorporated in the ODN sequence as described previously (29). Methidium was attached to the sugar–phosphate backbone either with a tetraethyleneglycol linker or with a longer spacer containing two triethyleneglycol spacers connected by a phosphate group.
Figure 1.
ODNs and duplex target sequences used in this study. 5′-Amino groups of the A and B strands were modified by conjugation as described in the text for some experiments.
The hairpin targets were created by insertion of a triethyleneglycol spacer between the complementary sequences (Fig. 1). Throughout this paper we designate the target duplex strand identical to the third strand A and that complementary to the third strand B; the third strand we designate a. Amino 5′-end-labeling of either the A or B strands in the target duplex was effected by reaction with BODIPY-TR-X-succinimidyl ester or using 7-diethylaminocoumarin-3-succinimidyl ester (D-6116 and D-1412, respectively; Molecular Probes, Eugene, OR) at pH 9.5 in NaCO3 buffer. All labeled ODNs were purified by HPLC on a Waters Delta Pak 5µ C18-300 RPC column.
Aliquots of concentrated stock solutions were diluted in water and the concentrations were determined as described elsewhere (30). Corrections were made for the contribution of intercalator conjugates (BODIPY-Texas Red or coumarin) to the absorption at 260 nm in the given ODNs.
Standard conditions for triplex formation
Except where indicated, each sample contained equal concentrations of the single-stranded (ss) intercalator conjugate and increasing concentrations of target. Target hairpin ODNs in 10 mM Tris–HCl, pH 8.0, were heated to 95°C for 3 min, followed by cooling on ice for 5–10 min. ss intercalator–ODN and 10 mM MgCl2 were added to each target and the samples incubated at room temperature for 1 h, followed by 4°C for 2 or 16 h to reach equilibrium. (No increase in the amount of triplex formation could be detected after the longer incubation periods.) Target double-helical ODNs were prepared by mixing equimolar quantities of A and B strands in 0.1 M NaCl, 10 mM Tris–HCl, pH 8.0, heating to 95°C, followed by slow annealing to room temperature for 1 h. The ss intercalator-ODN and 10 mM MgCl2 were added to the target on ice and equilibrium formation of complex was achieved at 4°C overnight.
Gel electrophoretic mobility shift assay
The experimental protocols were adapted from previous publications (31). Sucrose was added to the samples to a final concentration of 10% (w/v) before gel electrophoresis. The ds and ss ODN concentrations are given in the legends to the figures. Electrophoresis was carried out in 16% polyacrylamide gels (5% cross-linking) at 10 V/cm for 4 h in a 4°C cold room using 90 mM Tris–borate buffer, pH 8.0, containing 10 mM MgCl2. Gel temperatures were maintained at 8–10°C. Acridine fluorescence images of the intercalator strands in the wet gels were recorded with an ST6 CCD camera (Santa Barbara Instrument Group, Santa Barbara, CA) by epi-illumination excitation at 254 nm or direct illumination at 302 nm using either an emission longpass KV500 or bandpass (496 nm) SKF-9 filter, respectively (Schott, Mainz, Germany). The gels were subsequently stained and images recorded of the multi-stranded ODN bands using EtBr and a longpass 615 nm filter or of all of the bands using SybrGreenII or SybrGold (Molecular Probes) and a longpass KV450 nm filter. The digitized fluorescence images were quantitated using the image analysis program NIH-Image. Binding curves were plotted as the fraction of bound ss-Acr conjugate versus the concentration of the ds DNA. The equilibrium constant for the triplex was calculated from the following equation:
Kass = 1/Kd = [tr]/([ds][ss]) 1
where Kass is the triplex association constant, Kd is the triplex dissociation constant and [tr], [ds] and [ss] are the equilibrium concentrations of intermolecular triplex ODN, unbound double-helical ODN and ss ODN (or ODN–intercalator conjugate), respectively. [tr] was derived from the concentration of the retarded ss-intercalator conjugate. The quenching of the acridine fluorescence due to intercalation next to a guanine base in the triplex was measured both in gels and in solution (see Results). Apparent triplex dissociation constants (Kd) are given in the text.
Modification with OsO4-bipyridine
The 3′-end of the specific A target ODN was labeled with [32P]ddATP (Amersham) with terminal transferase (Boehringer). The 32P-labeled sample was run at 70°C in a 15% polyacrylamide–7 M urea denaturing gel and the band containing the pure 32P-labeled target ds ODN was cut out and electroeluted into a BioTrap (Schleicher & Schuell, Germany). Chemical modification with OsO4-bipyridine (1 mM) was performed as described previously (32). The modification was carried out on the triplex using a 5-fold excess of the third strand on ice in triplex-forming buffer (10 mM Tris–HCl, pH 8, 10 mM MgCl2) with addition of 20 mM NaCl for 30 min. The sequencing gel was dried and scanned with a phosphorimager (Molecular Dynamics, USA). The ds AB duplex and ss A strand were modified separately as controls.
Fluorescence and UV spectroscopy
The fluorescence emission and excitation spectra for the Acr-ODNs in the free and bound state were measured in an SLM-SPF spectrofluorometer (ISS, Urbana, IL) having a polarization accessory and thermostated cuvette holder. Samples at equilibrium prepared as described above were measured at 3.5°C.
In some experiments the fluorescence anisotropy, r, was measured:
r = (IVV – GIVH)/(IVV + 2GIVH) 2
where IVV is the fluorescence intensity measured with an emission polarizer aligned parallel to the vertical excitation polarization and IVH is the fluorescence intensity measured with a perpendicular alignment of the emission polarizer. G is a factor that accounts for the polarization detection bias of the spectrofluorometer.
Fluorescence resonance energy transfer experiments (FRET)
In order to determine the relative orientation of the strands in the triple strand complex we measured FRET between acridine attached to the 3′-end of the third strand as donor (ss-Acr) and Texas Red at the 5′-end of either the A or B strand in the duplex target as acceptor. FRET occurs if the donor emission spectrum and the acceptor absorption spectrum have a significant energy overlap and the dyes are in close proximity. The efficiency (E) of transfer is given by the formula
E = 1/[1 + (R/R0)6] 3
where R is the distance between the donor and acceptor moieties. The critical Förster distance R0 is defined as the distance at which the efficiency (E) of the transfer is 50% and was calculated from equations in the literature (33). Under FRET conditions excitation of the donor results in quenching of the donor fluorescence with concomitant sensitized acceptor emission. For conditions of excitation and emission of the electrophoretic gels of the FRET samples see Results.
RESULTS
Binding of the ODN-Acr probes to target ds ODNs
The target ODNs had an extra clamping GC pair on each of the 3′- and 5′-ends to promote stability of the duplex and to provide a potential intercalation site for the intercalating dye attached to the 3′- or 5′-ends of the ss ODN probe. At the concentrations used in these experiments the target hairpin ODNs can form both intra- (hairpin) and intermolecular duplexes after denaturation. The amount of each species formed under our experimental conditions was determined from a quantitative scan of the gels (see below). In both double-helical structures the Watson–Crick base pairs are identical. In the case of the intermolecular duplex target there was no evidence for higher order aggregation.
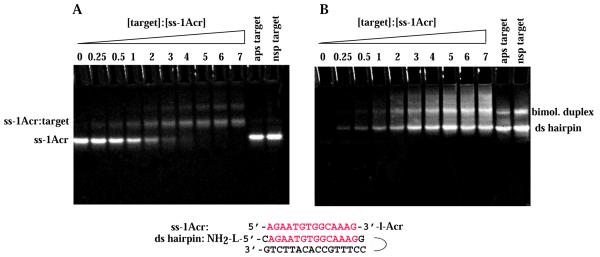
Electrophoretic gel mobility shift assays were used to assess the specific association of the probe ODN with the target ODNs. Figure 2 shows the results of such an assay using the ss-1Acr probe and three different hairpin target duplexes. In each lane a constant concentration of ss-1Acr of 27 µM (strands) was equilibrated with different concentrations of target duplex. The target ODNs were as follows: specific ds (sp), in which the sequence identical to the probe had the same (parallel) orientation (lanes 2–10); antiparallel (ap), in which the sequence identical to the probe was in the antiparallel orientation, (lane 11); non-specific (nsp), in which the base composition was the same but the sequence was randomized (lane 12) (see Fig. 1 for sequences). The target ODNs were in the form of hairpins (lower bands in Fig. 2B) and intermolecular duplexes (upper bands in Fig. 2B). The fluorescence of ss-1Acr was visualized (Fig. 2A) before EtBr staining. Specific association with the ds target was demonstrated by disappearance of the faster moving ss-1Acr monomer band accompanied by appearance of fluorescence in two retarded ds species at increasing target concentrations (upper bands, Fig. 2A). No complex was detected with either the ap (lane 11) or nsp (lane 12) targets at the same concentrations as the ds target (lane 7). The ds species were visualized in the same gel by post-staining with EtBr (Fig. 2B). Note that the upper acridine fluorescent bands in Figure 2A were slightly retarded compared to the unbound ds target in Figure 2B.
Figure 2.
Band shift assay of binding of ss-1Acr to the hairpin target by gel electrophoresis. (A) Fluorescence of the acridine conjugate. Fastest migrating band, ss-1Acr conjugate. Middle bands, ss-1Acr bound to the ds hairpin. Slowest migrating row of bands, ss-1Acr bound to the intermolecular duplex form of the ds target. Image of non-stained gel at >500 nm, epi-illumination excitation at 254 nm. (B) Gel from (A) stained with EtBr for detection of multi-stranded species. Fluorescence at > 615 nm, epi-illumination at 254 nm. ss-1Acr is not detected in this spectral region. Triplex bands (Acr stained upper bands in A) appear as retarded bands slightly above the excess ds hairpin and duplex ds targets. The concentration of ss-1Acr was 27 µM (strands) in all lanes. ds target concentrations are given as a factor of 27 µM (oligonucleotides) for each lane. Target concentration, sp hairpin: lane 1, 0; lane 2, 0.25; lane 3, 0.5; lane 4, 1; lane 5, 2; lane 6, 3; lane 7, 4; lane 8, 5; lane 9, 6; lane 10, 7. Target concentration, ap hairpin: lane 11, 4. Target concentration, nsp hairpin: lane 12, 4. Other conditions are indicated in Materials and Methods.
We observed that ss-1Acr was quenched by >50% upon complete binding (lane 8), most probably due to intercalation next to a guanine base between the 3′-terminal GC pair of the ds target (34). Integrated intensities of the ss-1Acr bands were used to calculate a binding curve and estimate a dissociation constant for the third strand of 42 ± 7 µM (strands). Similar results were obtained for triplex formation by ss-1Acr using an intermolecular AB duplex as the target and monitoring by gel retardation, but we observed little quenching of acridine fluorescence in the complex (data not shown). We assume that a different geometry exists at the terminal base pair of the hairpin adjacent to the loop and the terminal base pair of the duplex in which a 5′-amino group present on the B strand may lead to a perturbation of acridine intercalation.
The equilibrium solutions were also measured by fluorescence spectroscopy. The fluorescence anisotropy of the acridine emission at 505 nm (excitation 420 nm) increased from 0.100 for unbound ss-1Acr to 0.136 for probe bound to the ds target, while the anisotropies of ss-1Acr added to a 3.4-fold excess of the ap and nsp targets were 0.102 and 0.105, respectively. These data further support the conclusion that ss-1Acr forms a specific complex only with the ds target.
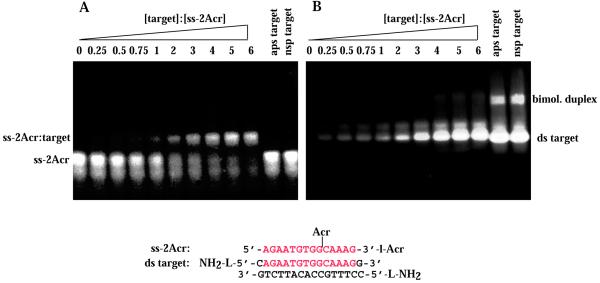
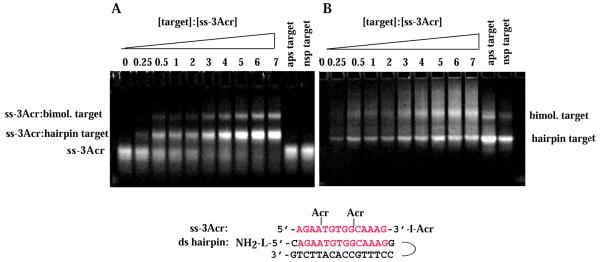
Gel electrophoresis retardation assays were also performed with the ODN conjugates ss-2Acr and ss-3Acr (Figs 1, 3 and 4). Both conjugates bound specifically to the ds target either in the intramolecular hairpin form or as the intermolecular duplex. Data are shown for the intermolecular ds target AB (Fig. 3) and the ds hairpin target (Fig. 4). We infer that intercalators included in the sugar–phosphate backbone did not prevent homologous recognition of the identical sequence. The estimated Kd for the ss-3Acr·ds hairpin triplex was 33 ± 9 µM (strands), a value within experimental error of that for the ss-1Acr·ds hairpin triplex. The affinity of ss-2Acr for the bimolecular Watson–Crick duplex was somewhat lower (Kd = 64 ± 10 µM). We did not investigate the influence on Kd of substituted versus non-substituted terminal amino groups in the targets.
Figure 3.
Band shift assay of binding of ss-2Acr to the intermolecular duplex target. The specific target was the ds AB duplex; control targets were the hairpins as in Figure 2. Conditions were as in Figure 2. (A) Acridine fluorescence. (B) Fluorescence of multi-strand species after staining with EtBr. The concentration of ss-2Acr was 28 µM in all lanes. ds target concentrations are given as a factor of 28 µM (oligonucleotides) for each lane. Target lanes: lane 1, 0; lane 2, 0.25; lane 3, 0.5; lane 4, 0.75; lane 5, 1; lane 6, 2; lane 7, 3; lane 8, 4; lane 9, 5; lane 10, 6. Lanes 11 and 12 as in Figure 2.
Figure 4.
Band shift assay of binding of ss-3Acr to the ds hairpin target. Conditions as in Figure 2. (A) Acridine fluorescence. (B) Fluorescence of multi-strand species after staining with EtBr. The concentration of ss-3Acr was 14 µM (strands) in all lanes. Concentrations of the ds targets are as in Figure 2.
Binding of the ODN-Met and ODN-pso probes to target ds ODNs
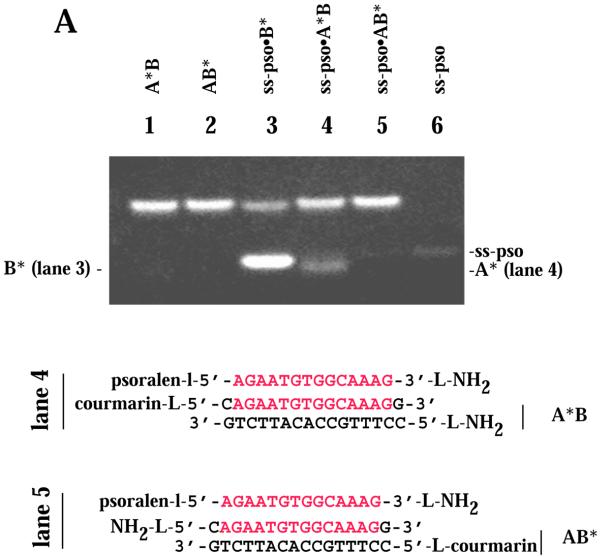
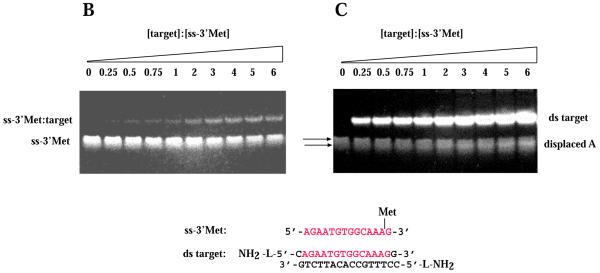
The nature of the intercalator in the conjugate governed the mode of DNA recognition with an ODN-intercalator conjugate. While acridine was found to stabilize triplex formation, other conjugates facilitated third strand invasion with displacement of the identical A strand from the complex. Non-denaturing gel electrophoretic mobility shift experiments indicated the presence of a displaced A strand upon binding of ss-pso, ss-2Met, ss-3′Met and ss-5′Met to the ds AB target (for sequences of the ODNs see Fig. 1). To prove displacement of the A strand by the ODN probe we labeled the 5′-end of the A or B strand with a coumarin fluorophore and detected the intrinsic fluorescence at 450 nm of the displaced single strand in the gel upon excitation at 254 nm. Data are shown for ss-pso in Figure 5A. The displaced homologous labeled A* strand is clearly seen in lane 4. Unlabeled A strand could also be visualized by staining the wet gel with SybrGold dye, as shown for binding of ss-3′Met to intermolecular target AB (Fig. 5B and C). ss-3′Met is retarded with increasing concentrations of AB target (Fig. 5B) but a new ss band appears slightly below the orginal ss probe after staining with SybrGold (Fig. 5C).
Figure 5.
Target strand displacement by ss-pso and ss-Met ODNs. (A) 5′,7-Diethylaminocoumarin-labeled A* or B* in the AB intermolecular target. ss-pso, 30 µM; ds AB, 140 µM. Lane 1, A*B duplex; lane 2, AB* duplex; lane 3, ss-pso·B* duplex with excess complementary B*; lane 4, ss-pso·A*B triplex; lane 5, ss-pso·AB* triplex; lane 6, ss-pso. (B and C) Titration of ss-3′Met with increasing concentrations of AB intermolecular target. (B) Methidium fluorescence. (C) Fluorescence of multi-strand species after staining with SybrGold. The concentration of ss-3′Met was 28 µM in all lanes. ds target concentrations are given as a factor of 28 µM (oligonucleotides) for each lane. Target lanes: lane 1, 0; lane 2, 0.25; lane 3, 0.5; lane 4, 0.75; lane 5, 1; lane 6, 2; lane 7, 3; lane 8, 4; lane 9, 5; lane 10, 6.
Chemical modification of the triplex formed with the ss-2Acr conjugate to test for strand displacement
In order to definitively demonstrate that strand displacement or D-loop formation does not occur upon binding of acridine conjugates to the intermolecular Watson–Crick duplex, we performed chemical modification experiments directed at the thymines of the A strand in the target duplex upon complexation with ss-2Acr. The A strand of the intermolecular AB duplex was radioactively labeled on the 3′-end with ddATP and accessibility of the thymines in the target examined as described previously (32). Piperidine digests of the target and complexes after reaction with OsO4-bipyridine were run on a sequencing polyacrylamide gel (Fig. 6). The ss A strand was modified with 1 mM OsO4-bipyridine as a control (lane 2) and showed clear modification of both thymines. In contrast, the ds target reacted for 30 min showed full protection (lane 3). The triplex samples contained 30 µM labeled ds target and 150 µM ss-2Acr strand. Taking into account the determined apparent association constant of the triplex, the 5-fold excess of the third strand should have ensured binding of the AB target of ∼75%. Figure 6 shows that the thymines from the A strand of the duplex upon triple strand complex formation were protected from modification to the same extent as thymines in the non-complexed ds target. Thus the homologous A strand was not displaced or looped out by formation of the triplex upon ss-2Acr binding. The data support a triplex structure involving two identical strands.
Figure 6.
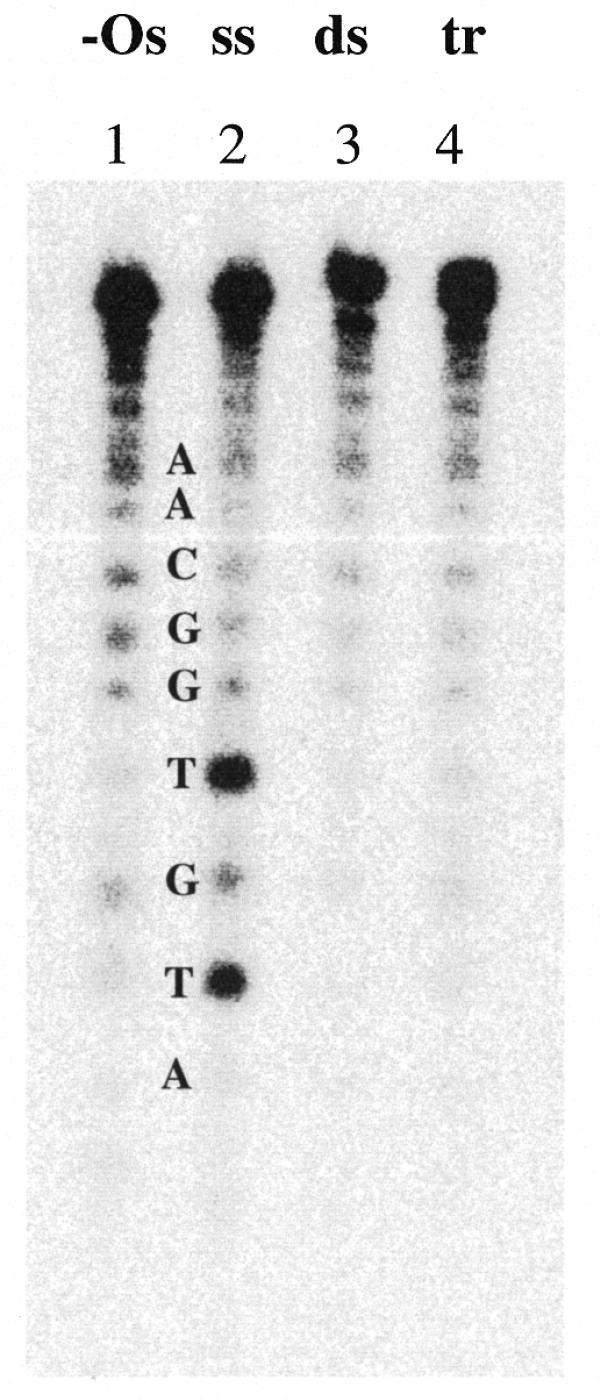
Chemical modification by OsO4-bipyridine of thymines in the A strand of the target. A sequencing gel after digestion of the samples with piperidine is presented. Lane 1, –Os, untreated A; lane 2, ss A strand reacted with OsO4-bipyridine; lane 3, ds AB duplex reacted with OsO4-bipyridine; lane 4, tr ss-2Acr·AB triplex reacted with OsO4-bipyridine. Other conditions were as in Materials and Methods.
FRET measurements to test the third strand orientation in the triplex
An R0 of ∼4.0 nm was calculated for the dye pair acridine and BODIPY-Texas Red. With this dye pair the parallel alignment of the ss-Acr strand to the identical A strand in the duplex target is expected to lead to maximum quenching of the acridine donor at the 3′-end of the third strand by BODIPY-TR attached to the 5′-end of the complementary B strand. On the other hand, only ∼20% quenching is to be expected in a three-stranded complex in which the BODIPY-TR was attached to the 5′-end of the A strand.
Samples containing a 3.5-fold excess of the AB target duplex with either the A or B strand 5′-labeled with BODIPY-TR, as well as control samples, were prepared as described above. The samples were analyzed by non-denaturing 16% PAGE (Fig. 7). The intrinsic acridine fluorescence was imaged in the gel (Fig. 7A). As a control a simple duplex was formed between ss-1Acr and excess complementary B-BODIPY-TR (B*) strand. Acridine emission monitored at 495 nm was fully quenched (Fig. 7A, lane 4). The residual fluorescence at 495 nm was comparable to the non-acridine-containing BODIPY-TR-labeled duplexes A*B and AB* (Fig. 7A, lanes 2 and 3). Binding (band shift) of ss-1Acr to the duplex target A*B to form a triplex (lane 5) as well as to the complementary unlabeled B strand to form a duplex (lane 7) was apparent. We note that under the experimental conditions used the different mobilities of the duplex and triplex were not resolved in the gel. Figure 7B shows the BODIPY-TR emission above 630 nm by excitation of the same gel at 366 nm. No A strand displacement was detected on ss-1Acr binding to the A*B duplex (Fig. 7B, lane 5). A comparison of the retarded A*B·ss-1Acr and AB*·ss-1Acr triplex bands in lanes 5 and 6 (Fig. 7A) demonstrated complete quenching of the acridine donor in the AB*·ss-1Acr complex, supporting a parallel orientation of the two identical strands in the triplex.
Figure 7.
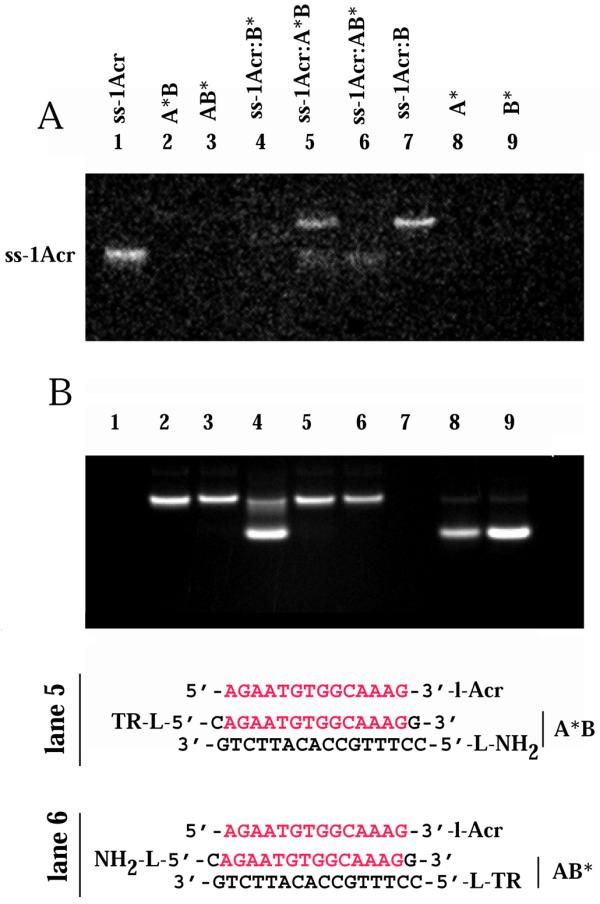
Non-denaturing gel band shift assay of the FRET samples. The concentration of the ss-1Acr strand was 28 µM, those of A, B, A-5′BODIPY-TR (A*) and B-5′BODIPY-TR (B*) were 100 µM. Lane 1, ss-1Acr; lane 2, A*B duplex; lane 3, AB* duplex; lane 4, ss-1Acr·B* duplex with excess B* strand; lane 5, ss-1Acr·A*B triplex; lane 6, ss-1Acr·AB* triplex; lane 7, ss-1Acr·B duplex; lane 8, ss A*; lane 9, ss B*. (A) Acridine fluorescence image of unstained gel at 496 nm, detected with a bandpass SKF-9 filter and excitation at 302 nm. (B) BODIPY-Texas Red fluorescence image of the same gel as in (A). Emission at > 630 nm, excitation at 366 nm.
We deduce from the amount of free ss-1Acr in Figure 7A (lanes 5 and 6) that the affinity of the ss-1Acr oligonucleotide for the BODIPY-TR-labeled duplex was weaker than that for the unlabeled AB target. This is apparent from the presence of bands corresponding to free ss-1Acr in Figure 7A (lanes 5 and 6). We can estimate the fraction of bound ss-Acr in the triplex from integration of the gel bands. The fluorescence of free (unquenched) ss-1Acr is taken from lane 1 and the fluorescence of the bound (quenched) form from lane 4. Using these assumptions we obtained a value for the amount of ss-1Acr bound in the triplex with the AB* duplex of 57%. Assuming an equivalent percentage bound to the A*B duplex and calculating the emission of the triplex in lane 5, we obtain a quenching of 25% in comparison with the emission of free ss-1Acr. This is in a good agreement with the estimations for FRET efficiency of 20% for a 16 bp acridine–BODIPY-TR distance based on R0 ≈ 4 nm.
DISCUSSION
Prior to this study only intramolecular fold-back synthetic hairpins (21,23,24,35) had been used to test the theoretical structure prediction that any arbitrary nucleotide sequence could form a triple-helical DNA in which the identical strands are oriented in a parallel fashion (14). The present experiments were aimed at determining whether a protein-free parallel intermolecular triplex could be formed. We investigated a number of 14 nt dye-conjugate probes and four 16 bp duplex targets. Our evidence from gel mobility shift assays, fluorescence anisotropy and FRET measurements demonstrates that protein-independent triplex formation occurs only with a ds target in which the homologous probe and target strands are in the parallel orientation.
In Figures 2–4 we demonstrate, by gel retardation, triplex formation of acridine labeled ODNs targeted to AB duplexes. The emission of ss-1Acr was partially quenched upon formation of a triplex with the specific hairpin target (Fig. 2), whereas the ss-2Acr and ss-3Acr probes remained fluorescent upon binding to either target (Figs 3 and 4). Dissociation constants evaluated from the gels for probes with between one and three acridine intercalators confirmed that the intercalators attached to the sugar–phosphate backbone with the flexible pentamethylene linker intercalated into the triplex structure without significant reduction in target sequence specificity. The binding affinity of Acr-labeled ODNs did not depend significantly on the number of attached acridine molecules, in contrast to binding of free intercalator dye to the intramolecular triplex, in which two dye molecules stabilize but three dye molecules destabilize the short triplex (24).
From theoretical considerations the parallel GCG triplet was predicted to be the most stable of all triplets because the G of the third strand can form three hydrogen bonds to the GC Watson–Crick pair (14). In fact, parallel orientation of two dG strands in a triplex formed by one dC and two dG strands has been verified by FTIR and Raman spectroscopy (36).
Previous studies with alternating purine/pyrimidine sequences in parallel and anti-parallel intramolecular triplex constructs showed a consistently higher stability for the anti-parallel sequences (Tm values higher by 10–17°C) except for ATA versus AAT, for which the stabilities were comparable (37,38). However, specific antiparallel triplexes have not been achieved previously with purely mixed sequences. We demonstrate in this study that the presence of only three pyrimidine bases in the 14 nt DNA sequence 5′-AGAATGTGGCAAAG-3′ results in a thermodynamic preference for the parallel triplex and a high selectivity of recognition of the specific ds target in comparison with the ap target.
Chemical modification analysis also supported the conclusion that triplex formation occurred with our Acr-ODN conjugates rather than strand displacement in the duplex. As shown in Figure 6, the A target strand remained equally resistant to OsO4 modification under saturating triplex-forming concentrations of the conjugate ss-2Acr as the same strand in the target duplex.
All of the data presented in Figures 2–4 and 6 were consistent with formation of an intermolecular triplex in which the identical strands lie in the parallel orientation. To further substantiate this model for the structure we undertook FRET measurements on the triplex formed with ss-1Acr, in which the acridine moiety was on the 5′-end of the ODN and the target was labeled with BODIPY-Texas Red on the 3′-end of either the A or B strand. In the parallel triplex BODIPY-TR and acridine would be at the same end only when the B strand is labeled whereas the dyes will be on opposite ends when the target A strand carries the BODIPY-TR. The results of these experiments are shown in Figure 7.
Comparing the acridine fluorescence of the retarded bands in lanes 4–6 of Figure 7A, one notes that significant and similar quenching occurred in the ss-1Acr·AB* triplex as in the ss-1Acr·B* duplex, whereas much less quenching occurred in the ss-1Acr·A*C triplex. These are exactly the results we would have predicted for a parallel orientation of the two identical strands in the triplex.
From previous experiments with EtBr stabilization of intramolecular parallel triplexes we reasoned that other intercalators than acridine might promote greater stability of the parallel intermolecular triplex (24). We synthesized three different ODNs with a methidium moiety at different positions on both a shorter linker and a longer linker (see Materials and Methods and Fig. 1). None of these ODNs promoted triplex formation, but instead caused strand exchange (Fig. 5B and C). An additional ODN with a psoralen linked to the 3′-end (Fig. 1) showed triplex formation under some conditions, but at equilibrium also resulted in strand exchange (Fig. 5A). Therefore, we conclude that these intercalator conjugates may transiently promote triplex formation during recognition of the target duplex but because of a more stable intercalation geometry in the duplex than the triplex helix the final equilibrium state may be the strand exchanged duplex.
CONCLUSIONS
The use of a ss ODN-intercalator conjugate enabled us to observe spectroscopically and isolate on gels an intermolecular parallel triplex with the 5′-AGAATGTGGCAAAG-3′ sequence from the Zn finger repeat of the human KRAB protein gene at physiological ionic strength and pH and temperatures up to 20°C. Parallel triplex formation was shown to occur in a sequence-specific manner whereby no complex formation could be demonstrated with two control sequences, one having the homologous sequence in the antiparallel orientation and another with a non-specific nucleotide sequence of the same base composition.
Acridine attached to either end as well as to the sugar–phosphate backbone between bases of the ss ODN probe was shown to stabilize the parallel triplex. Increasing the number of acridine moieties did not significantly increase or decrease this effect. Further evidence for the involvement of both homologous strands in the triplex structure, rather than displacement of the A strand by the homologous ss-Acr conjugate, was obtained from OsO4-bipyridine modification experiments in which we found full protection of the thymines of the target under triplex-forming conditions. However, ODNs labeled with other intercalators such as 5′-psoralen or methidium internally or one base from either end promoted strand exchange.
The parallel orientation of the acridine-labeled ODN probe and the identical strand in the target was confirmed by FRET experiments in non-denaturing gels using BODIPY-Texas Red-labeled target duplexes.
Our data show that mixed irregular base sequences can form protein-independent intermolecular parallel triplexes. Nevertheless, the triplex dissociation constants are high and attempts to improve the stability with intercalators other than acridine resulted in duplex strand displacement. Thus, the goal of utilizing parallel triplex formation involving practically any mixed sequence for in vivo diagnostics or clinical applications is not yet realizable.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Gudrun Heim for excellent technical assistance, Dr E. Palecek for recommendations concerning the optimal experimental conditions for the OsO4 modification and Drs V.B. Zhurkin and R. Clegg for helpful discussions. This study was commenced with support by a European Union PECO grant, supplementary agreement ERB CIPD-CT94-0627 to contract ERB CHRX-CT93-0177, and completed with support by the North Atlantic Treaty Organization, NATO Linkage Grant SA.HTECH (LG 971252)WK. We thank both organizations for promoting international cooperation on basic scientific research problems.
References
- 1.Ebbinghaus S.W., Fortinberry,H. and Gamper,H.B.,Jr (1999) Inhibition of transcription elongation in the HER-2/neu coding sequence by triplex-directed covalent modification of the template strand. Biochemistry, 38, 619–628. [DOI] [PubMed] [Google Scholar]
- 2.Floris R., Scaggiante,B., Manzini,G., Quadrifoglio,F. and Xodo,L.E. (1999) Effect of cations on purine·purine·pyrimidine triple helix formation in mixed-valence salt solutions. Eur. J. Biochem., 260, 801–809. [DOI] [PubMed] [Google Scholar]
- 3.Mills M., Arimondo,P.B., Lacroix,L., Garestier,T., Helene,C., Klump,H. and Mergny,J.L. (1999) Energetics of strand-displacement reactions in triple helices: a spectroscopic study. J. Mol. Biol., 291, 1035–1054. [DOI] [PubMed] [Google Scholar]
- 4.Porumb H., Gousset,H. and Taillandier,E. (1999) Parallel and antiparallel triple helices with G,A-containing third strands. Electrophoresis, 20, 511–513. [DOI] [PubMed] [Google Scholar]
- 5.Sedelnikova O.A., Panyutin,I.G., Luu,A.N. and Neumann,R.D. (1999) The stability of DNA triplexes inside cells as studied by iodine-125 radioprinting. Nucleic Acids Res., 27, 3844–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishnan I. and Patel,D.J. (1994) DNA triplexes: solution structures, hydration sites, energetics, interactions, and function. Biochemistry, 33, 11405–11416. [DOI] [PubMed] [Google Scholar]
- 7.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 8.Fox K.R. (2000) Targeting DNA with triplexes. Curr. Med. Chem., 7, 17–37. [DOI] [PubMed] [Google Scholar]
- 9.Francois J.C., Lacoste,J., Lacroix,L. and Mergny,J.L. (2000) Design of antisense and triplex-forming oligonucleotides. Methods Enzymol., 313, 74–95. [DOI] [PubMed] [Google Scholar]
- 10.Giovannangeli C. and Helene,C. (1997) Progress in developments of triplex-based strategies. Antisense Nucleic Acid Drug Dev., 7, 413–421. [DOI] [PubMed] [Google Scholar]
- 11.Helene C., Giovannangeli,C., Guieysse-Peugeot,A.L. and Praseuth,D. (1997) Sequence-specific control of gene expression by antigene and clamp oligonucleotides. CIBA Found. Symp., 209, 94–102. [DOI] [PubMed] [Google Scholar]
- 12.Vasquez K.M. and Wilson,J.H. (2000) Triplex-directed site-specific genome modification. Methods Mol. Biol., 133, 183–200. [DOI] [PubMed] [Google Scholar]
- 13.Howard-Flanders P., West,S.C. and Stasiak,A.J. (1984) Role of RecA protein spiral filaments in genetic recombination. Nature, 309, 215–220. [DOI] [PubMed] [Google Scholar]
- 14.Zhurkin V.B., Raghunathan,G., Ulyanov,N.B., Camerini-Otero,R.D. and Jernigan,R.L. (1994) A parallel DNA triplex as a model for the intermediate in homologous recombination. J. Mol. Biol., 239, 181–200. [DOI] [PubMed] [Google Scholar]
- 15.Kim M.G., Zhurkin,V.B., Jernigan,R.L. and Camerini-Otero,R.D. (1995) Probing the structure of a putative intermediate in homologous recombination: the third strand in the parallel DNA triplex is in contact with the major groove of the duplex. J. Mol. Biol., 247, 874–889. [DOI] [PubMed] [Google Scholar]
- 16.Rao B.J. and Radding,C.M. (1993) Homologous recognition promoted by RecA protein via non-Watson-Crick bonds between identical DNA strands. Proc. Natl Acad. Sci. USA, 90, 6646–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao B.J. and Radding,C.M. (1995) RecA protein mediates homologous recognition via non-Watson-Crick bonds in base triplets. Phil. Trans. R. Soc. B, 347, 5–12. [DOI] [PubMed] [Google Scholar]
- 18.Bertucat G., Lavery,R. and Prevost,C. (1999) A molecular model for RecA-promoted strand exchange via parallel triple-stranded helices. Biophys. J., 77, 1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagneaux C., Porumb,H., Liquier,J., Takahashi,M. and Taillandier,E. (1995) Conformations of three-stranded DNA structures formed in presence and in absence of the RecA protein. J. Biomol. Struct. Dyn., 13, 465–470. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh P., Camerini-Otero,C.S. and Camerini-Otero,R.D. (1992) The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl Acad. Sci. USA, 89, 6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shchyolkina A.K., Timofeev,E.N., Borisova,O.F., Il’icheva,I.A., Minyat,E.E., Khomyakova,E.B. and Florentiev,V.L. (1994) The R-form of DNA does exist. FEBS Lett., 339, 113–118. [DOI] [PubMed] [Google Scholar]
- 22.Wilson W.D., Tanious,F.A., Mizan,S., Yao,S., Kiselyov,A.S., Zon,G. and Strekowski,L. (1993) DNA triple-helix specific intercalators as antigene enhancers: unfused aromatic cations. Biochemistry, 32, 10614–10621. [DOI] [PubMed] [Google Scholar]
- 23.Borisova O.F., Shchyolkina,A.K., Timofeev,E.N., Tsybenko,S., Mirzabekov,A.D. and Florentiev,V.L. (1995) Stabilization of parallel (recombinant) triplex with propidium iodide. J. Biomol. Struct. Dyn., 13, 15–27. [DOI] [PubMed] [Google Scholar]
- 24.Shchyolkina A.K. and Borisova,O.F. (1997) Stabilizing and destabilizing effects of intercalators on DNA triplexes. FEBS Lett., 419, 27–31. [DOI] [PubMed] [Google Scholar]
- 25.Silver G.C., Nguyen,C.H., Boutorine,A.S., Bisagni,E., Garestier,T. and Helene,C. (1997) Conjugates of oligonucleotides with triplex-specific intercalating agents. Stabilization of triple-helical DNA in the promoter region of the gene for the alpha-subunit of interleukin 2 (IL-2R alpha). Bioconjug. Chem., 8, 15–22. [DOI] [PubMed] [Google Scholar]
- 26.Asseline U., Bonfils,E., Dupret,D. and Thuong,N.T. (1996) Synthesis and binding properties of oligonucleotides covalently linked to an acridine derivative: new study of the influence of the dye attachment site. Bioconjug. Chem., 7, 369–379. [DOI] [PubMed] [Google Scholar]
- 27.Kukreti S., Sun,J.S., Garestier,T. and Helene,C. (1997) Extension of the range of DNA sequences available for triple helix formation: stabilization of mismatched triplexes by acridine-containing oligonucleotides. Nucleic Acids Res., 25, 4264–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellefroid E.J., Marine,J.C., Ried,T., Lecocq,P.J., Riviere,M., Amemiya,C., Poncelet,D.A., Coulie,P.G., de Jong,P., Szpirer,C., Ward,D.C. and Martial,J.A. (1993) Clustered organization of homologous KRAB zink-finger genes with enchanced expression in human T lymphoid cells. EMBO J., 12, 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timofeev E.N., Smirnov,I.P., Haff,L.A., Tishchenko,E.I., Mirzabekov,A.D. and Florentiev,V.L. (1996) Methidium intercalator inserted into synthetic oligonucleotides. Tetrahedron Lett., 37, 8467–8470. [Google Scholar]
- 30.Minchenkova L.E., Shchyolkina,A.K., Chernov,B.K. and Ivanov,V.I. (1986) CC/GG contacts facilitate the B to A transition of DNA in solution. Nucleic Acids Res., 22, 463–475. [DOI] [PubMed] [Google Scholar]
- 31.Orson F.M., Kinsey,B.M. and McShan,W.M. (1994) Linkage structures strongly influence the binding cooperativity of DNA intercalators conjugated to triplex forming oligonucleotides. Nucleic Acids Res., 22, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palecek E. (1992) Probing DNA structure with osmium tetroxide complexes in vitro. Methods Enzymol., 212, 140–155. [DOI] [PubMed] [Google Scholar]
- 33.Jares-Erijman E.A. and Jovin,T.M. (1996) Determination of DNA helical handedness by fluorescence resonance energy transfer. J. Mol. Biol., 257, 596–617. [DOI] [PubMed] [Google Scholar]
- 34.Mergny J.-L., Boutorine,A.S., Garestier,T., Belloc,F., Rougee,M., Bulychev,N.V., Koshkin,A.A., Bourson,J., Lebedev,A.V., Valeur,B., Thuong,N.T. and Helene,C. (1994) Fluorescence energy transfer as a probe for nucleic acid structures and sequences. Nucleic Acids Res., 22, 920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dagneaux C., Shchyolkina,A.K., Liquier,J., Florentiev,V.L. and Taillandier,E. (1995) A triple helix obtained by specific recognition of all 4 bases in duplex DNA can adopt a collapsed or an extended form. C. R. Acad. Sci. III, 318, 559–562. [PubMed] [Google Scholar]
- 36.Ouali M., Letellier,R., Adnet,F., Liquier,J., Sun,J., Lavery,R. and Taillandier,E. (1993) A possible family of B-like triple helix structures: comparison with the Arnott A-like triple helix. Biochemistry, 32, 2098–2103. [DOI] [PubMed] [Google Scholar]
- 37.Shchyolkina A.K., Borisova,O.F., Minyat,E.E., Timofeev,E.N., Il’icheva,I.A., Khomyakova,E.B. and Florentiev,V.L. (1995) Parallel purine-pyrimidine-purine triplex: experimental evidence for existence. FEBS Lett., 367, 81–84. [DOI] [PubMed] [Google Scholar]
- 38.Shchyolkina A.K., Borisova,O.F., Timofeev,E.N., Il’icheva,I.A., Minyat,E.E., Khomyakova,E.B., Florentiev,V.L. and Jovin,T.M. (1995) Intramolecular parallel (recombinant) triplexes formed by oligonucleotides with non-nucleotide linkers. J. Biomol. Struct. Dyn., 12, a214. [Google Scholar]